Abstract
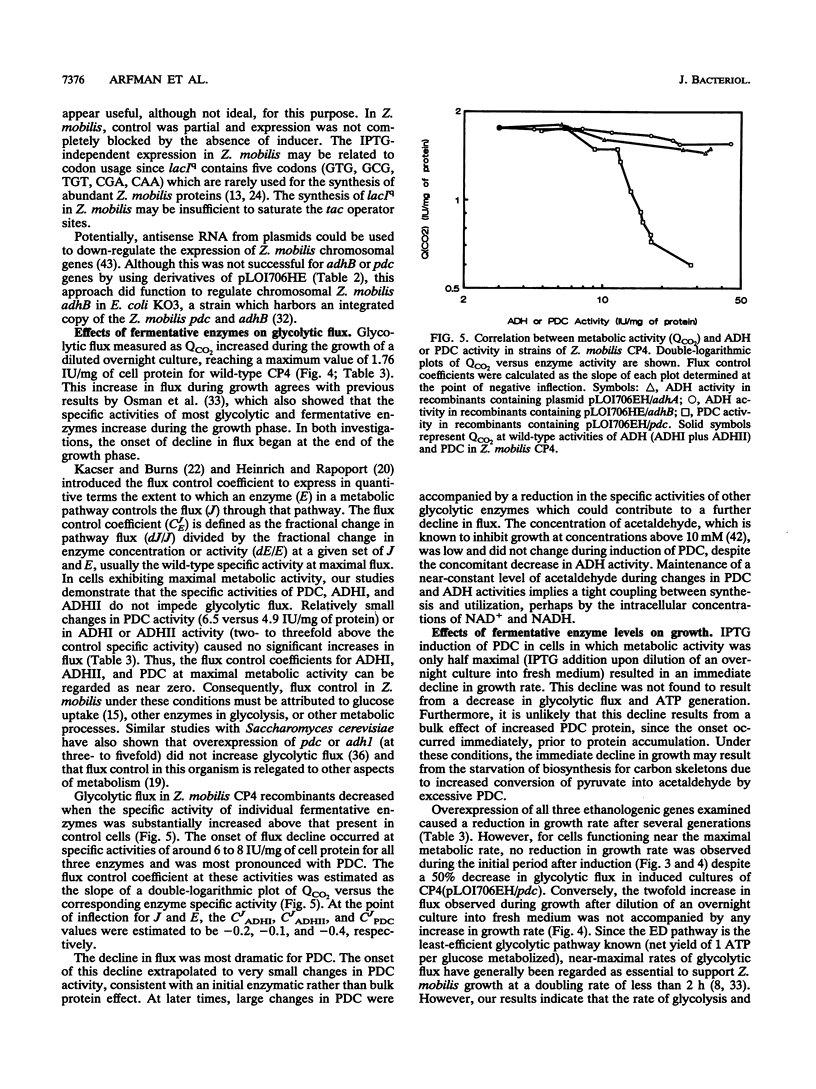
The Zymomonas mobilis genes encoding alcohol dehydrogenase I (adhA), alcohol dehydrogenase II (adhB), and pyruvate decarboxylase (pdc) were overexpressed in Escherichia coli and Z. mobilis by using a broad-host-range vector containing the tac promoter and the lacIq repressor gene. Maximal IPTG (isopropyl-beta-D-thiogalactopyranoside) induction of these plasmid-borne genes in Z. mobilis resulted in a 35-fold increase in alcohol dehydrogenase I activity, a 16.7-fold increase in alcohol dehydrogenase II activity, and a 6.3-fold increase in pyruvate decarboxylase activity. Small changes in the activities of these enzymes did not affect glycolytic flux in cells which are at maximal metabolic activity, indicating that flux under these conditions is controlled at some other point in metabolism. Expression of adhA, adhB, or pdc at high specific activities (above 8 IU/mg of cell protein) resulted in a decrease in glycolytic flux (negative flux control coefficients), which was most pronounced for pyruvate decarboxylase. Growth rate and flux are imperfectly coupled in this organism. Neither a twofold increase in flux nor a 50% decline from maximal flux caused any immediate change in growth rate. Thus, the rates of biosynthesis and growth in this organism are not limited by energy generation in rich medium.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aldrich H. C., McDowell L., Barbosa M. F., Yomano L. P., Scopes R. K., Ingram L. O. Immunocytochemical localization of glycolytic and fermentative enzymes in Zymomonas mobilis. J Bacteriol. 1992 Jul;174(13):4504–4508. doi: 10.1128/jb.174.13.4504-4508.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An H., Scopes R. K., Rodriguez M., Keshav K. F., Ingram L. O. Gel electrophoretic analysis of Zymomonas mobilis glycolytic and fermentative enzymes: identification of alcohol dehydrogenase II as a stress protein. J Bacteriol. 1991 Oct;173(19):5975–5982. doi: 10.1128/jb.173.19.5975-5982.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnell W. O., Yi K. C., Conway T. Sequence and genetic organization of a Zymomonas mobilis gene cluster that encodes several enzymes of glucose metabolism. J Bacteriol. 1990 Dec;172(12):7227–7240. doi: 10.1128/jb.172.12.7227-7240.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrow K. D., Collins J. G., Norton R. S., Rogers P. L., Smith G. M. 31P nuclear magnetic resonance studies of the fermentation of glucose to ethanol by Zymomonas mobilis. J Biol Chem. 1984 May 10;259(9):5711–5716. [PubMed] [Google Scholar]
- Conway T., Fliege R., Jones-Kilpatrick D., Liu J., Barnell W. O., Egan S. E. Cloning, characterization and expression of the Zymononas mobilis eda gene that encodes 2-keto-3-deoxy-6-phosphogluconate aldolase of the Entner-Doudoroff pathway. Mol Microbiol. 1991 Dec;5(12):2901–2911. doi: 10.1111/j.1365-2958.1991.tb01850.x. [DOI] [PubMed] [Google Scholar]
