Abstract
Cells of budding yeast organize their cytoskeleton in a highly polarized manner during vegetative growth. Selection of a site for polarization requires a group of proteins including a Ras-like GTPase, Bud1, and its regulators. Another group of proteins, which includes a Rho-like GTPase (Cdc42), its guanine nucleotide exchange factor (Cdc24), and Bem1, is necessary for organization of the actin cytoskeleton and for cell polarization. We have proposed previously that the Bud1 protein, through its GTPase cycle, determines the localization of one or more of the cell polarity proteins to the bud site. Herein we demonstrate that Bud1 directly interacts with Cdc24 and Bem1: Bud1 in its GTP-bound form associates preferentially with Cdc24, whereas the GDP-bound form of Bud1 associates with Bem1. We also present subcellular fractionation data for Bud1 that is consistent with the idea that Bud1 can travel between the site for budding on the plasma membrane and the cytosol. We propose that Bud1 can exist in two active states for association with different partners and that the switch from Bud1–GTP to Bud1–GDP provides a regulatory device for ordered assembly of a macromolecular complex at the bud site.
Cells of the budding yeast Saccharomyces cerevisiae are highly polarized during their vegetative cell cycle and during mating. During vegetative growth, all cell surface growth takes place in the bud, and cytoskeletal elements are oriented toward this new bud (1, 2). The process of bud initiation is thought to involve several distinct molecular events. (i) A specific site for budding is selected. (ii) Assembly of components required for bud formation takes place at the chosen site to restrict cell growth to that position. (iii) The actin cytoskeleton is organized toward that site and secretion is targeted to it. The choice of budding site determines the axis for cell polarity and the cell division plane. The site chosen for polarization is dependent upon cell type: a and α cells exhibit an axial pattern, in which mother and daughter cells choose new bud sites adjacent to the previous bud site. In contrast, a/α cells exhibit a bipolar pattern, in which mother and daughter cells choose new bud sites at either end of the cell (3–5).
Three different classes of yeast genes are required for proper bud site selection in the different cell types. BUD3, BUD4, AXL1, and AXL2/BUD10 are required only in a and α cells and are thought to be involved in recognizing or constituting an axial landmark in these cells (3, 6–10). BUD6, BUD7, BUD8, and BUD9 are required only in a/α cells and are thought to be involved in recognizing or constituting the bipolar landmarks in this cell type (11). In contrast, BUD1, BUD2, and BUD5 are required for bud site selection in all cell types: mutants defective in these genes exhibit a random budding pattern in a, α, and a/α cells (3, 12–14). It has been proposed that Bud1, Bud2, and Bud5 function as general bud site selection machinery that brings other proteins to the cell-type-specific landmarks in a, α, and a/α cells (3).
Bud1 (also known as Rsr1) has strong sequence similarity to the Ras family of proteins (12) and is indeed a GTPase (13, 15). Bud2 has sequence similarity to GTPase-activating proteins and has been shown to activate GTP hydrolysis by Bud1 (13). Bud5 has sequence similarity to the guanine nucleotide exchange protein Cdc25 (14, 16) and catalyzes GDP–GTP exchange for Bud1 in vitro (17). The general bud site selection machinery (Bud1, Bud2, and Bud5) thus makes up a functional GTPase module, a GTPase and its regulatory proteins.
A group of genes that includes CDC42, CDC24, and BEM1 is required for organizing the actin cytoskeleton toward the chosen site. Conditional mutants defective in these genes fail to form a bud, instead exhibiting unpolarized cell surface growth at nonpermissive temperature (18–23). Thus these genes appear to be involved in polarity establishment (1, 2). CDC42 and CDC24 encode a Rho-like GTPase and its exchange factor, respectively (18, 24); BEM1 encodes a protein with two SH3 domains (22). A variety of genetic and physiological observations led to the hypothesis that the Bud1 GTPase recruits proteins such as Cdc24, Cdc42, or Bem1 to the bud site by associating with them in a guanine nucleotide-dependent manner (3, 13).
Herein we have used an in vitro binding assay to test directly whether Bud1 can interact with these proteins. We show that Bud1 in its GTP-bound form associates preferentially with Cdc24, whereas the GDP-bound form of Bud1 associates with Bem1. We also present in vivo and in vitro analysis of a mutant Bud1 protein altered in its presumed effector domain that provides support for the functional relevance of some of these in vitro interactions. We also provide biochemical evidence suggesting that Bud1 can exist in both membrane and cytosolic fractions. Finally, we propose a model for how the Bud1 GTPase cycle directs cell polarity during budding that accommodates these and other observations.
MATERIALS AND METHODS
Media, Growth Conditions, and Yeast Strains.
Standard yeast culture media were prepared essentially as described (25). Standard procedures were used for yeast transformation (25). Strains with the temperature-sensitive cdc24 mutation (Y147) transformed with 2-μm plasmid (pRS425) carrying BUD1, bud1T35A, or vector alone were streaked on SD-leu plates containing 1 M sorbitol and incubated at 25°C or 36°C.
A yeast strain carrying the bud1T35A allele (HPY172) was constructed by two-step gene replacement (26). Genotypes of yeast strains are the following: HPY172, MATa ura3–52 lys2–801 ade2–101 his3Δ200 leu2Δ1 trp1Δ63 bud1T35A; HPY22, isogenic BUD1 strain; Y147, MATa cdc24–4 ura3 leu2–3,112 his3 (12).
Yeast strains expressing hemagglutinin (HA) epitope-tagged Bud1, Bud1G12V, and Bud1K16N were constructed by two-step gene replacement (26) using plasmids pHP659, pHP660, and pHP655, respectively. Genotypes of yeast strains are the following: HPY164–1 (Bud1-HA), MATa ura3–52 his3-Δ1 leu2 trp1Δ63 prb1-1122 pep4–3 prc1–407 BUD1-HA; HPY166–1 (Bud1G12V-HA), isogenic to HPY164-1 except bud1G12V-HA; HPY167 (Bud1K16N), isogenic to HPY164-1 except bud1K16N-HA.
