Abstract
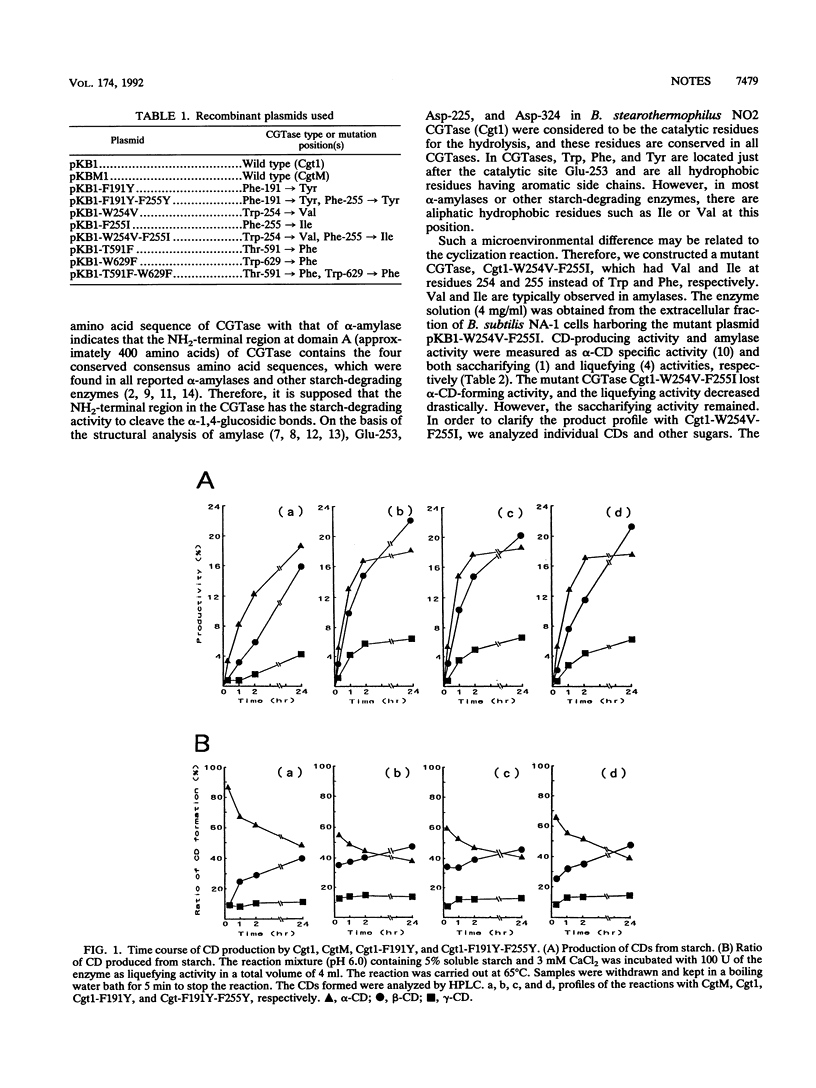
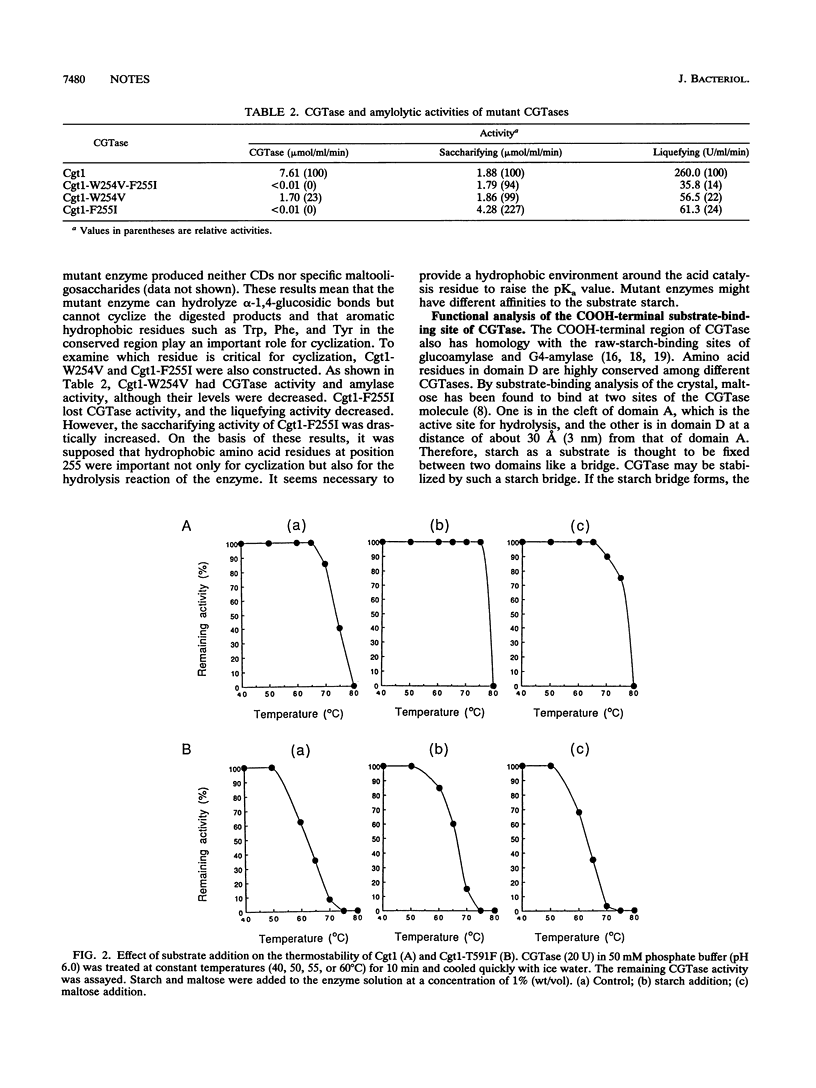
Cyclodextrin glucanotransferase (CGTase; EC 2.4.1.19) produces cyclodextrin from starch. The CGTase molecule is composed of four globular domains, A, B, C, and D. In order to gain better understanding of the amylolytic and cyclization mechanisms of CGTase, mutant CGTases were constructed from a CGTase gene (cgt1) of Bacillus stearothermophilus NO2. Cgt1-F191Y (Phe at position 191 was replaced by Tyr), Cgt1-F191Y-F255Y, Cgt1-W254V-F255I, Cgt1-W254V, and Cgt1-F255I were constructed for the analysis of the NH2-terminal region. It was revealed that amino acids surrounding a spiral amylose are important for cyclization characteristics and that hydrophobic amino acids just after the Glu catalytic site play an important role in the hydrolysis characteristics of the enzyme. Mutant CGTases Cgt1-T591F and Cgt1-W629F were also constructed to study the role of a second substrate-binding site in domain D, and it was suggested that substrate binding at both domains A and D stabilized the enzyme and optimized cyclodextrin production.
Full text
PDF

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aiba S., Kitai K., Imanaka T. Cloning and Expression of Thermostable alpha-Amylase Gene from Bacillus stearothermophilus in Bacillus stearothermophilus and Bacillus subtilis. Appl Environ Microbiol. 1983 Nov;46(5):1059–1065. doi: 10.1128/aem.46.5.1059-1065.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binder F., Huber O., Böck A. Cyclodextrin-glycosyltransferase from Klebsiella pneumoniae M5a1: cloning, nucleotide sequence and expression. Gene. 1986;47(2-3):269–277. doi: 10.1016/0378-1119(86)90070-3. [DOI] [PubMed] [Google Scholar]
- Hofmann B. E., Bender H., Schulz G. E. Three-dimensional structure of cyclodextrin glycosyltransferase from Bacillus circulans at 3.4 A resolution. J Mol Biol. 1989 Oct 20;209(4):793–800. doi: 10.1016/0022-2836(89)90607-4. [DOI] [PubMed] [Google Scholar]