- Conway T., Ingram L. O. Phosphoglycerate kinase gene from Zymomonas mobilis: cloning, sequencing, and localization within the gap operon. J Bacteriol. 1988 Apr;170(4):1926–1933. doi: 10.1128/jb.170.4.1926-1933.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conway T., Osman Y. A., Konnan J. I., Hoffmann E. M., Ingram L. O. Promoter and nucleotide sequences of the Zymomonas mobilis pyruvate decarboxylase. J Bacteriol. 1987 Mar;169(3):949–954. doi: 10.1128/jb.169.3.949-954.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conway T., Sewell G. W., Ingram L. O. Glyceraldehyde-3-phosphate dehydrogenase gene from Zymomonas mobilis: cloning, sequencing, and identification of promoter region. J Bacteriol. 1987 Dec;169(12):5653–5662. doi: 10.1128/jb.169.12.5653-5662.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conway T., Sewell G. W., Osman Y. A., Ingram L. O. Cloning and sequencing of the alcohol dehydrogenase II gene from Zymomonas mobilis. J Bacteriol. 1987 Jun;169(6):2591–2597. doi: 10.1128/jb.169.6.2591-2597.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimarco A. A., Romano A. H. d-Glucose Transport System of Zymomonas mobilis. Appl Environ Microbiol. 1985 Jan;49(1):151–157. doi: 10.1128/aem.49.1.151-157.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fürste J. P., Pansegrau W., Frank R., Blöcker H., Scholz P., Bagdasarian M., Lanka E. Molecular cloning of the plasmid RP4 primase region in a multi-host-range tacP expression vector. Gene. 1986;48(1):119–131. doi: 10.1016/0378-1119(86)90358-6. [DOI] [PubMed] [Google Scholar]
- Heinrich R., Rapoport T. A. A linear steady-state treatment of enzymatic chains. General properties, control and effector strength. Eur J Biochem. 1974 Feb 15;42(1):89–95. doi: 10.1111/j.1432-1033.1974.tb03318.x. [DOI] [PubMed] [Google Scholar]
- Kacser H., Burns J. A. The control of flux. Symp Soc Exp Biol. 1973;27:65–104. [PubMed] [Google Scholar]
- Keshav K. F., Yomano L. P., An H. J., Ingram L. O. Cloning of the Zymomonas mobilis structural gene encoding alcohol dehydrogenase I (adhA): sequence comparison and expression in Escherichia coli. J Bacteriol. 1990 May;172(5):2491–2497. doi: 10.1128/jb.172.5.2491-2497.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Mackenzie K. F., Eddy C. K., Ingram L. O. Modulation of alcohol dehydrogenase isoenzyme levels in Zymomonas mobilis by iron and zinc. J Bacteriol. 1989 Feb;171(2):1063–1067. doi: 10.1128/jb.171.2.1063-1067.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mejia J. P., Burnett M. E., An H., Barnell W. O., Keshav K. F., Conway T., Ingram L. O. Coordination of expression of Zymomonas mobilis glycolytic and fermentative enzymes: a simple hypothesis based on mRNA stability. J Bacteriol. 1992 Oct;174(20):6438–6443. doi: 10.1128/jb.174.20.6438-6443.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neale A. D., Scopes R. K., Kelly J. M., Wettenhall R. E. The two alcohol dehydrogenases of Zymomonas mobilis. Purification by differential dye ligand chromatography, molecular characterisation and physiological roles. Eur J Biochem. 1986 Jan 2;154(1):119–124. doi: 10.1111/j.1432-1033.1986.tb09366.x. [DOI] [PubMed] [Google Scholar]