Plasmids.
To overexpress wild-type Bud1 in yeast, a SacI–SalI fragment carrying BUD1 from pPB290 (12) was cloned into pRS425 (2-μm LEU2), generating pHP674-1. To overexpress Bud1T35A in yeast, a SacI–SalI fragment carrying the bud1T35A allele from pHP656 was cloned into pRS425, generating pHP675-1.
The plasmid expressing HA-epitope-tagged Bud1 (pHP659) was constructed by site-directed mutagenesis, resulting in insertion of the 9-amino acid HA epitope after residue 202 of Bud1. Plasmids pHP660 and pHP655 are identical to pHP659 except that they carry mutations affecting residue 12 (Bud1G12V) or 16 (Bud1K16N), respectively. These mutations were introduced by substituting a SacI–ClaI fragment carrying each mutation from YEp(rsr1val12) or YEp(rsr1asn16) (27).
To express Bud1 as a glutathione S-transferase (GST) fusion protein in Escherichia coli, full-length Bud1 was cloned into the vector pGEX, generating plasmid pRS4 (gift of H. Maruta, Ludwig Institute for Cancer Research, Victoria, Australia). GST–Bud1T35A was expressed by using plasmid pHP649, which is the same as pRS4 except for a single amino acid substitution at residue 35 generated by PCR. To express six-histidine-tagged Cdc24, an NcoI–SpeI fragment was cloned into the vector pTrcHisA (Invitrogen) to form plasmid pHP611. The plasmid carries the coding sequence of Cdc24 from residues 153 to the end (residue 854) and thus contains the GEF domain, the pleckstrin homology domain, and a region for binding to Bem1 (28). To express Cdc24 as a maltose-binding protein (MBP) fusion protein, a HincII–HincII fragment was cloned into the vector pMAL-c (New England Biolabs). MBP–Cdc24 carries approximately half of the coding sequence of Cdc24, from residues 472 to 854. To express six-His-tagged Bem1, a KpnI–KpnI fragment containing BEM1 (22) was cloned into pTrcHisA to form pHP610. pHP610 carries the coding sequence of Bem1 from residues 44 to 551 (the end), which includes its two SH3 domains.
Antibodies.
Cdc24-specific antibodies were raised in rabbits (Cocalico Biologicals, Reamstown, PA) using as immunogen a GST–Cdc24 fusion that contained amino acids 472–854 of Cdc24 and then affinity-purified on a nitrocellulose blot of MBP–Cdc24 as described in ref. 29. Bem1-specific antibodies in rabbits were previously described in Pringle et al. (30). Monoclonal antibodies against a HA epitope were purchased from Babco (Richmond, CA).
Purification of Proteins.
All proteins used in this study were purified from the protease-deficient E. coli strain NB42 (gift of Peter Jackson, Stanford University). Bud1 and Bud1T35A were purified as GST fusion proteins as described (31). Cdc24 and Bem1 were purified as six-histidine-tagged proteins on a column containing iminodiacetic acid immobilized on Sepharose-6B (Sigma) coupled to Co2+ as described in the manufacturer’s protocol. MBP–Cdc24 was expressed using the vector pMAL-c (New England Biolabs) and purified on an amylose column essentially as described in ref. 32.
In Vitro Binding Assays.
Guanine nucleotide loading of GST–Bud1 and in vitro binding reactions were performed essentially as described in ref. 32 with slight modifications. Guanosine 5′-[γ-thio]triphosphate (GTP[γS])- and GDP-bound GST–Bud1 were incubated for 40 min on ice with each binding partner in 100 μl of binding buffer (20 mM Tris⋅HCl, pH 7.5/85 mM NaCl/6 mM MgCl2/10% glycerol/0.6 mM GDP or GTP[γS]). After incubation, glutathione-Sepharose was added and centrifuged to collect GST–Bud1. After washing three times with 1 ml of wash buffer (10 mM Tris⋅HCl, pH 7.6/10% glycerol/5 mM MgCl2/1 mM dithiothreitol/0.1% Triton X-100/0.1 mM phenylmethylsulfonyl fluoride), bound proteins were eluted from glutathione-Sepharose using elution buffer containing 10 mM reduced glutathione and analyzed by SDS/PAGE. Association of each binding candidate with GST–Bud1 was detected by immunoblotting with affinity-purified antibodies specific for each binding protein.
To determine association of Bud1 with MBP–Cdc24, the in vitro binding reaction was performed as described above. After incubation, amylose resin was added and centrifuged to collect MBP–Cdc24.
Budding Pattern.
The budding pattern of logarithmic-phase cells was determined by Calcofluor staining of bud scars as described in ref. 29. About 400 cells were analyzed for each strain.
Cell Fractionation.
Cell fractionation experiments were performed by using techniques described by Goud et al. (33) with slight modifications. Briefly, 50 OD600 units of cells grown at 30°C were collected, washed with water, and resuspended in 0.5 ml of lysis buffer (50 mM Hepes, pH 7.6/50 mM KCl/1 mM EGTA/1 mM MgCl2/1 mM dithiothreitol/10% glycerol) with protease inhibitors [0.5 mM phenylmethylsulfonyl fluoride/1:1000 dilution of a stock of aprotinin (1 mg/ml)/1:1000 dilution of a stock of leupeptin (1 mg/ml)/1:1000 dilution of a stock of pepstatin (1 mg/ml)]. All of the following procedures were performed at 4°C. The cells were then lysed by using glass beads in a bead beater. Cell lysates were spun at 500 × g for 4 min, and the supernatants were subsequently centrifuged at 10,000 × g for 10 min to produce pellet (P) and supernatant (S) fractions. The pellets were then resuspended in the same volume of lysis buffer as the supernatants. Aliquots of the pellet fractions (P) were then treated with NaCl (final 500 mM), Triton X-100 (final 2%), or SDS (final 1%) before centrifuging at 10,000 × g again as described in ref. 34.
