Abstract
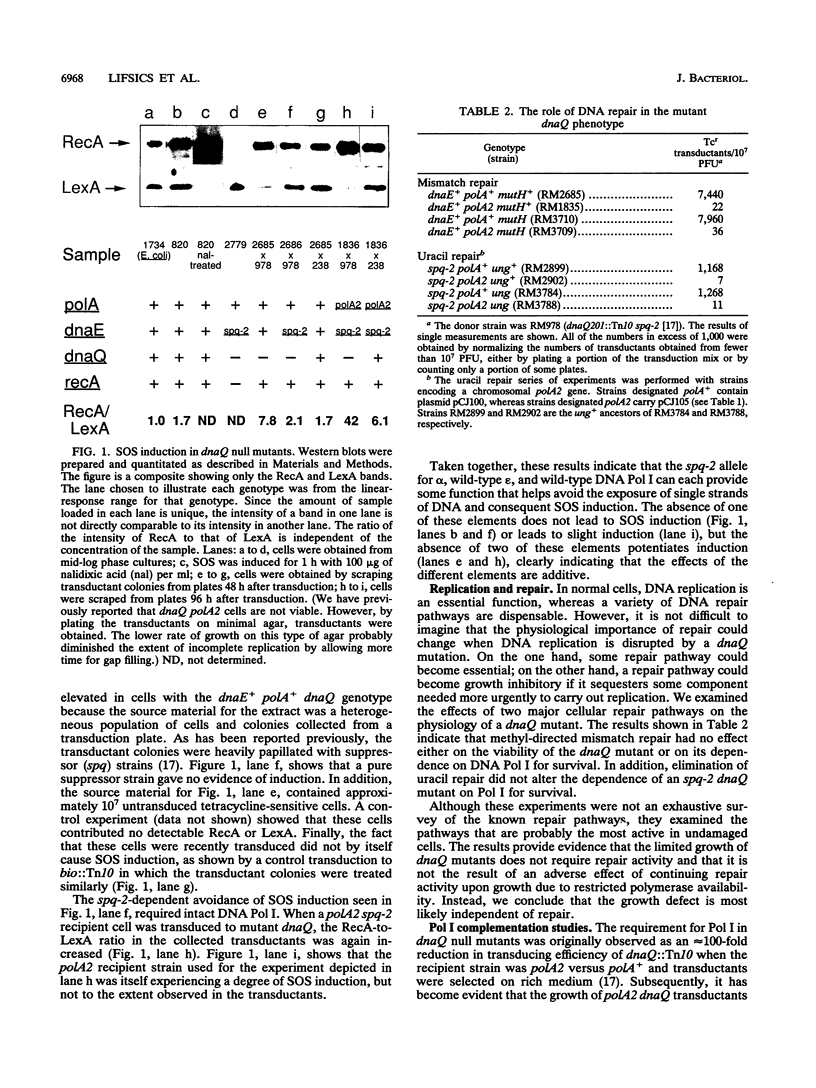
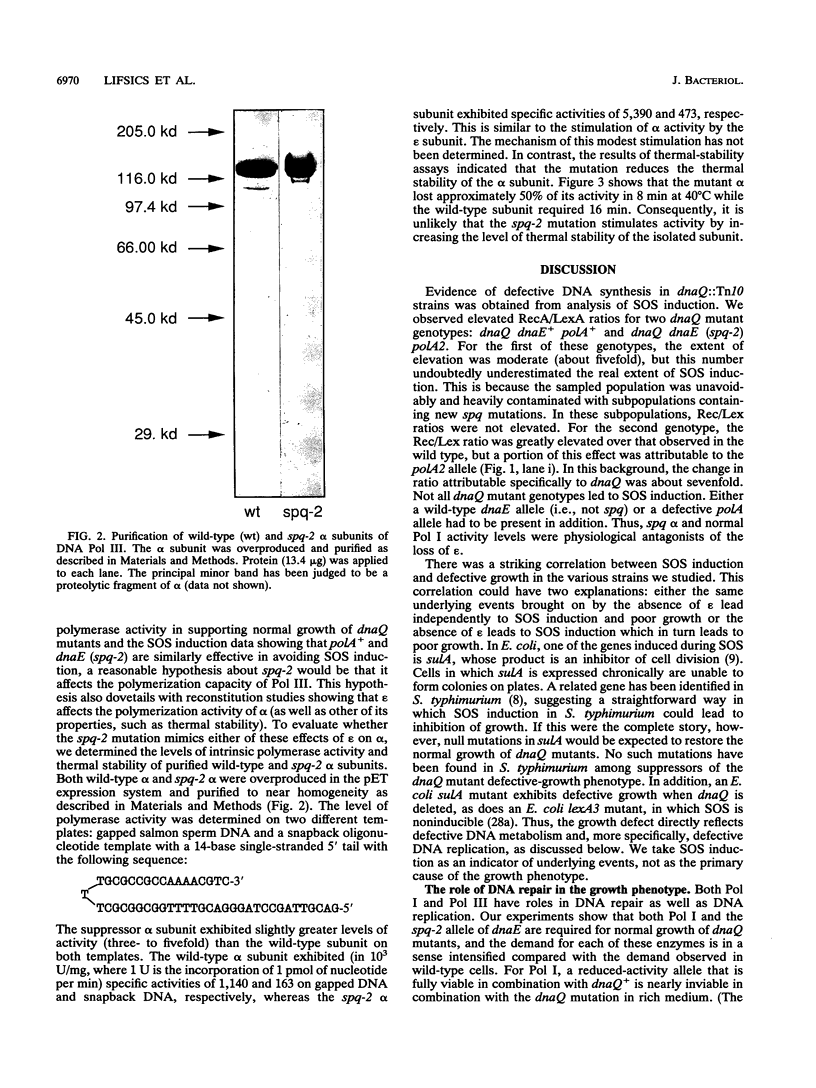
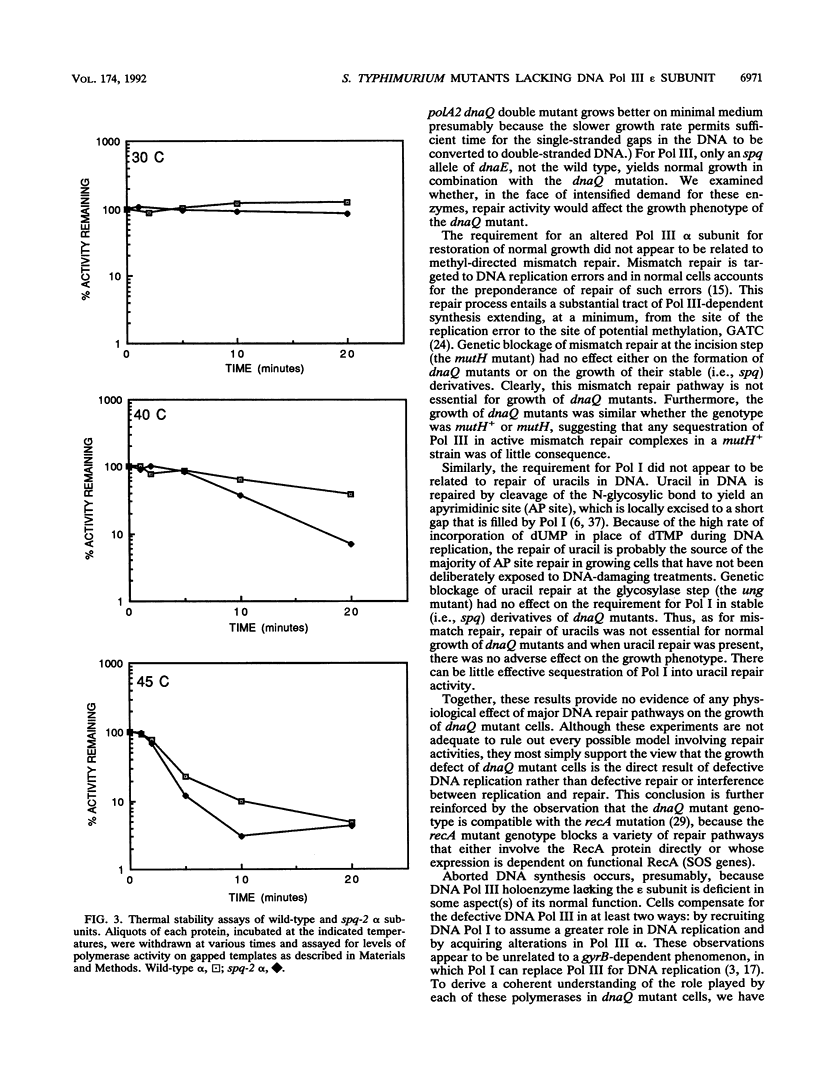
In Salmonella typhimurium, dnaQ null mutants (encoding the epsilon editing subunit of DNA polymerase III [Pol III]) exhibit a severe growth defect when the genetic background is otherwise wild type. Suppression of the growth defect requires both a mutation affecting the alpha (polymerase) subunit of DNA polymerase III and adequate levels of DNA polymerase I. In the present paper, we report on studies that clarify the nature of the physiological defect imposed by the loss of epsilon and the mechanism of its suppression. Unsuppressed dnaQ mutants exhibited chronic SOS induction, indicating exposure of single-stranded DNA in vivo, most likely as gaps in double-stranded DNA. Suppression of the growth defect was associated with suppression of SOS induction. Thus, Pol I and the mutant Pol III combined to reduce the formation of single-stranded DNA or accelerate its maturation to double-stranded DNA. Studies with mutants in major DNA repair pathways supported the view that the defect in DNA metabolism in dnaQ mutants was at the level of DNA replication rather than of repair. The requirement for Pol I was satisfied by alleles of the gene for Pol I encoding polymerase activity or by rat DNA polymerase beta (which exhibits polymerase activity only). Consequently, normal growth is restored to dnaQ mutants when sufficient polymerase activity is provided and this compensatory polymerase activity can function independently of Pol III. The high level of Pol I polymerase activity may be required to satisfy the increased demand for residual DNA synthesis at regions of single-stranded DNA generated by epsilon-minus pol III. The emphasis on adequate polymerase activity in dnaQ mutants is also observed in the purified alpha subunit containing the suppressor mutation, which exhibits a modestly elevated intrinsic polymerase activity relative to that of wild-type alpha.