- Neale A. D., Scopes R. K., Wettenhall R. E., Hoogenraad N. J. Pyruvate decarboxylase of Zymomonas mobilis: isolation, properties, and genetic expression in Escherichia coli. J Bacteriol. 1987 Mar;169(3):1024–1028. doi: 10.1128/jb.169.3.1024-1028.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta K., Beall D. S., Mejia J. P., Shanmugam K. T., Ingram L. O. Genetic improvement of Escherichia coli for ethanol production: chromosomal integration of Zymomonas mobilis genes encoding pyruvate decarboxylase and alcohol dehydrogenase II. Appl Environ Microbiol. 1991 Apr;57(4):893–900. doi: 10.1128/aem.57.4.893-900.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawluk A., Scopes R. K., Griffiths-Smith K. Isolation and properties of the glycolytic enzymes from Zymomonas mobilis. The five enzymes from glyceraldehyde-3-phosphate dehydrogenase through to pyruvate kinase. Biochem J. 1986 Aug 15;238(1):275–281. doi: 10.1042/bj2380275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saiki R. K., Gelfand D. H., Stoffel S., Scharf S. J., Higuchi R., Horn G. T., Mullis K. B., Erlich H. A. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science. 1988 Jan 29;239(4839):487–491. doi: 10.1126/science.2448875. [DOI] [PubMed] [Google Scholar]
- Schaaff I., Heinisch J., Zimmermann F. K. Overproduction of glycolytic enzymes in yeast. Yeast. 1989 Jul-Aug;5(4):285–290. doi: 10.1002/yea.320050408. [DOI] [PubMed] [Google Scholar]
- Scholz P., Haring V., Wittmann-Liebold B., Ashman K., Bagdasarian M., Scherzinger E. Complete nucleotide sequence and gene organization of the broad-host-range plasmid RSF1010. Gene. 1989 Feb 20;75(2):271–288. doi: 10.1016/0378-1119(89)90273-4. [DOI] [PubMed] [Google Scholar]
- Scopes R. K., Testolin V., Stoter A., Griffiths-Smith K., Algar E. M. Simultaneous purification and characterization of glucokinase, fructokinase and glucose-6-phosphate dehydrogenase from Zymomonas mobilis. Biochem J. 1985 Jun 15;228(3):627–634. doi: 10.1042/bj2280627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutcliffe J. G. Complete nucleotide sequence of the Escherichia coli plasmid pBR322. Cold Spring Harb Symp Quant Biol. 1979;43(Pt 1):77–90. doi: 10.1101/sqb.1979.043.01.013. [DOI] [PubMed] [Google Scholar]
- Wecker M. S., Zall R. R. Production of Acetaldehyde by Zymomonas mobilis. Appl Environ Microbiol. 1987 Dec;53(12):2815–2820. doi: 10.1128/aem.53.12.2815-2820.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lorenzo V., Herrero M., Jakubzik U., Timmis K. N. Mini-Tn5 transposon derivatives for insertion mutagenesis, promoter probing, and chromosomal insertion of cloned DNA in gram-negative eubacteria. J Bacteriol. 1990 Nov;172(11):6568–6572. doi: 10.1128/jb.172.11.6568-6572.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]