RESULTS
Bud1–GTP Associates with Cdc24 in Vitro.
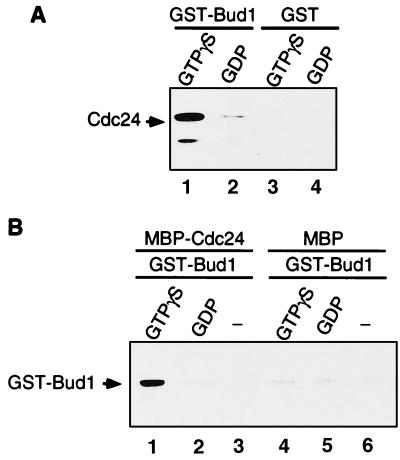
To test directly whether Bud1 interacts with Cdc24, Bud1 was purified as a GST fusion protein, and Cdc24 was purified as a six-histidine-tagged protein or a MBP fusion protein from E. coli. To test association of Bud1 and Cdc24, GST–Bud1 was preloaded with the nonhydrolyzable GTP analogue, GTP[γS], or with GDP and incubated with six-histidine-tagged Cdc24. GST–Bud1 was collected with glutathione-Sepharose beads, and the association of Cdc24 was determined by immunoblotting eluents from the beads with antibodies specific to Cdc24. As shown in Fig. 1A, Cdc24 bound preferentially to GTP[γS]-bound Bud1 but did not to GST. In a complementary experiment, we examined binding of GST–Bud1 to an MBP–Cdc24 fusion protein that contains residues 472–854 of Cdc24. MBP–Cdc24 was collected by addition of amylose resin, and the association of Bud1 was determined by immunoblotting using anti-GST antibodies. GTP[γS]-bound Bud1 preferentially interacted with MBP–Cdc24 but not with MBP (Fig. 1B). These results show that Cdc24 interacts preferentially with GTP[γS]-bound Bud1 and that the Bud1-binding site on Cdc24 lies between residues 472 and 854.
Figure 1.
Cdc24 binds preferentially to GTP[γS]-bound Bud1 in vitro. (A) Association of GST–Bud1 and six-histidine-tagged Cdc24. Purified GST–Bud1 and GST (final concentration, approximately 750 nM) were preincubated with 1 mM GDP or GTP[γS] and then incubated with six-histidine-tagged Cdc24 (final concentration, 15 nM). Cdc24 was incubated with either GTP[γS]-bound Bud1 (lane 1) or GDP-bound Bud1 (lane 2), or with GST that had been preincubated with GTP[γS] (lane 3) or GDP (lane 4). Proteins bound to glutathione-Sepharose beads were collected and analyzed by SDS/PAGE. Approximately equal amounts of GST–Bud1 and GST proteins were recovered for each reaction as judged by Coomassie blue staining of the gel (data not shown). Association of Cdc24 was determined by immunoblotting with polyclonal antibodies against Cdc24. (B) Association of GST–Bud1 and MBP–Cdc24 in vitro. MBP–Cdc24 (750 nM) was incubated either with GST–Bud1 (75 nM) preloaded with GTP[γS] (lane 1) or GDP (lane 2), or with GST–Bud1 that had not been preincubated with any nucleotides (lane 3). MBP (750 nM) was also incubated with GST–Bud1 (75 nM) preloaded either with GTP[γS] (lane 4) or GDP (lane 5), or with GST–Bud1 that had not been preincubated with any nucleotides (lane 6). Proteins bound to amylose resin were collected and analyzed by SDS/PAGE. Approximately equal amounts of MBP–Cdc24 and MBP were recovered for each reaction as judged by Coomassie blue staining of the gel (data not shown). Association of GST–Bud1 was determined by immunoblotting with polyclonal antibodies against GST.
The Putative Effector Domain Mutant of Bud1 Bud1T35A Does Not Bind to Cdc24.
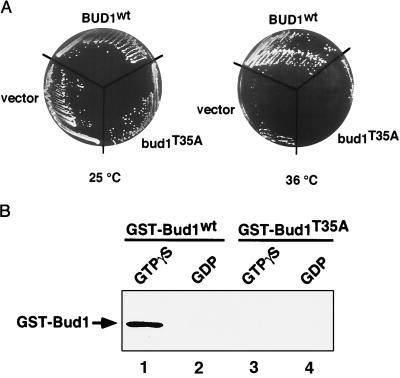
In the case of Ras, residues involved in interacting with a target molecule, the effector, have been defined genetically and biochemically and are localized to the so-called effector region, which contains residues 32–40 (35). Bud1 is identical to H-ras in this region (12). To determine whether this region is also important for Bud1 function and whether Cdc24 is an effector of Bud1, we constructed a mutant form of Bud1 with a T → A substitution at position 35 in the putative effector region of Bud1. Cells with the bud1T35A mutant allele grew normally at 16–37°C, but more than 90% of cells exhibited a random budding pattern (data not shown), which is the same phenotype of a bud1 deletion mutant (12). Since overproduction of Bud1 (Rsr1) can suppress a temperature-sensitive cdc24 mutation (12), presumably because of interaction between Bud1 and Cdc24, we tested whether overexpression of bud1T35A can suppress the cdc24ts mutation. Overexpression of bud1T35A failed to suppress the cdc24ts mutation (Fig. 2A), suggesting that Bud1T35A was unable to interact with Cdc24. We tested directly whether Bud1T35A can interact with Cdc24 by purifying the mutant protein as a GST fusion protein from E. coli. Bud1T35A was unable to interact with Cdc24 regardless of whether it was bound to GTP[γS] or to GDP (Fig. 2B). These results indicate that the binding of Cdc24 to the GTP-bound form of Bud1 is functionally relevant since their association in vitro is eliminated by the bud1T35A mutation, which inactivates BUD1 function in vivo. These results also indicate that the C-terminal half of Cdc24 interacts with the effector region of Bud1.
