Abstract
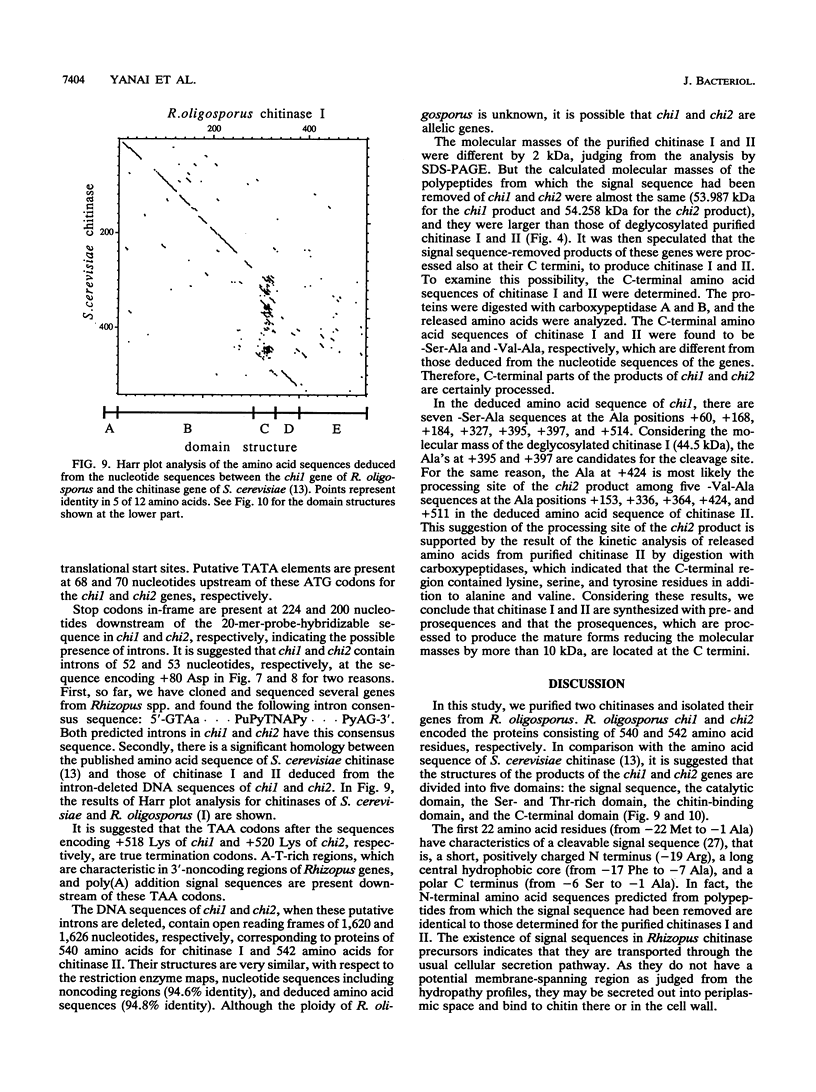
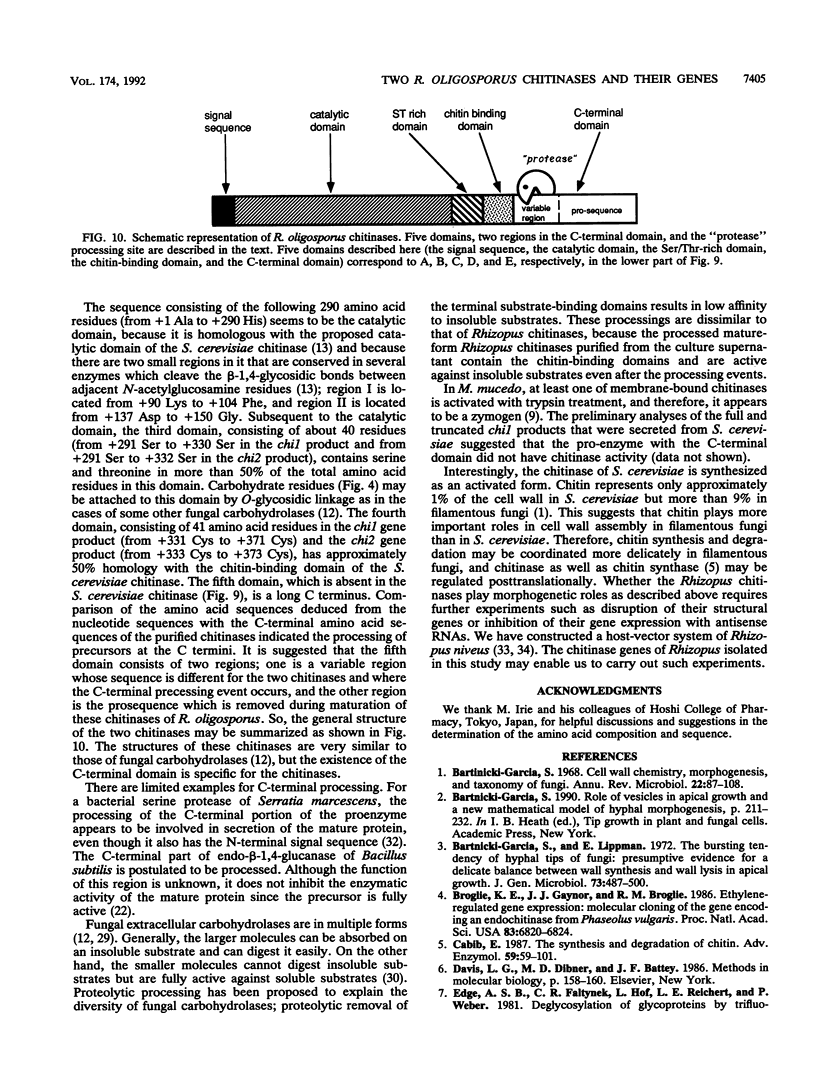
Two chitinases were purified from Rhizopus oligosporus, a filamentous fungus belonging to the class Zygomycetes, and designated chitinase I and chitinase II. Their N-terminal amino acid sequences were determined, and two synthetic oligonucleotide probes corresponding to these amino acid sequences were synthesized. Southern blot analyses of the total genomic DNA from R. oligosporus with these oligonucleotides as probes indicated that one of the two genes encoding these two chitinases was contained in a 2.9-kb EcoRI fragment and in a 3.6-kb HindIII fragment and that the other one was contained in a 2.9-kb EcoRI fragment and in a 11.5-kb HindIII fragment. Two DNA fragments were isolated from the phage bank of R. oligosporus genomic DNA with the synthetic oligonucleotides as probes. The restriction enzyme analyses of these fragments coincided with the Southern blot analyses described above and the amino acid sequences deduced from their nucleotide sequences contained those identical to the determined N-terminal amino acid sequences of the purified chitinases, indicating that each of these fragments contained a gene encoding chitinase (designated chi 1 and chi 2, encoding chitinase I and II, respectively). The deduced amino acid sequences of these two genes had domain structures similar to that of the published sequence of chitinase of Saccharomyces cerevisiae, except that they had an additional C-terminal domain. Furthermore, there were significant differences between the molecular weights experimentally determined with the two purified enzymes and those deduced from the nucleotide sequences for both genes. Analysis of the N- and C-terminal amino acid sequences of both chitinases and comparison of them with the amino acid sequences deduced from the nucleotide sequences revealed posttranslational processing not only at the N-terminal signal sequences but also at the C-terminal domains. It is concluded that these chitinases are synthesized with pre- and prosequences in addition to the mature enzyme sequences and that the prosequences are located at the C terminal.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bartnicki-Garcia S. Cell wall chemistry, morphogenesis, and taxonomy of fungi. Annu Rev Microbiol. 1968;22:87–108. doi: 10.1146/annurev.mi.22.100168.000511. [DOI] [PubMed] [Google Scholar]