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bochner B. R., Huang H. C., Schieven G. L., Ames B. N. Positive selection for loss of tetracycline resistance. J Bacteriol. 1980 Aug;143(2):926–933. doi: 10.1128/jb.143.2.926-933.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenowitz S., Kwack S., Goodman M. F., O'Donnell M., Echols H. Specificity and enzymatic mechanism of the editing exonuclease of Escherichia coli DNA polymerase III. J Biol Chem. 1991 Apr 25;266(12):7888–7892. [PubMed] [Google Scholar]
- Bryan S. K., Moses R. E. Map location of the pcbA mutation and physiology of the mutant. J Bacteriol. 1984 Apr;158(1):216–221. doi: 10.1128/jb.158.1.216-221.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullas L. R., Ryu J. I. Salmonella typhimurium LT2 strains which are r- m+ for all three chromosomally located systems of DNA restriction and modification. J Bacteriol. 1983 Oct;156(1):471–474. doi: 10.1128/jb.156.1.471-474.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derbyshire V., Freemont P. S., Sanderson M. R., Beese L., Friedman J. M., Joyce C. M., Steitz T. A. Genetic and crystallographic studies of the 3',5'-exonucleolytic site of DNA polymerase I. Science. 1988 Apr 8;240(4849):199–201. doi: 10.1126/science.2832946. [DOI] [PubMed] [Google Scholar]
- Duncan B. K. Isolation of insertion, deletion, and nonsense mutations of the uracil-DNA glycosylase (ung) gene of Escherichia coli K-12. J Bacteriol. 1985 Nov;164(2):689–695. doi: 10.1128/jb.164.2.689-695.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engler M. J., Bessman M. J. Characterization of a mutator DNA polymerase I from Salmonella typhimurium. Cold Spring Harb Symp Quant Biol. 1979;43(Pt 2):929–935. doi: 10.1101/sqb.1979.043.01.102. [DOI] [PubMed] [Google Scholar]
- Freudl R., Braun G., Honoré N., Cole S. T. Evolution of the enterobacterial sulA gene: a component of the SOS system encoding an inhibitor of cell division. Gene. 1987;52(1):31–40. doi: 10.1016/0378-1119(87)90392-1. [DOI] [PubMed] [Google Scholar]