Figure 2.
Bud1 effector domain mediates interaction between Bud1 and Cdc24. (A) Overexpression of Bud1T35A fails to suppress a temperature-sensitive cdc24 mutation. Strains with the temperature-sensitive cdc24 mutation (Y147) transformed with 2-μm plasmid (pRS425) carrying BUD1, bud1T35A, or vector alone were streaked on SD-Leu plate containing 1 M sorbitol and incubated at 25°C or 36°C for 3 days. (B) The Bud1T35A protein fails to interact with Cdc24 in vitro. MBP–Cdc24 (750 nM) was incubated with wild-type GST–Bud1 (15 nM) preloaded with GTP[γS] (lane 1) or GDP (lane 2), or with GST–Bud1T35A (15 nM) preloaded with GTP[γS] (lane 3) or GDP (lane 4). Proteins were analyzed as in Fig. 1B. GST–Bud1T35A was expressed by using plasmid HP649, which is the same as pRS4 (15) except for a single amino acid substitution at residue 35.
Bud1–GDP Associates with Bem1.
The Bem1 protein, like Cdc24 and Cdc42, is important for establishment of cell polarity and organization of actin (14, 21, 22). We tested whether Bem1 can interact with wild-type Bud1 and Bud1T35A. As shown in Fig. 3, Bem1 associated preferentially with Bud1–GDP in comparison with GST–Bud1–GTP[γS] or with nucleotide-free GST–Bud1 (compare lane 2 with lanes 1 and 7). In contrast, Bem1 interacted with GST–Bud1T35A preloaded with GTP[γS] or GDP with almost equal efficiency but not with nucleotide-free GST–Bud1T35A (Fig. 3, lanes 3, 4, and 8). Bem1 did not interact with GST or glutathione-Sepharose (Fig. 3, lanes 5, 6, and 9). The binding of Bud1T35A to both GTP and GDP was not noticeably different from that of wild-type Bud1 at the concentrations tested (data not shown). These results demonstrate that the GDP-bound form of Bud1 specifically interacts with Bem1 and that Bud1T35A behaves the same as the GDP-bound form of Bud1 even when bound to GTP.
Figure 3.
Bem1 binds preferentially to GDP-bound Bud1 in vitro. Purified GST–Bud1, GST–Bud1T35A, and GST (approximately 750 nM) were preincubated with 1 mM GDP or GTP[γS] and then incubated with six-histidine-tagged Bem1 (15 nM). Bem1 was incubated with GTP[γS]–Bud1 (lane 1), GDP–Bud1 (lane 2), GTP[γS]–Bud1T35A (lane 3), GDP–Bud1T35A (lane 4), GST preincubated with GTP[γS] (lane 5), or GDP (lane 6) or with GST–Bud1 (lane 7) or GST–Bud1T35A (lane 8) that had not been preloaded with any nucleotides. Proteins bound to glutathione-Sepharose beads were collected and analyzed by SDS/PAGE. Association of Bem1 was determined by immunoblotting with polyclonal antibodies against Bem1 (ref. 30 and Kathy Corrado and J.R.P., unpublished results). A negative control is shown in lane 9, in which no GST–Bud1 or GST was added.
Bud1 Can Exist Both in the Membrane and Cytosolic Fractions.
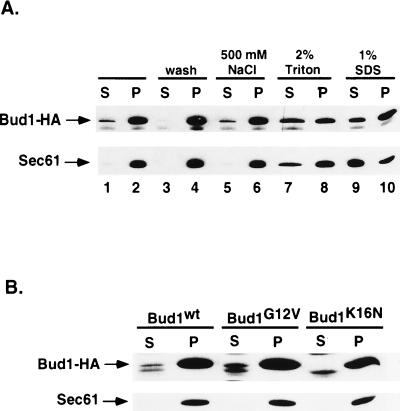
We proposed previously that Bud1 brings proteins important for polarity establishment such as Cdc24 to the bud site on the plasma membrane. We therefore wished to know whether Bud1 is present in both the cytosol and plasma membrane compartments. Bud1 contains the sequence Cys-Thr-Ile-Leu at its C terminus (12), a signal for isoprenylation that is believed to be important for association of Bud1 with the membrane. To determine whether Bud1 is associated with a subcellular membrane, cell fractionation was performed by centrifugation. Although most Bud1 (≈90%) was found in the particulate fraction that contains plasma membrane components as well as other dense material, a significant amount of Bud1 (≈10%) was found in the soluble fraction (Fig. 4A). This observation is consistent with the idea that Bud1 can travel between the site for budding on the plasma membrane and the cytosol. We further examined the nature of the interaction between Bud1 and the particulate (presumably membranous) fraction by treating this fraction with reagents that are known to release peripheral or integral membrane proteins (34). Bud1 was efficiently solubilized from the particulate fractions by treatment with 2% Triton X-100 or 1% SDS. Treatment with 0.5 M NaCl had little effect (Fig. 4A), which is similar to what has been observed for Cdc42 (37) and Sec61 (36), an endoplasmic reticulum membrane protein used as a control. These results suggest that Bud1 is tightly associated with cellular membranes. Since GTP- or GDP-bound Bud1 interacts with different partners, we further tested whether mutant forms of Bud1 that are thought to be locked into GTP- or GDP-bound forms partition differently in soluble and particulate fractions. Interestingly, we found that Bud1G12V [analogous to constitutively activated RasVal12 (27) and thus presumably in a GTP-bound form in vivo] was present in both fractions as observed for the wild-type protein, whereas Bud1K16N [analogous to a dominant negative RasAsn16 (27) and thus presumably in a GDP-bound form in vivo] was present only in the particulate fraction (Fig. 4B). These observations suggest that Bud1–GTP can travel between the bud site on the plasma membrane and the cytosol but that Bud1–GDP is mainly associated with the plasma membrane.