- Kimura K., Kataoka S., Nakamura A., Takano T., Kobayashi S., Yamane K. Functions of the COOH-terminal region of cyclodextrin glucanotransferase of alkalophilic Bacillus sp. #1011: relation to catalyzing activity and pH stability. Biochem Biophys Res Commun. 1989 Jun 30;161(3):1273–1279. doi: 10.1016/0006-291x(89)91380-6. [DOI] [PubMed] [Google Scholar]
- Klein C., Schulz G. E. Structure of cyclodextrin glycosyltransferase refined at 2.0 A resolution. J Mol Biol. 1991 Feb 20;217(4):737–750. doi: 10.1016/0022-2836(91)90530-j. [DOI] [PubMed] [Google Scholar]
- Kuriki T., Imanaka T. Nucleotide sequence of the neopullulanase gene from Bacillus stearothermophilus. J Gen Microbiol. 1989 Jun;135(6):1521–1528. doi: 10.1099/00221287-135-6-1521. [DOI] [PubMed] [Google Scholar]
- Lejeune A., Sakaguchi K., Imanaka T. A spectrophotometric assay for the cyclization activity of cyclomaltohexaose (alpha-cyclodextrin) glucanotransferase. Anal Biochem. 1989 Aug 15;181(1):6–11. doi: 10.1016/0003-2697(89)90385-0. [DOI] [PubMed] [Google Scholar]
- MacGregor E. A., Svensson B. A super-secondary structure predicted to be common to several alpha-1,4-D-glucan-cleaving enzymes. Biochem J. 1989 Apr 1;259(1):145–152. doi: 10.1042/bj2590145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuura Y., Kusunoki M., Harada W., Kakudo M. Structure and possible catalytic residues of Taka-amylase A. J Biochem. 1984 Mar;95(3):697–702. doi: 10.1093/oxfordjournals.jbchem.a134659. [DOI] [PubMed] [Google Scholar]
- PULLEY A. O., FRENCH D. Studies on the Schardinger dextrins. XI. The isolation of new Schardinger dextrins. Biochem Biophys Res Commun. 1961 May 15;5:11–15. doi: 10.1016/0006-291x(61)90071-7. [DOI] [PubMed] [Google Scholar]
- Svensson B., Jespersen H., Sierks M. R., MacGregor E. A. Sequence homology between putative raw-starch binding domains from different starch-degrading enzymes. Biochem J. 1989 Nov 15;264(1):309–311. doi: 10.1042/bj2640309. [DOI] [PMC free article] [PubMed] [Google Scholar]


