Abstract
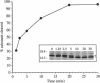
PilD, originally isolated as an essential component for the biogenesis of the type IV pili of Pseudomonas aeruginosa, is a unique endopeptidase responsible for processing the precursors of the P. aeruginosa pilin subunits. It is also required for the cleavage of the leader peptides from the Pdd proteins, which are essential components of an extracellular secretion pathway specific for the export of a number of P. aeruginosa hydrolytic enzymes and toxins. Substrates for PilD are initially synthesized with short, i.e., 6- to 8-amino-acid-long, leader peptides with a net basic charge and share a high degree of amino acid homology through the first 16 to 30 residues at the amino terminus. In addition, they all have a phenylalanine residue at the +1 site relative to the cleavage site, which is N methylated prior to assembly into the oligomeric structures. In this study, the kinetics of leader peptide cleavage from the precursor of the P. aeruginosa pilin subunit by PilD was determined in vitro. The rates of cleavage were compared for purified enzyme and substrate as well as for enzyme and substrate contained within total membranes extracted from P. aeruginosa strains overexpressing the cloned pilD or pilA genes. Optimal conditions were obtained only when both PilD and substrate were contained within total membranes. PilD catalysis of P. aeruginosa prepilin followed normal Michaelis-Menten kinetics, with a measured apparent Km of approximately 650 microM, and a kcat of 180 min-1. The kinetics of PilD processing of another type IV pilin precursor, that from Neisseria gonorrhoeae with a 7-amino-acid-long leader peptide, were essentially the same as that measured for wild-type P. aeruginosa prepilin. Quite different results were obtained for a number of prepilin substrates containing substitutions at the conserved phenylalanine at the +1 position relative to the cleavage site, which were previously shown to be well tolerated in vivo. Substitutions of methionine, serine, and cysteine for phenylalanine show that Km values remain close to that measured for wild-type substrate, while kcat and kcat/Km values were significantly decreased. This indicates that while the affinity of enzyme for substrate is relatively unaffected by the substitutions, the maximum rate of catalysis favors a phenylalanine at this position. Interesting, PilD cleavage of one mutated pillin (asparagine) resulted in a lower Km value of 52.5 microM, which indicates a higher affinity for the enzyme, as well as a lower kcat value of 6.1 min m(-1). This suggests that it may be feasible to design peptide inhibitors of PilD.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Albano M., Breitling R., Dubnau D. A. Nucleotide sequence and genetic organization of the Bacillus subtilis comG operon. J Bacteriol. 1989 Oct;171(10):5386–5404. doi: 10.1128/jb.171.10.5386-5404.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bally M., Ball G., Badere A., Lazdunski A. Protein secretion in Pseudomonas aeruginosa: the xcpA gene encodes an integral inner membrane protein homologous to Klebsiella pneumoniae secretion function protein PulO. J Bacteriol. 1991 Jan;173(2):479–486. doi: 10.1128/jb.173.2.479-486.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bally M., Filloux A., Akrim M., Ball G., Lazdunski A., Tommassen J. Protein secretion in Pseudomonas aeruginosa: characterization of seven xcp genes and processing of secretory apparatus components by prepilin peptidase. Mol Microbiol. 1992 May;6(9):1121–1131. doi: 10.1111/j.1365-2958.1992.tb01550.x. [DOI] [PubMed] [Google Scholar]
- Bodey G. P., Bolivar R., Fainstein V., Jadeja L. Infections caused by Pseudomonas aeruginosa. Rev Infect Dis. 1983 Mar-Apr;5(2):279–313. doi: 10.1093/clinids/5.2.279. [DOI] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Breitling R., Dubnau D. A membrane protein with similarity to N-methylphenylalanine pilins is essential for DNA binding by competent Bacillus subtilis. J Bacteriol. 1990 Mar;172(3):1499–1508. doi: 10.1128/jb.172.3.1499-1508.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DAVIS B. D., MINGIOLI E. S. Mutants of Escherichia coli requiring methionine or vitamin B12. J Bacteriol. 1950 Jul;60(1):17–28. doi: 10.1128/jb.60.1.17-28.1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalbey R. E. Leader peptidase. Mol Microbiol. 1991 Dec;5(12):2855–2860. doi: 10.1111/j.1365-2958.1991.tb01844.x. [DOI] [PubMed] [Google Scholar]
- Dev I. K., Harvey R. J., Ray P. H. Inhibition of prolipoprotein signal peptidase by globomycin. J Biol Chem. 1985 May 25;260(10):5891–5894. [PubMed] [Google Scholar]
- Dev I. K., Ray P. H., Novak P. Minimum substrate sequence for signal peptidase I of Escherichia coli. J Biol Chem. 1990 Nov 25;265(33):20069–20072. [PubMed] [Google Scholar]
- Dev I. K., Ray P. H. Signal peptidases and signal peptide hydrolases. J Bioenerg Biomembr. 1990 Jun;22(3):271–290. doi: 10.1007/BF00763168. [DOI] [PubMed] [Google Scholar]
- Dums F., Dow J. M., Daniels M. J. Structural characterization of protein secretion genes of the bacterial phytopathogen Xanthomonas campestris pathovar campestris: relatedness to secretion systems of other gram-negative bacteria. Mol Gen Genet. 1991 Oct;229(3):357–364. doi: 10.1007/BF00267456. [DOI] [PubMed] [Google Scholar]