Figure 4.
Subcellular fractionation and solubilization of Bud1 from the particulate fraction. (A) Subcellular fractionation and solubilization of wild-type Bud1. Cells of yeast strain expressing HA-epitope-tagged Bud1 were broken open by bead beating and centrifuged at 500 × g to produce whole cell extract (wce); the wce was subsequently centrifuged at 10,000 × g to produce pellect (P) and supernatant (S) fractions (lanes 1 and 2). Aliquots of the pellet fractions (P) were washed with the same lysis buffer (lanes 3 and 4) or treated with 500 mM NaCl (lanes 5 and 6), 2% Triton (lanes 7 and 8), and 1% SDS (lanes 9 and 10) before centrifuging at 10,000 × g again (34). Both pellet and supernatant fractions from equal amounts of cells (OD600 unit) were loaded on 10% polyacrylamide gel and subjected to immunoblot analysis using monoclonal antibodies against HA epitope to detect HA-tagged Bud1. As a control for fractionation, identical samples were also subjected to immunoblot analysis using antibodies against the yeast endoplasmic reticulum membrane protein Sec61 (36). (B) Subcellular fractionation of Bud1G12V and Bud1K16N. Cells of yeast strain expressing HA-epitope-tagged Bud1G12V or Bud1K16N were treated in the same way as described in A.
DISCUSSION
Bud1, Bud2, and Bud5 form a GTPase module that is required for determining cell polarization toward certain preferred sites in budding yeast. A key issue in understanding how these proteins commit the yeast cell to utilize a specific site for polarization is to identify the proteins with which the components of this module interact. Of special interest is to identify the proteins that interact with the GTPase Bud1. A variety of genetic and physiological observations led to the proposal that Bud1 might interact with proteins that are important for cell polarity and that are thought to organize the actin cytoskeleton (3, 13). Our studies have focused on two of these proteins, Cdc24 and Bem1. We found that Bud1 protein binds to these proteins in vitro and that it appears to exist in two functional states, one GTP bound, which interacts with Cdc24, and the other GDP bound, which interacts with Bem1. Analysis of mutant forms of Bud1 indicates that the in vitro binding is functionally significant in vivo.
Our findings demonstrate the coupling of two GTPase cycles in the process of determining cell polarity: Bud1 directly associates with Cdc24, a guanine nucleotide exchange protein for Cdc42. In vivo studies with mammalian cells have led to the proposal that a GTPase cascade involving the ordered function of Cdc42, Rac, and Rho proteins governs cytoskeletal organization (38). The GTPase cascade that we describe functions upstream of a possible cascade involving Cdc42 and the other yeast Rho proteins (39).
We previously proposed that association of Bud1 with other proteins necessary for cell polarity guides these proteins to a proper cellular site (13). Having established that Bud1 does indeed interact with this class of proteins, we propose a possible sequence of events in which Bud1 guides a macromolecular assembly process in which hydrolysis of GTP provides a monitor of progress through the different steps.
Association of Bud1–GTP with Cdc24.
We have used two different methods to show that Bud1 associates with Cdc24 in a GTP-dependent manner in vitro. In one case, we recovered proteins bound to a GST–Bud1 fusion protein containing all 272 residues of Bud1. In the second case, we recovered proteins associated with an MBP–Cdc24 fusion protein. Both analyses demonstrated that Bud1–GTP preferentially associated with Cdc24. Zheng et al. (17) also demonstrated binding of Cdc24 to GTP-bound Bud1 in vitro by using full-length Bud1 and Cdc24 proteins.
Our studies provide further information on the regions of Cdc24 and Bud1 that are important for this interaction. Because the MBP–Cdc24 fusion protein contains only the C-terminal half of Cdc24 (residues 472–854), it is apparent that this part of the protein, which lacks the catalytic domain (28), is sufficient for interaction with Bud1. A further indication of the residues that are important for binding of Cdc24 to Bud1 comes from analysis of the Bud1T35A mutant. This mutant form of Bud1 was designed to test the idea that Bud1 interacts with an effector such as Cdc24 in the same manner as H-ras with its effector, raf. The residue Thr-35 is necessary for binding of H-ras to raf (32) and is also required for binding of Bud1 to Cdc24. We furthermore observed that this mutant form of Bud1 does not provide Bud1 function in vivo: the bud1T35A mutant exhibits a random budding pattern and is unable to suppress the growth defect of a cdc24ts mutant at nonpermissive temperature. Similar in vivo observations were recently reported by Michelitch and Chant (41). These observations indicate that Thr-35 is part of the effector region of Bud1, necessary for interaction with Cdc24.
Association of Bud1–GDP with Bem1.
In contrast to binding of Cdc24, Bem1 binds preferentially to the GDP-bound form of Bud1. In addition, it binds equally well to the Bud1T35A protein when this mutant protein is bound to GTP[γS] or to GDP. The ability of Bud1T35A to interact with Bem1 indicates that Bud1T35A is not grossly misfolded and instead suggests that it may be locked into a conformation like that of the GDP-bound state. In contrast to the situation for binding of Bud1 and Cdc24, we cannot yet evaluate the physiological significance of the in vitro binding of Bud1 and Bem1: we will need to search for mutants of Bud1 or Bem1 that specifically affect their interaction in vitro and then examine their phenotype in vivo. Despite this gap in our knowledge, we consider the binding of Bud1–GDP to Bem1 to be striking and thought-provoking. Below we propose that their association plays a crucial role in a series of morphogenetic events leading to actin organization.
