Abstract
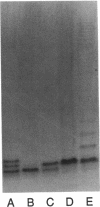
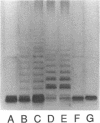
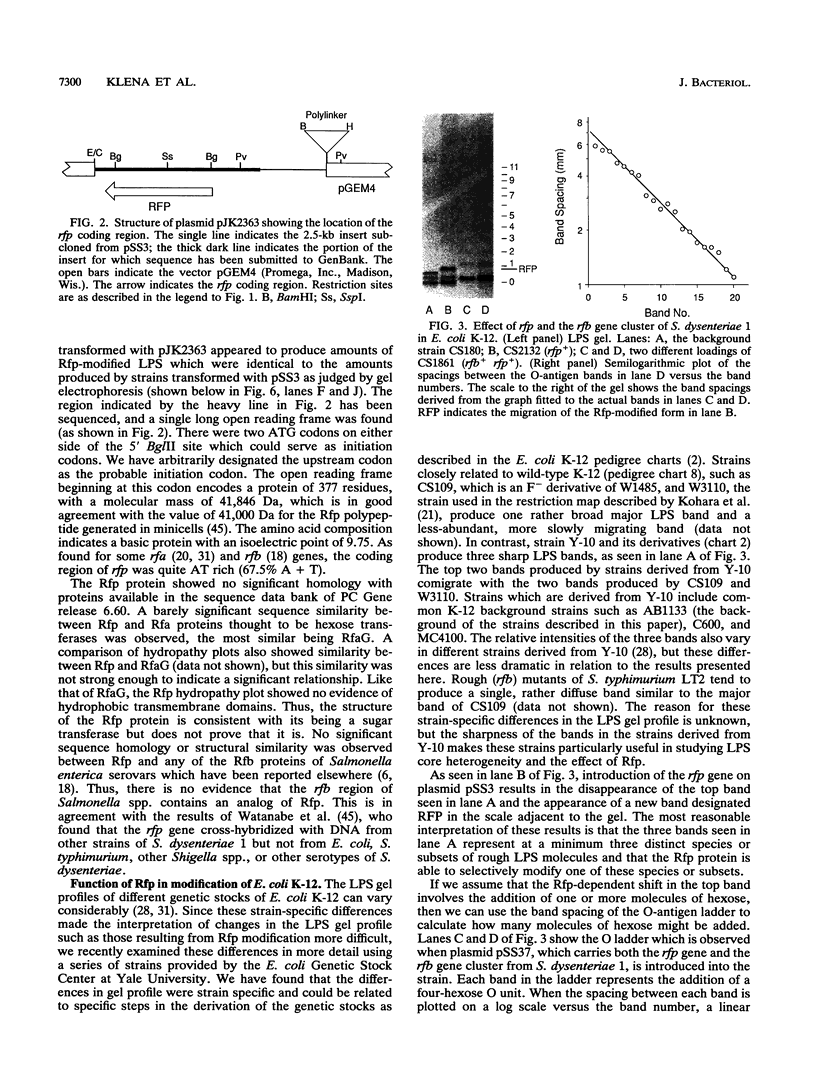
The rfp gene of Shigella dysenteriae 1 and the rfa genes of Escherichia coli K-12 and Salmonella typhimurium LT2 have been studied to determine their relationship to lipopolysaccharide (LPS) core heterogeneity and their role in the attachment of O antigen to LPS. It has been inferred from the nucleotide sequence that the rfp gene encodes a protein of 41,864 Da which has a structure similar to that of RfaG protein. Expression of this gene in E. coli K-12 results in the loss of one of the three bands seen in gel analysis of the LPS and in the appearance of a new, more slowly migrating band. This is consistent with the hypothesis that Rfp is a sugar transferase which modifies a subset of core molecules so that they become substrates for attachment of S. dysenteriae O antigen. A shift in gel migration of the bands carrying S. dysenteriae O antigen and disappearance of the Rfp-modified band in strains producing O antigen suggest that the core may be trimmed or modified further before attachment of O antigen. Mutation of rfaL results in a loss of the rough LPS band which appears to be modified by Rfp and prevents the appearance of the Rfp-modified band. Thus, RfaL protein is involved in core modification and is more than just a component of the O-antigen ligase. The products of rfaK and rfaQ also appear to be involved in modification of the core prior to attachment of O antigen, and the sites of rfaK modification are different in E. coli K-12 and S. typhimurium. In contrast, mutations in rfaS and rfaZ result in changes in the LPS core but do not affect the attachment of O antigen. We propose that these genes are involved in an alternative pathway for the synthesis of rough LPS species which are similar to lipooligosaccharides of other species and which are not substrates for O-antigen attachment. All of these studies indicate that the apparent heterogeneity of E. coli K-12 LPS observed on gels is not an artifact but instead a reflection of functional differences among LPS species.