- Eisenthal R., Cornish-Bowden A. The direct linear plot. A new graphical procedure for estimating enzyme kinetic parameters. Biochem J. 1974 Jun;139(3):715–720. doi: 10.1042/bj1390715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fürste J. P., Pansegrau W., Frank R., Blöcker H., Scholz P., Bagdasarian M., Lanka E. Molecular cloning of the plasmid RP4 primase region in a multi-host-range tacP expression vector. Gene. 1986;48(1):119–131. doi: 10.1016/0378-1119(86)90358-6. [DOI] [PubMed] [Google Scholar]
- Hart M. E., Champlin F. R. Susceptibility to hydrophobic molecules and phospholipid composition in Pasteurella multocida and Actinobacillus lignieresii. Antimicrob Agents Chemother. 1988 Sep;32(9):1354–1359. doi: 10.1128/aac.32.9.1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He S. Y., Lindeberg M., Chatterjee A. K., Collmer A. Cloned Erwinia chrysanthemi out genes enable Escherichia coli to selectively secrete a diverse family of heterologous proteins to its milieu. Proc Natl Acad Sci U S A. 1991 Feb 1;88(3):1079–1083. doi: 10.1073/pnas.88.3.1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson K., Parker M. L., Lory S. Nucleotide sequence and transcriptional initiation site of two Pseudomonas aeruginosa pilin genes. J Biol Chem. 1986 Nov 25;261(33):15703–15708. [PubMed] [Google Scholar]
- Kaufman M. R., Seyer J. M., Taylor R. K. Processing of TCP pilin by TcpJ typifies a common step intrinsic to a newly recognized pathway of extracellular protein secretion by gram-negative bacteria. Genes Dev. 1991 Oct;5(10):1834–1846. doi: 10.1101/gad.5.10.1834. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Marrs C. F., Schoolnik G., Koomey J. M., Hardy J., Rothbard J., Falkow S. Cloning and sequencing of a Moraxella bovis pilin gene. J Bacteriol. 1985 Jul;163(1):132–139. doi: 10.1128/jb.163.1.132-139.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKern N. M., O'Donnell I. J., Inglis A. S., Stewart D. J., Clark B. L. Amino acid sequence of pilin from Bacteroides nodosus (strain 198), the causative organism of ovine footrot. FEBS Lett. 1983 Nov 28;164(1):149–153. doi: 10.1016/0014-5793(83)80039-8. [DOI] [PubMed] [Google Scholar]
- Meyer T. F., Billyard E., Haas R., Storzbach S., So M. Pilus genes of Neisseria gonorrheae: chromosomal organization and DNA sequence. Proc Natl Acad Sci U S A. 1984 Oct;81(19):6110–6114. doi: 10.1073/pnas.81.19.6110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohan S., Aghion J., Guillen N., Dubnau D. Molecular cloning and characterization of comC, a late competence gene of Bacillus subtilis. J Bacteriol. 1989 Nov;171(11):6043–6051. doi: 10.1128/jb.171.11.6043-6051.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunn D. N., Lory S. Components of the protein-excretion apparatus of Pseudomonas aeruginosa are processed by the type IV prepilin peptidase. Proc Natl Acad Sci U S A. 1992 Jan 1;89(1):47–51. doi: 10.1073/pnas.89.1.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunn D. N., Lory S. Product of the Pseudomonas aeruginosa gene pilD is a prepilin leader peptidase. Proc Natl Acad Sci U S A. 1991 Apr 15;88(8):3281–3285. doi: 10.1073/pnas.88.8.3281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunn D., Bergman S., Lory S. Products of three accessory genes, pilB, pilC, and pilD, are required for biogenesis of Pseudomonas aeruginosa pili. J Bacteriol. 1990 Jun;172(6):2911–2919. doi: 10.1128/jb.172.6.2911-2919.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pugsley A. P., Dupuy B. An enzyme with type IV prepilin peptidase activity is required to process components of the general extracellular protein secretion pathway of Klebsiella oxytoca. Mol Microbiol. 1992 Mar;6(6):751–760. doi: 10.1111/j.1365-2958.1992.tb01525.x. [DOI] [PubMed] [Google Scholar]
- Reyss I., Pugsley A. P. Five additional genes in the pulC-O operon of the gram-negative bacterium Klebsiella oxytoca UNF5023 which are required for pullulanase secretion. Mol Gen Genet. 1990 Jul;222(2-3):176–184. doi: 10.1007/BF00633815. [DOI] [PubMed] [Google Scholar]
- Schägger H., von Jagow G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal Biochem. 1987 Nov 1;166(2):368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- Shaw C. E., Taylor R. K. Vibrio cholerae O395 tcpA pilin gene sequence and comparison of predicted protein structural features to those of type 4 pilins. Infect Immun. 1990 Sep;58(9):3042–3049. doi: 10.1128/iai.58.9.3042-3049.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson D. A., Ramphal R., Lory S. Genetic analysis of Pseudomonas aeruginosa adherence: distinct genetic loci control attachment to epithelial cells and mucins. Infect Immun. 1992 Sep;60(9):3771–3779. doi: 10.1128/iai.60.9.3771-3779.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strom M. S., Lory S. Amino acid substitutions in pilin of Pseudomonas aeruginosa. Effect on leader peptide cleavage, amino-terminal methylation, and pilus assembly. J Biol Chem. 1991 Jan 25;266(3):1656–1664. [PubMed] [Google Scholar]
- Strom M. S., Nunn D., Lory S. Multiple roles of the pilus biogenesis protein pilD: involvement of pilD in excretion of enzymes from Pseudomonas aeruginosa. J Bacteriol. 1991 Feb;173(3):1175–1180. doi: 10.1128/jb.173.3.1175-1180.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods D. E., Straus D. C., Johanson W. G., Jr, Berry V. K., Bass J. A. Role of pili in adherence of Pseudomonas aeruginosa to mammalian buccal epithelial cells. Infect Immun. 1980 Sep;29(3):1146–1151. doi: 10.1128/iai.29.3.1146-1151.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]