The Bem1 protein has a wealth of binding partners. In particular, it has recently been shown to associate with Ste20, Cdc24, Far1, and actin (17, 28, 42, 43). Binding of Bem1 and Cdc24 has been demonstrated in vivo by the two-hybrid system (28) and in vitro, in which case binding is sensitive to Ca2+ (17). The fission yeast homologues of Cdc24 and Bem1 (Scd1 and Scd2, respectively) have also been shown to associate with each other in vitro (40).
It has been generally accepted that the GTP-bound state of GTPases such as ras is the active form. This is clearly the case for oncogenic forms of ras (for review, see ref. 44) and for other activated forms of ras (45–47). Our observations indicate that Bud1 also has a second active form, the GDP-bound form. We stress that this interaction between the GDP-bound form of Bud1 and Bem1 is novel. Proteins that bind to GDP-bound forms of GTPases are known, but there is no reason to think that Bem1 is either a guanine nucleotide exchange protein or a guanine nucleotide dissociation inhibitor (GDI). GDI proteins interact with prenylated membrane-bound forms of GTPases (48), but Bem1 interacts in vitro with unprenylated Bud1 protein synthesized in E. coli. Another example of the GDP-bound form of a GTPase having a binding partner distinct from an exchange protein or GDI has recently been described for the Ran protein, which is involved in uptake of proteins into the nucleus (49). The examples of Ran and Bud1 make it clear that GTPases such as Ras and Rho family members have two active forms and should not be viewed exclusively as an ON/OFF switch in which only the GTP-bound form is functional.
A Scheme for Bud-Site Assembly.
We had earlier proposed that the Bud1 GTPase cycle is involved in localizing proteins necessary for cell polarity to a cellular landmark (13). The finding that mutants defective in the GTPase itself (Bud1) exhibit the same phenotype as mutants defective in the GTPase-activating protein (Bud2) or in the guanine nucleotide exchange factor (Bud5) led to the proposal that Bud1 must cycle between GTP- and GDP-bound forms to function (13). Based on in vitro binding and subcellular fractionation data for Bud1, we now propose a scheme by which these proteins interact with each other and with the bud-site landmark to establish a position for polarization of the actin cytoskeleton (Fig. 5). We have incorporated the following new information into this scheme: Bud1–GTP binds to Cdc24 (this paper and ref. 17), Bud1–GDP binds to Bem1 (this paper), and Cdc24 binds to Bem1 (17, 28).
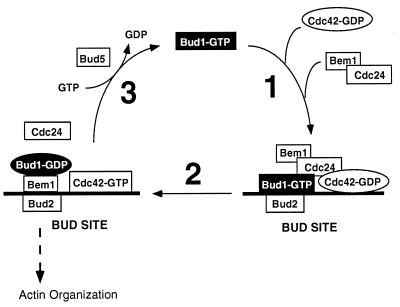
Figure 5.
Scheme for function of a GTPase cascade in determination of yeast cell polarity. In this model, the GTPase Bud1 determines the localization to the bud site of proteins that organize the actin cytoskeleton—Bem1, Cdc24, and the GTPase Cdc42. First, a tetrapartite complex is formed at the bud site, at which the Bud2, the GTPase-activating protein for Bud1, appears to be located (H.-O.P., unpublished results). Bud2 stimulates conversion of Bud1–GTP to Bud1–GDP, which allows a reconfiguration of components at the bud site and activation of Cdc24. This guanine nucleotide exchange factor, Cdc24, then stimulates formation of Cdc42–GTP, which is presumably the active species for organizing the actin cytoskeleton. Bud1–GDP is converted to Bud1–GTP by its guanine-nucleotide exchange factor, Bud5. Multiple rounds of this cycle may result in a critical concentration of Bem1 and Cdc42–GTP at the presumptive bud site, which stimulates polarization of the actin cytoskeleton toward that point (see text for details).
The end result of the bud-site selection process is the localization of Bem1 and the active species of Cdc42 (Cdc42-GTP) at a specific cellular location. These proteins are known to be localized at the nascent budding site (30, 37). We propose that in step 1 (Fig. 5), association of Bud1–GTP and its binding partners would yield a complex of Bud1–GTP, Cdc24, Cdc42-GDP, and Bem1 at the cellular landmark that defines the bud site (7). Bem1 might join the complex through its association with Cdc24. Cdc42–GDP might join the complex through its association with its guanine nucleotide exchange factor, Cdc24 (24), or through the direct interaction with Bud1–GTP (H.-O.P., unpublished results). In step 2, several critical events could occur in a coupled process that leads to activation of Cdc42. In one scenario, Bud1–GTP is converted to Bud1–GDP through action of its GTPase-activating protein, Bud2 (which is localized to the bud site; H.-O.P., unpublished results). Bud1, now in its GDP-bound state, no longer associates with Cdc24 and instead becomes a binding partner for Bem1. Dissociation of Bem1 from Cdc24 may allow Cdc24 to become active and, therefore, convert Cdc42–GDP to Cdc42–GTP. Conversion of Bud1–GDP to Bud1–GTP by Bud5 in step 3 would allow recycling of Bud1 and further shuttling of Cdc24, Cdc42, and Bem1 to the bud site. The cycle is thus proposed to result in localization of a critical level of Cdc42–GTP and Bem1 proteins to the bud site, which then organizes the cytoskeleton through unknown mechanisms.
