Abstract
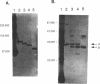
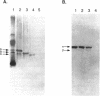
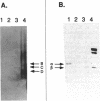
Four isozymes of bile salt hydrolase (BSH) have been purified from the cytosol of cells of Lactobacillus sp. strain 100-100. The four proteins were designated BSH A, B, C, and D. They eluted from anion-exchange high-pressure liquid chromatography columns at 0.15, 0.18, 0.21, and 0.25 M NaCl, respectively. They are catalytically similar, except that the Vmax of BSH D is about 10-fold lower than those of the other three isozymes. All four proteins consist of one or two polypeptides. The peptides have molecular weights of 42,000 and 38,000 and are designated alpha and beta, respectively. The approximate native molecular weights of BSH A, B, C, and D are 115,000, 105,000, 95,000, and 80,000, respectively. The native proteins are probably trimers; the four isozymes are the array of possible subunit combinations alpha 3, alpha 2 beta 1, alpha 1 beta 2, and beta 3 for A, B, C, and D, respectively. The two subunits are antigenically distinct. Polyclonal antibodies raised against BSH A (all alpha peptide) react in Western blots (immunoblots) only with proteins containing the alpha peptide; such antibodies raised against BSH D (all beta peptide) react only with proteins containing the beta peptide. The amino acid compositions of the two peptides differ. This is the first report of a bacterium that makes four BSH isozymes.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Dickinson A. B., Gustafsson B. E., Norman A. Determination of bile acid conversion potencies of intestinal bacteria by screening in vitro and subsequent establishment in germfree rats. Acta Pathol Microbiol Scand B Microbiol Immunol. 1971;79(5):691–698. doi: 10.1111/j.1699-0463.1971.tb00098.x. [DOI] [PubMed] [Google Scholar]
- Feighner S. D., Dashkevicz M. P. Subtherapeutic levels of antibiotics in poultry feeds and their effects on weight gain, feed efficiency, and bacterial cholyltaurine hydrolase activity. Appl Environ Microbiol. 1987 Feb;53(2):331–336. doi: 10.1128/aem.53.2.331-336.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopal-Srivastava R., Hylemon P. B. Purification and characterization of bile salt hydrolase from Clostridium perfringens. J Lipid Res. 1988 Aug;29(8):1079–1085. [PubMed] [Google Scholar]
- Hylemon P. B., Sherrod J. A. Multiple forms of 7-alpha-hydroxysteroid dehydrogenase in selected strains of Bacteroides fragilis. J Bacteriol. 1975 May;122(2):418–424. doi: 10.1128/jb.122.2.418-424.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hylemon P. B., Stellwag E. J. Bile acid biotransformation rates of selected gram-positive and gram-negative intestinal anaerobic bacteria. Biochem Biophys Res Commun. 1976 Apr 19;69(4):1088–1094. doi: 10.1016/0006-291x(76)90484-8. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lundeen S. G., Savage D. C. Characterization and purification of bile salt hydrolase from Lactobacillus sp. strain 100-100. J Bacteriol. 1990 Aug;172(8):4171–4177. doi: 10.1128/jb.172.8.4171-4177.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsudaira P. Sequence from picomole quantities of proteins electroblotted onto polyvinylidene difluoride membranes. J Biol Chem. 1987 Jul 25;262(21):10035–10038. [PubMed] [Google Scholar]
- Midtvedt T. Microbial bile acid transformation. Am J Clin Nutr. 1974 Nov;27(11):1341–1347. doi: 10.1093/ajcn/27.11.1341. [DOI] [PubMed] [Google Scholar]
- Nair P. P., Gordon M., Reback J. The enzymatic cleavage of the carbon-nitrogen bond in 3-alpha, 7-alpha, 12-alpha-trihydroxy-5-beta-cholan-24-oylglycine. J Biol Chem. 1967 Jan 10;242(1):7–11. [PubMed] [Google Scholar]
- Oakley B. R., Kirsch D. R., Morris N. R. A simplified ultrasensitive silver stain for detecting proteins in polyacrylamide gels. Anal Biochem. 1980 Jul 1;105(2):361–363. doi: 10.1016/0003-2697(80)90470-4. [DOI] [PubMed] [Google Scholar]
- Stellwag E. J., Hylemon P. B. Purification and characterization of bile salt hydrolase from Bacteroides fragilis subsp. fragilis. Biochim Biophys Acta. 1976 Nov 8;452(1):165–176. doi: 10.1016/0005-2744(76)90068-1. [DOI] [PubMed] [Google Scholar]
- Tannock G. W., Dashkevicz M. P., Feighner S. D. Lactobacilli and bile salt hydrolase in the murine intestinal tract. Appl Environ Microbiol. 1989 Jul;55(7):1848–1851. doi: 10.1128/aem.55.7.1848-1851.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towbin H., Staehelin T., Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci U S A. 1979 Sep;76(9):4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]