Full text
PDF









Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Austin E. A., Graves J. F., Hite L. A., Parker C. T., Schnaitman C. A. Genetic analysis of lipopolysaccharide core biosynthesis by Escherichia coli K-12: insertion mutagenesis of the rfa locus. J Bacteriol. 1990 Sep;172(9):5312–5325. doi: 10.1128/jb.172.9.5312-5325.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachmann B. J. Linkage map of Escherichia coli K-12, edition 8. Microbiol Rev. 1990 Jun;54(2):130–197. doi: 10.1128/mr.54.2.130-197.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastin D. A., Romana L. K., Reeves P. R. Molecular cloning and expression in Escherichia coli K-12 of the rfb gene cluster determining the O antigen of an E. coli O111 strain. Mol Microbiol. 1991 Sep;5(9):2223–2231. doi: 10.1111/j.1365-2958.1991.tb02152.x. [DOI] [PubMed] [Google Scholar]
- Clementz T., Raetz C. R. A gene coding for 3-deoxy-D-manno-octulosonic-acid transferase in Escherichia coli. Identification, mapping, cloning, and sequencing. J Biol Chem. 1991 May 25;266(15):9687–9696. [PubMed] [Google Scholar]
- Engleberg N. C., Howe D. C., Rogers J. E., Arroyo J., Eisenstein B. I. Characterization of a Legionella pneumophila gene encoding a lipoprotein antigen. Mol Microbiol. 1991 Aug;5(8):2021–2029. doi: 10.1111/j.1365-2958.1991.tb00824.x. [DOI] [PubMed] [Google Scholar]
- Farquhar M. G. Progress in unraveling pathways of Golgi traffic. Annu Rev Cell Biol. 1985;1:447–488. doi: 10.1146/annurev.cb.01.110185.002311. [DOI] [PubMed] [Google Scholar]
- Fellay R., Frey J., Krisch H. Interposon mutagenesis of soil and water bacteria: a family of DNA fragments designed for in vitro insertional mutagenesis of gram-negative bacteria. Gene. 1987;52(2-3):147–154. doi: 10.1016/0378-1119(87)90041-2. [DOI] [PubMed] [Google Scholar]
- Hale T. L., Guerry P., Seid R. C., Jr, Kapfer C., Wingfield M. E., Reaves C. B., Baron L. S., Formal S. B. Expression of lipopolysaccharide O antigen in Escherichia coli K-12 hybrids containing plasmid and chromosomal genes from Shigella dysenteriae 1. Infect Immun. 1984 Nov;46(2):470–475. doi: 10.1128/iai.46.2.470-475.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haraguchi G. E., Hull R. A., Krallmann-Wenzel U., Hull S. I. Molecular cloning and expression of the O4 polysaccharide gene cluster from Escherichia coli. Microb Pathog. 1989 Feb;6(2):123–132. doi: 10.1016/0882-4010(89)90015-6. [DOI] [PubMed] [Google Scholar]
- Hitchcock P. J., Leive L., Mäkelä P. H., Rietschel E. T., Strittmatter W., Morrison D. C. Lipopolysaccharide nomenclature--past, present, and future. J Bacteriol. 1986 Jun;166(3):699–705. doi: 10.1128/jb.166.3.699-705.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holst O., Röhrscheidt-Andrzejewski E., Brade H. Isolation and characterisation of 3-deoxy-D-manno-2-octulopyranosonate 7-(2-aminoethyl phosphate) from the inner core region of Escherichia coli K-12 and Salmonella minnesota lipopolysaccharides. Carbohydr Res. 1990 Sep 5;204:93–102. doi: 10.1016/0008-6215(90)84024-o. [DOI] [PubMed] [Google Scholar]
- Holst O., Zähringer U., Brade H., Zamojski A. Structural analysis of the heptose/hexose region of the lipopolysaccharide from Escherichia coli K-12 strain W3100. Carbohydr Res. 1991 Aug 20;215(2):323–335. doi: 10.1016/0008-6215(91)84031-9. [DOI] [PubMed] [Google Scholar]