Our subcellular fractionation data on Bud1 suggest that some Bud1–GTP is soluble before it forms a complex at the bud site whereas Bud1–GDP is always associated with the plasma membrane. These observations are consistent with the view that Bud1 shuttles between the bud site on the plasma membrane and the cytosol to bring proteins essential for bud formation to the proper bud site. Michelitch and Chant (41) recently reported that Bud1 is present only in the pellet fraction, which differs from our observation. It is possible that the HA antibody that we used is more sensitive than the polyclonal antibody used by Michelitch and Chant (41).
Recent studies with fission yeast (40) and mammalian cells (38) show that Ras proteins control not only signal transduction pathways but also morphogenesis. In fission yeast, two-hybrid analysis and in vitro experiments indicate the existence of interactions between Ras and homologues of Cdc24, Cdc42, and Bem1 that are involved in morphogenesis. As noted by Chang et al. (40), the Bud1 protein of budding yeast can be viewed as a specialized form of Ras that is exclusively dedicated to morphogenesis. In fission yeast and in mammalian systems, the same Ras protein functions in both signal transduction and morphogenesis. Despite this difference between budding yeast and these other organisms, Ras-like proteins and their binding partners are conserved. Thus the mechanism by which Bud1 functions in organizing the actin cytoskeleton may also be conserved.
Acknowledgments
We thank P. Walter, F. Banuett, M. Peter, H. Bourne, P. J. Kang, A. Sil, R. Tabtiang, S. Sanders, and D. Morgan for valuable discussions and comments on the manuscript; H. Maruta and R. Ruggieri for plasmids; P. Jackson for an E. coli strain; D. Kellogg for GST antibody; K. Corrado for Bem1 antibody; and S. Sanders for Sec61 antibody. This work was supported by research grants from the National Institutes of Health to I.H. (GM48052) and J.R.P. (GM31006). E.B. was supported by a fellowship from the Damon Runyon–Walter Winchell Cancer Research Fund.
ABBREVIATIONS
- HA
hemagglutinin
- GST
glutathione S-transferase
- MBP
maltose-binding protein
- GTP[γS]
guanosine 5′-[γ-thio]triphosphate
References
- 1.Pringle J R, Lillie S H, Adams A E M, Jacobs C W, Haarer B K, Coleman K G, Robinson J S, Bloom L, Preston R A. Yeast Cell Biology. New York: Liss; 1986. pp. 47–80. [Google Scholar]
- 2.Drubin D. Cell. 1991;65:1093–1096. doi: 10.1016/0092-8674(91)90001-f. [DOI] [PubMed] [Google Scholar]
- 3.Chant J, Herskowitz I. Cell. 1991;65:1203–1212. doi: 10.1016/0092-8674(91)90015-q. [DOI] [PubMed] [Google Scholar]
- 4.Freifelder D. J Bacteriol. 1960;80:567–568. doi: 10.1128/jb.80.4.567-568.1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hicks J B, Strathern J N, Herskowitz I. Genetics. 1977;85:373–393. doi: 10.1093/genetics/85.3.373b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fujita A, Oka C, Arikawa Y, Katagai T, Tonouchi A, Kuhara S, Misumi Y. Nature (London) 1994;372:567–570. doi: 10.1038/372567a0. [DOI] [PubMed] [Google Scholar]
- 7.Chant J, Mischke M, Mitchell E, Herskowitz I, Pringle J R. J Cell Biol. 1995;129:767–778. doi: 10.1083/jcb.129.3.767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Roemer T, Madden K, Chang J, Snyder M. Genes Dev. 1996;10:777–793. doi: 10.1101/gad.10.7.777. [DOI] [PubMed] [Google Scholar]
- 9.Halme A, Michelitch M, Mitchell E L, Chant J. Curr Biol. 1996;6:570–579. doi: 10.1016/s0960-9822(02)00543-2. [DOI] [PubMed] [Google Scholar]
- 10.Sanders S, Herskowitz I. J Cell Biol. 1996;134:413–427. doi: 10.1083/jcb.134.2.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zahner J, Harkins H I, Pringle J R. Mol Cell Biol. 1996;16:1857–1870. doi: 10.1128/mcb.16.4.1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bender A, Pringle J R. Proc Natl Acad Sci USA. 1989;86:9976–9980. doi: 10.1073/pnas.86.24.9976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Park H-O, Chant J, Herskowitz I. Nature (London) 1993;365:269–274. doi: 10.1038/365269a0. [DOI] [PubMed] [Google Scholar]
- 14.Chant J, Corrado K, Pringle J R, Herskowitz I. Cell. 1991;65:1213–1224. doi: 10.1016/0092-8674(91)90016-r. [DOI] [PubMed] [Google Scholar]
- 15.Holden J L, Nur-E-Kamal M S A, Fabri L, Nice E, Hammacher A, Maruta H. J Biol Chem. 1991;266:16992–16995. [PubMed] [Google Scholar]
- 16.Powers S, Gonzales E, Christensen T, Cubert J, Broek D. Cell. 1991;65:1225–1231. doi: 10.1016/0092-8674(91)90017-s. [DOI] [PubMed] [Google Scholar]
- 17.Zheng Y, Bender A, Cerione R A. J Biol Chem. 1995;270:626–630. doi: 10.1074/jbc.270.2.626. [DOI] [PubMed] [Google Scholar]