- Broglie K. E., Gaynor J. J., Broglie R. M. Ethylene-regulated gene expression: molecular cloning of the genes encoding an endochitinase from Phaseolus vulgaris. Proc Natl Acad Sci U S A. 1986 Sep;83(18):6820–6824. doi: 10.1073/pnas.83.18.6820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabib E. The synthesis and degradation of chitin. Adv Enzymol Relat Areas Mol Biol. 1987;59:59–101. doi: 10.1002/9780470123058.ch2. [DOI] [PubMed] [Google Scholar]
- Edge A. S., Faltynek C. R., Hof L., Reichert L. E., Jr, Weber P. Deglycosylation of glycoproteins by trifluoromethanesulfonic acid. Anal Biochem. 1981 Nov 15;118(1):131–137. doi: 10.1016/0003-2697(81)90168-8. [DOI] [PubMed] [Google Scholar]
- Horiuchi H., Yanai K., Okazaki T., Takagi M., Yano K. Isolation and sequencing of a genomic clone encoding aspartic proteinase of Rhizopus niveus. J Bacteriol. 1988 Jan;170(1):272–278. doi: 10.1128/jb.170.1.272-278.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones J. D., Grady K. L., Suslow T. V., Bedbrook J. R. Isolation and characterization of genes encoding two chitinase enzymes from Serratia marcescens. EMBO J. 1986 Mar;5(3):467–473. doi: 10.1002/j.1460-2075.1986.tb04235.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karn J., Matthes H. W., Gait M. J., Brenner S. A new selective phage cloning vector, lambda 2001, with sites for XbaI, BamHI, HindIII, EcoRI, SstI and XhoI. Gene. 1984 Dec;32(1-2):217–224. doi: 10.1016/0378-1119(84)90049-0. [DOI] [PubMed] [Google Scholar]
- Kuranda M. J., Robbins P. W. Chitinase is required for cell separation during growth of Saccharomyces cerevisiae. J Biol Chem. 1991 Oct 15;266(29):19758–19767. [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Metraux J. P., Burkhart W., Moyer M., Dincher S., Middlesteadt W., Williams S., Payne G., Carnes M., Ryals J. Isolation of a complementary DNA encoding a chitinase with structural homology to a bifunctional lysozyme/chitinase. Proc Natl Acad Sci U S A. 1989 Feb;86(3):896–900. doi: 10.1073/pnas.86.3.896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molano J., Durán A., Cabib E. A rapid and sensitive assay for chitinase using tritiated chitin. Anal Biochem. 1977 Dec;83(2):648–656. doi: 10.1016/0003-2697(77)90069-0. [DOI] [PubMed] [Google Scholar]