- Higashitani N., Higashitani A., Roth A., Horiuchi K. SOS induction in Escherichia coli by infection with mutant filamentous phage that are defective in initiation of complementary-strand DNA synthesis. J Bacteriol. 1992 Mar;174(5):1612–1618. doi: 10.1128/jb.174.5.1612-1618.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joyce C. M., Grindley N. D. Method for determining whether a gene of Escherichia coli is essential: application to the polA gene. J Bacteriol. 1984 May;158(2):636–643. doi: 10.1128/jb.158.2.636-643.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kukral A. M., Strauch K. L., Maurer R. A., Miller C. G. Genetic analysis in Salmonella typhimurium with a small collection of randomly spaced insertions of transposon Tn10 delta 16 delta 17. J Bacteriol. 1987 May;169(5):1787–1793. doi: 10.1128/jb.169.5.1787-1793.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel T. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc Natl Acad Sci U S A. 1985 Jan;82(2):488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuwabara N., Uchida H. Functional cooperation of the dnaE and dnaN gene products in Escherichia coli. Proc Natl Acad Sci U S A. 1981 Sep;78(9):5764–5767. doi: 10.1073/pnas.78.9.5764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancy E. D., Lifsics M. R., Kehres D. G., Maurer R. Isolation and characterization of mutants with deletions in dnaQ, the gene for the editing subunit of DNA polymerase III in Salmonella typhimurium. J Bacteriol. 1989 Oct;171(10):5572–5580. doi: 10.1128/jb.171.10.5572-5580.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancy E. D., Lifsics M. R., Munson P., Maurer R. Nucleotide sequences of dnaE, the gene for the polymerase subunit of DNA polymerase III in Salmonella typhimurium, and a variant that facilitates growth in the absence of another polymerase subunit. J Bacteriol. 1989 Oct;171(10):5581–5586. doi: 10.1128/jb.171.10.5581-5586.