- 18.Johnson D I, Pringle J R. J Cell Biol. 1990;111:143–152. doi: 10.1083/jcb.111.1.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Adams A E, Johnson D I, Longnecker R M, Sloat B F, Pringle J R. J Cell Biol. 1990;111:131–42. doi: 10.1083/jcb.111.1.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miyamoto S, Ohya Y, Sano Y, Sakaguchi S, Iida H, Anraku Y. Biochem Biophys Res Commun. 1991;181:604–610. doi: 10.1016/0006-291x(91)91233-3. [DOI] [PubMed] [Google Scholar]
- 21.Bender A, Pringle J R. Mol Cell Biol. 1991;11:1295–1305. doi: 10.1128/mcb.11.3.1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chenevert J, Corrado K, Bender A, Pringle J, Herskowitz I. Nature (London) 1992;356:77–79. doi: 10.1038/356077a0. [DOI] [PubMed] [Google Scholar]
- 23.Sloat B, Adams A, Pringle J R. J Cell Biol. 1981;89:395–405. doi: 10.1083/jcb.89.3.395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zheng Y, Cerione R, Bender A. J Biol Chem. 1994;269:2369–2372. [PubMed] [Google Scholar]
- 25.Sherman F, Fink G R, Hicks J B. Laboratory Course Manual for Methods in Yeast Genetics. Plainview, NY: Cold Spring Harbor Lab. Press; 1986. [Google Scholar]
- 26.Rothstein R. In: Guide to Yeast Genetics and Molecular Biology. Guthrie C, Fink G R, editors. San Diego: Academic; 1991. pp. 281–301. [Google Scholar]
- 27.Ruggieri R, Bender A, Matsui Y, Powers S, Takai Y, Pringle J R, Matsumoto K. Mol Cell Biol. 1992;12:758–766. doi: 10.1128/mcb.12.2.758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Peterson J, Zheng Y, Bender L, Myers A, Cerione R, Bender A. J Cell Biol. 1994;127:1395–1406. doi: 10.1083/jcb.127.5.1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pringle J R, Preston R A, Adams A E M, Sterns T, Drubin D G, Haarer B K, Jones E W. Methods Cell Biol. 1989;31:357–435. doi: 10.1016/s0091-679x(08)61620-9. [DOI] [PubMed] [Google Scholar]
- 30.Pringle J R, Bi E, Harkiins H A, Zahner J E, De Virgilio C, Chant J, Corrado K, Fares H. Cold Spring Harbor Symp Quant Biol. 1995;55:729–744. doi: 10.1101/sqb.1995.060.01.079. [DOI] [PubMed] [Google Scholar]
- 31.Kellogg D R, Kikuchi A, Fujii-Nakata T, Turck C, Murray A W. J Cell Biol. 1995;130:661–673. doi: 10.1083/jcb.130.3.661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vojtek A B, Hollenberg S M, Cooper J A. Cell. 1993;74:205–214. doi: 10.1016/0092-8674(93)90307-c. [DOI] [PubMed] [Google Scholar]
- 33.Goud B, Salminen A, Walworth N C, Novick P J. Cell. 1988;53:753–768. doi: 10.1016/0092-8674(88)90093-1. [DOI] [PubMed] [Google Scholar]
- 34.Deshaies R J, Schekman R. Mol Cell Biol. 1990;10:6024–6035. doi: 10.1128/mcb.10.11.6024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bourne H R, Sanders D A, McCormick F. Nature (London) 1991;349:117–126. doi: 10.1038/349117a0. [DOI] [PubMed] [Google Scholar]
- 36.Stirling C J, Rothblatt J, Hosobuchi M, Deshaies R, Schekman R. Mol Biol Cell. 1992;3:129. doi: 10.1091/mbc.3.2.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ziman M, Preuss D, Mulholland J, O’Brien J M, Botstein D, Johnson D I. Mol Biol Cell. 1993;4:1307–1316. doi: 10.1091/mbc.4.12.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nobes C D, Hall A. Cell. 1995;81:53–62. doi: 10.1016/0092-8674(95)90370-4. [DOI] [PubMed] [Google Scholar]
- 39.Chant J, Stowers L. Cell. 1995;81:1–4. doi: 10.1016/0092-8674(95)90363-1. [DOI] [PubMed] [Google Scholar]
- 40.Chang E C, Barr M, Wang Y, Jung V, Xu H-P, Wigler M H. Cell. 1994;79:131–141. doi: 10.1016/0092-8674(94)90406-5. [DOI] [PubMed] [Google Scholar]
- 41.Michelitch M, Chant J. Curr Biol. 1996;6:446–454. doi: 10.1016/s0960-9822(02)00512-2. [DOI] [PubMed] [Google Scholar]
- 42.Leeuw T, Fourest-Lieuvin A, Wu C, Chenevert J, Clark K, Whiteway M, Thomas D Y, Leberer E. Science. 1995;270:1210–1213. doi: 10.1126/science.270.5239.1210. [DOI] [PubMed] [Google Scholar]
- 43.Lyons D M, Mahanty S K, Choi K -Y, Manandhar M, Elion E A. Mol Cell Biol. 1996;16:4095–4106. doi: 10.1128/mcb.16.8.4095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lowy D R, Willumsen B M. Annu Rev Biochem. 1993;62:851–891. doi: 10.1146/annurev.bi.62.070193.004223. [DOI] [PubMed] [Google Scholar]
- 45.Beitel G J, Clark S G, Horvitz H R. Nature (London) 1990;348:503–509. doi: 10.1038/348503a0. [DOI] [PubMed] [Google Scholar]
- 46.Fortini M E, Simon M A, Rubin G M. Nature (London) 1992;355:559–561. doi: 10.1038/355559a0. [DOI] [PubMed] [Google Scholar]
- 47.Han M, Sternberg P W. Cell. 1990;63:921–931. doi: 10.1016/0092-8674(90)90495-z. [DOI] [PubMed] [Google Scholar]
- 48.Ando S, Kaibuchi K, Sasaki T, Hiraoka K, Nishiyama T, Mizuno T, Asada M, Nunoi H, Matsuda I, Matsuura Y. J Biol Chem. 1992;267:25709–25713. [PubMed] [Google Scholar]
- 49.Nehrbass U, Blobel G. Science. 1996;272:120–122. doi: 10.1126/science.272.5258.120. [DOI] [PubMed] [Google Scholar]