- Oakley B. R., Kirsch D. R., Morris N. R. A simplified ultrasensitive silver stain for detecting proteins in polyacrylamide gels. Anal Biochem. 1980 Jul 1;105(2):361–363. doi: 10.1016/0003-2697(80)90470-4. [DOI] [PubMed] [Google Scholar]
- Parsons T. J., Bradshaw H. D., Jr, Gordon M. P. Systemic accumulation of specific mRNAs in response to wounding in poplar trees. Proc Natl Acad Sci U S A. 1989 Oct;86(20):7895–7899. doi: 10.1073/pnas.86.20.7895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polacheck I., Rosenberger R. F. Distribution of autolysins in hyphae of Aspergillus nidulans: evidence for a lipid-mediated attachment to hyphal walls. J Bacteriol. 1978 Sep;135(3):741–747. doi: 10.1128/jb.135.3.741-747.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbins P. W., Albright C., Benfield B. Cloning and expression of a Streptomyces plicatus chitinase (chitinase-63) in Escherichia coli. J Biol Chem. 1988 Jan 5;263(1):443–447. [PubMed] [Google Scholar]
- Robson L. M., Chambliss G. H. Cloning of the Bacillus subtilis DLG beta-1,4-glucanase gene and its expression in Escherichia coli and B. subtilis. J Bacteriol. 1986 Feb;165(2):612–619. doi: 10.1128/jb.165.2.612-619.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinshi H., Mohnen D., Meins F. Regulation of a plant pathogenesis-related enzyme: Inhibition of chitinase and chitinase mRNA accumulation in cultured tobacco tissues by auxin and cytokinin. Proc Natl Acad Sci U S A. 1987 Jan;84(1):89–93. doi: 10.1073/pnas.84.1.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silhavy T. J., Benson S. A., Emr S. D. Mechanisms of protein localization. Microbiol Rev. 1983 Sep;47(3):313–344. doi: 10.1128/mr.47.3.313-344.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutter R. P. Mutations affecting sexual development in Phycomyces blakesleeanus. Proc Natl Acad Sci U S A. 1975 Jan;72(1):127–130. doi: 10.1073/pnas.72.1.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe H., Katoh H., Ishii M., Komoda Y., Sanda A., Takizawa Y., Ohgi K., Irie M. Primary structure of a ribonuclease from bovine brain. J Biochem. 1988 Dec;104(6):939–945. doi: 10.1093/oxfordjournals.jbchem.a122587. [DOI] [PubMed] [Google Scholar]
- Yanagida N., Uozumi T., Beppu T. Specific excretion of Serratia marcescens protease through the outer membrane of Escherichia coli. J Bacteriol. 1986 Jun;166(3):937–944. doi: 10.1128/jb.166.3.937-944.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]