- Jansson P. E., Lindberg A. A., Lindberg B., Wollin R. Structural studies on the hexose region of the core in lipopolysaccharides from Enterobacteriaceae. Eur J Biochem. 1981 Apr;115(3):571–577. doi: 10.1111/j.1432-1033.1981.tb06241.x. [DOI] [PubMed] [Google Scholar]
- Jiang X. M., Neal B., Santiago F., Lee S. J., Romana L. K., Reeves P. R. Structure and sequence of the rfb (O antigen) gene cluster of Salmonella serovar typhimurium (strain LT2). Mol Microbiol. 1991 Mar;5(3):695–713. doi: 10.1111/j.1365-2958.1991.tb00741.x. [DOI] [PubMed] [Google Scholar]
- Klena J. D., Pradel E., Schnaitman C. A. Comparison of lipopolysaccharide biosynthesis genes rfaK, rfaL, rfaY, and rfaZ of Escherichia coli K-12 and Salmonella typhimurium. J Bacteriol. 1992 Jul;174(14):4746–4752. doi: 10.1128/jb.174.14.4746-4752.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohara Y., Akiyama K., Isono K. The physical map of the whole E. coli chromosome: application of a new strategy for rapid analysis and sorting of a large genomic library. Cell. 1987 Jul 31;50(3):495–508. doi: 10.1016/0092-8674(87)90503-4. [DOI] [PubMed] [Google Scholar]
- Lesse A. J., Campagnari A. A., Bittner W. E., Apicella M. A. Increased resolution of lipopolysaccharides and lipooligosaccharides utilizing tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis. J Immunol Methods. 1990 Jan 24;126(1):109–117. doi: 10.1016/0022-1759(90)90018-q. [DOI] [PubMed] [Google Scholar]
- Lindberg A. A., Holme T. Influence of O side chains on the attachment of the Felix O-1 bacteriophage to Salmonella bacteria. J Bacteriol. 1969 Aug;99(2):513–519. doi: 10.1128/jb.99.2.513-519.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLachlan P. R., Kadam S. K., Sanderson K. E. Cloning, characterization, and DNA sequence of the rfaLK region for lipopolysaccharide synthesis in Salmonella typhimurium LT2. J Bacteriol. 1991 Nov;173(22):7151–7163. doi: 10.1128/jb.173.22.7151-7163.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manoil C. Analysis of protein localization by use of gene fusions with complementary properties. J Bacteriol. 1990 Feb;172(2):1035–1042. doi: 10.1128/jb.172.2.1035-1042.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker C. T., Kloser A. W., Schnaitman C. A., Stein M. A., Gottesman S., Gibson B. W. Role of the rfaG and rfaP genes in determining the lipopolysaccharide core structure and cell surface properties of Escherichia coli K-12. J Bacteriol. 1992 Apr;174(8):2525–2538. doi: 10.1128/jb.174.8.2525-2538.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker C. T., Pradel E., Schnaitman C. A. Identification and sequences of the lipopolysaccharide core biosynthetic genes rfaQ, rfaP, and rfaG of Escherichia coli K-12. J Bacteriol. 1992 Feb;174(3):930–934. doi: 10.1128/jb.174.3.930-934.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pradel E., Parker C. T., Schnaitman C. A. Structures of the rfaB, rfaI, rfaJ, and rfaS genes of Escherichia coli K-12 and their roles in assembly of the lipopolysaccharide core. J Bacteriol. 1992 Jul;174(14):4736–4745. doi: 10.1128/jb.174.14.4736-4745.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pradel E., Schnaitman C. A. Effect of rfaH (sfrB) and temperature on expression of rfa genes of Escherichia coli K-12. J Bacteriol. 1991 Oct;173(20):6428–6431. doi: 10.1128/jb.173.20.6428-6431.