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maki H., Kornberg A. Proofreading by DNA polymerase III of Escherichia coli depends on cooperative interaction of the polymerase and exonuclease subunits. Proc Natl Acad Sci U S A. 1987 Jul;84(13):4389–4392. doi: 10.1073/pnas.84.13.4389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maki H., Kornberg A. The polymerase subunit of DNA polymerase III of Escherichia coli. II. Purification of the alpha subunit, devoid of nuclease activities. J Biol Chem. 1985 Oct 25;260(24):12987–12992. [PubMed] [Google Scholar]
- Maurer R., Osmond B. C., Botstein D. Genetic analysis of DNA replication in bacteria: dnaB mutations that suppress dnaC mutations and dnaQ mutations that suppress dnaE mutations in Salmonella typhimurium. Genetics. 1984 Sep;108(1):25–38. doi: 10.1093/genetics/108.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McHenry C. S. DNA polymerase III holoenzyme of Escherichia coli. Annu Rev Biochem. 1988;57:519–550. doi: 10.1146/annurev.bi.57.070188.002511. [DOI] [PubMed] [Google Scholar]
- Modrich P. Mechanisms and biological effects of mismatch repair. Annu Rev Genet. 1991;25:229–253. doi: 10.1146/annurev.ge.25.120191.001305. [DOI] [PubMed] [Google Scholar]
- Rayssiguier C., Thaler D. S., Radman M. The barrier to recombination between Escherichia coli and Salmonella typhimurium is disrupted in mismatch-repair mutants. Nature. 1989 Nov 23;342(6248):396–401. doi: 10.1038/342396a0. [DOI] [PubMed] [Google Scholar]
- Sassanfar M., Roberts J. W. Nature of the SOS-inducing signal in Escherichia coli. The involvement of DNA replication. J Mol Biol. 1990 Mar 5;212(1):79–96. doi: 10.1016/0022-2836(90)90306-7. [DOI] [PubMed] [Google Scholar]
- Schmieger H. Phage P22-mutants with increased or decreased transduction abilities. Mol Gen Genet. 1972;119(1):75–88. doi: 10.1007/BF00270447. [DOI] [PubMed] [Google Scholar]
- Scott J. R. Regulation of plasmid replication. Microbiol Rev. 1984 Mar;48(1):1–23. doi: 10.1016/b978-0-12-048850-6.50006-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slater S. C., Maurer R. Requirements for bypass of UV-induced lesions in single-stranded DNA of bacteriophage phi X174 in Salmonella typhimurium. Proc Natl Acad Sci U S A. 1991 Feb 15;88(4):1251–1255. doi: 10.1073/pnas.88.4.1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloane D. L., Goodman M. F., Echols H. The fidelity of base selection by the polymerase subunit of DNA polymerase III holoenzyme. Nucleic Acids Res. 1988 Jul 25;16(14A):6465–6475. doi: 10.1093/nar/16.14.6465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studier F. W., Rosenberg A. H., Dunn J. J., Dubendorff J. W. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- Studwell P. S., O'Donnell M. Processive replication is contingent on the exonuclease subunit of DNA polymerase III holoenzyme. J Biol Chem. 1990 Jan 15;265(2):1171–1178. [PubMed] [Google Scholar]
- Stukenberg P. T., Studwell-Vaughan P. S., O'Donnell M. Mechanism of the sliding beta-clamp of DNA polymerase III holoenzyme. J Biol Chem. 1991 Jun 15;266(17):11328–11334. [PubMed] [Google Scholar]
- Sweasy J. B., Loeb L. A. Mammalian DNA polymerase beta can substitute for DNA polymerase I during DNA replication in Escherichia coli. J Biol Chem. 1992 Jan 25;267(3):1407–1410. [PubMed] [Google Scholar]
- Tanabe K., Bohn E. W., Wilson S. H. Steady-state kinetics of mouse DNA polymerase beta. Biochemistry. 1979 Jul 24;18(15):3401–3406. doi: 10.1021/bi00582a029. [DOI] [PubMed] [Google Scholar]
- Towbin H., Staehelin T., Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci U S A. 1979 Sep;76(9):4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner H. R., Demple B. F., Deutsch W. A., Kane C. M., Linn S. Apurinic/apyrimidinic endonucleases in repair of pyrimidine dimers and other lesions in DNA. Proc Natl Acad Sci U S A. 1980 Aug;77(8):4602–4606. doi: 10.1073/pnas.77.8.4602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witkin E. M. RecA protein in the SOS response: milestones and mysteries. Biochimie. 1991 Feb-Mar;73(2-3):133–141. doi: 10.1016/0300-9084(91)90196-8. [DOI] [PubMed] [Google Scholar]