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raina S., Georgopoulos C. The htrM gene, whose product is essential for Escherichia coli viability only at elevated temperatures, is identical to the rfaD gene. Nucleic Acids Res. 1991 Jul 25;19(14):3811–3819. doi: 10.1093/nar/19.14.3811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley L. W., Junio L. N., Libaek L. B., Schoolnik G. K. Plasmid-encoded expression of lipopolysaccharide O-antigenic polysaccharide in enteropathogenic Escherichia coli. Infect Immun. 1987 Sep;55(9):2052–2056. doi: 10.1128/iai.55.9.2052-2056.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roncero C., Casadaban M. J. Genetic analysis of the genes involved in synthesis of the lipopolysaccharide core in Escherichia coli K-12: three operons in the rfa locus. J Bacteriol. 1992 May;174(10):3250–3260. doi: 10.1128/jb.174.10.3250-3260.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roncero C., Sanderson K. E., Casadaban M. J. Analysis of the host ranges of transposon bacteriophages Mu, MuhP1, and D108 by use of lipopolysaccharide mutants of Salmonella typhimurium LT2. J Bacteriol. 1991 Aug;173(16):5230–5233. doi: 10.1128/jb.173.16.5230-5233.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnaitman C. A., Parker C. T., Klena J. D., Pradel E. L., Pearson N. B., Sanderson K. E., MacClachlan P. R. Physical maps of the rfa loci of Escherichia coli K-12 and Salmonella typhimurium. J Bacteriol. 1991 Dec;173(23):7410–7411. doi: 10.1128/jb.173.23.7410-7411.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturm S., Jann B., Jann K., Fortnagel P., Timmis K. N. Genetic and biochemical analysis of Shigella dysenteriae 1 O antigen polysaccharide biosynthesis in Escherichia coli K-12: 9 kb plasmid of S. dysenteriae 1 determines addition of a galactose residue to the lipopolysaccharide core. Microb Pathog. 1986 Jun;1(3):299–306. doi: 10.1016/0882-4010(86)90055-0. [DOI] [PubMed] [Google Scholar]
- Sturm S., Jann B., Jann K., Fortnagel P., Timmis K. N. Genetic and biochemical analysis of Shigella dysenteriae 1 O antigen polysaccharide biosynthesis in Escherichia coli K-12: structure and functions of the rfb gene cluster. Microb Pathog. 1986 Jun;1(3):307–324. doi: 10.1016/0882-4010(86)90056-2. [DOI] [PubMed] [Google Scholar]
- Sturm S., Timmis K. N. Cloning of the rfb gene region of Shigella dysenteriae 1 and construction of an rfb-rfp gene cassette for the development of lipopolysaccharide-based live anti-dysentery vaccines. Microb Pathog. 1986 Jun;1(3):289–297. doi: 10.1016/0882-4010(86)90054-9. [DOI] [PubMed] [Google Scholar]
- Valvano M. A., Crosa J. H. Molecular cloning and expression in Escherichia coli K-12 of chromosomal genes determining the O7 lipopolysaccharide antigen of a human invasive strain of E. coli O7:K1. Infect Immun. 1989 Mar;57(3):937–943. doi: 10.1128/iai.57.3.937-943.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe H., Nakamura A., Timmis K. N. Small virulence plasmid of Shigella dysenteriae 1 strain W30864 encodes a 41,000-dalton protein involved in formation of specific lipopolysaccharide side chains of serotype 1 isolates. Infect Immun. 1984 Oct;46(1):55–63. doi: 10.1128/iai.46.1.55-63.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe H., Timmis K. N. A small plasmid in Shigella dysenteriae 1 specifies one or more functions essential for O antigen production and bacterial virulence. Infect Immun. 1984 Jan;43(1):391–396. doi: 10.1128/iai.43.1.391-396.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]