Abstract
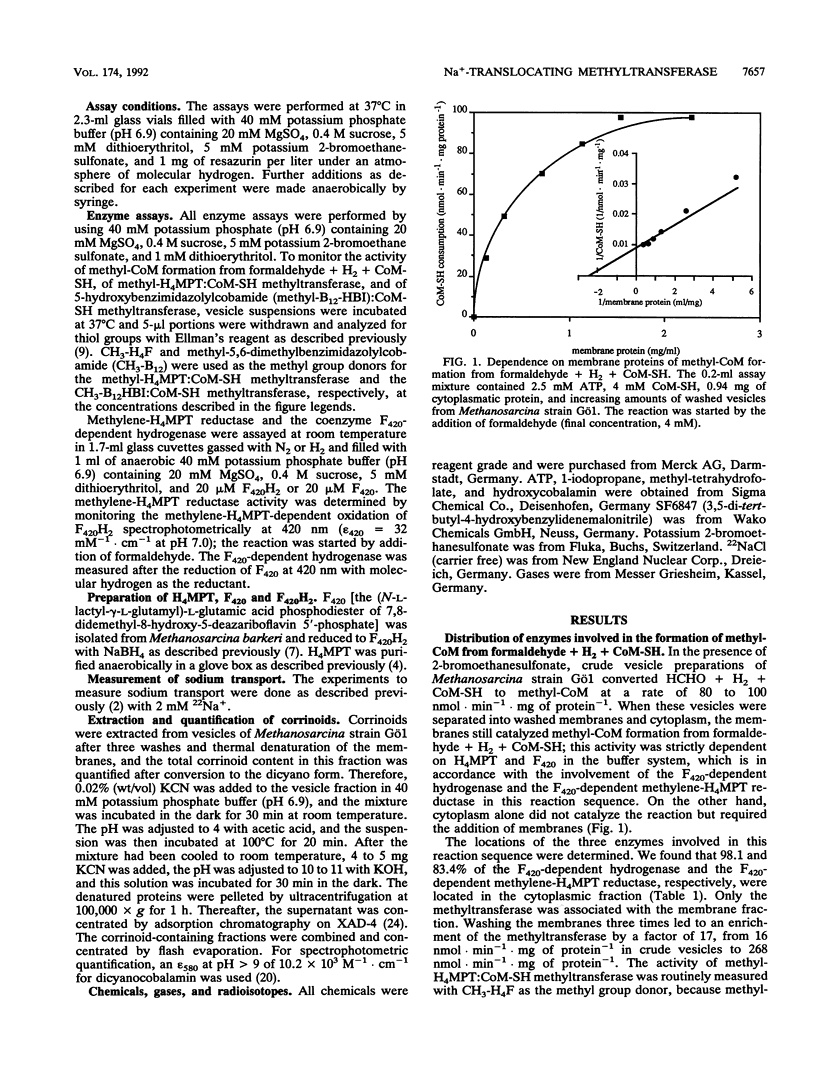
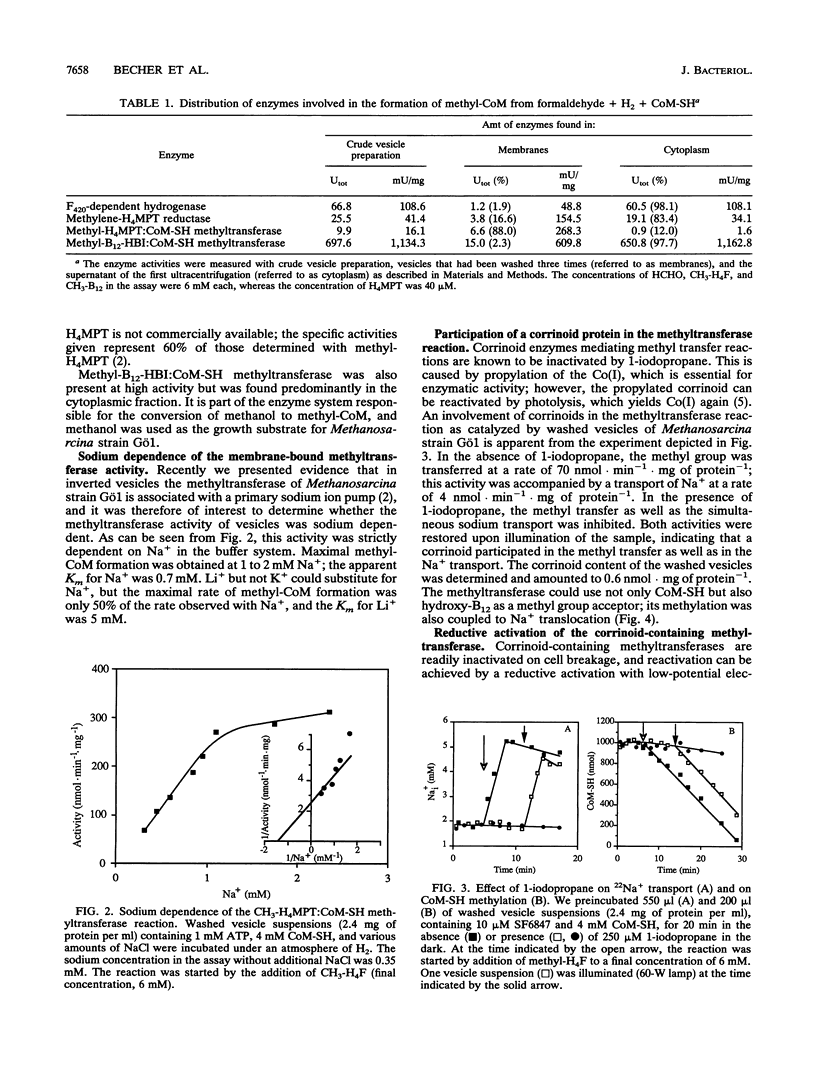
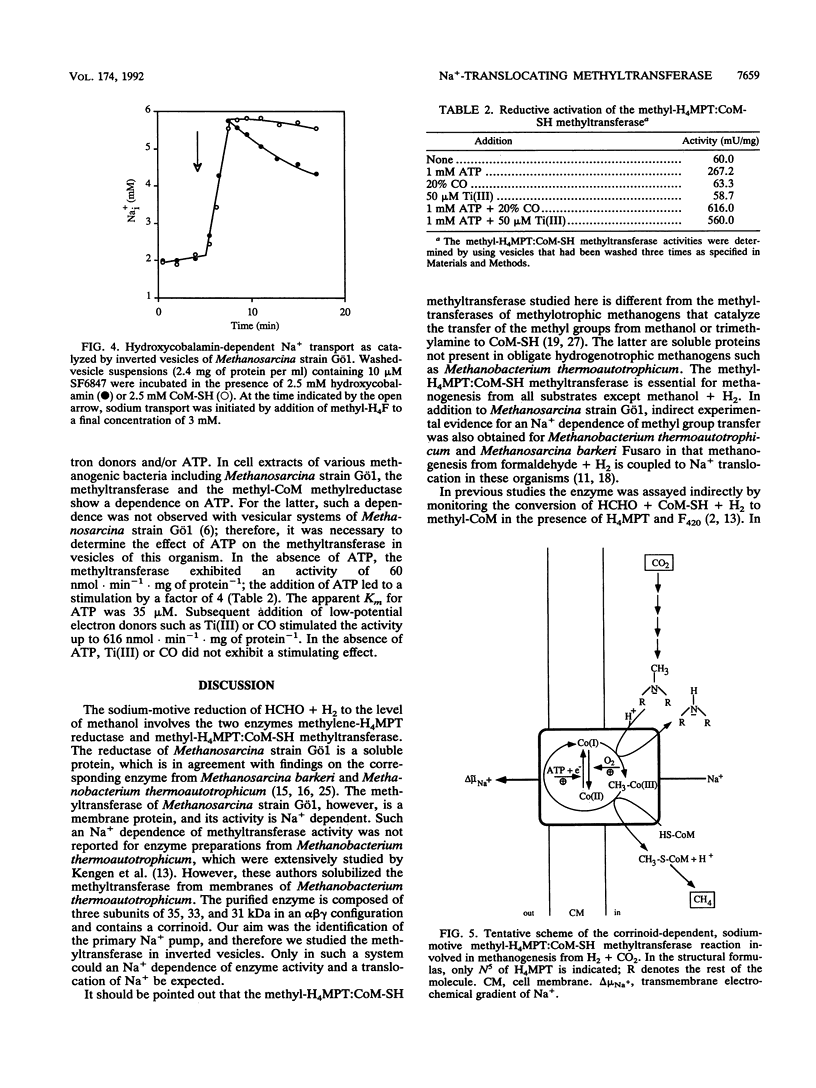
To determine the cellular localization of components of the methyltransferase system, we separated cell extracts of Methanosarcina strain Gö1 into cytoplasmic and inverted-vesicle fractions. Measurements demonstrated that 83% of the methylene-tetrahydromethanopterin reductase activity resided in the cytoplasm whereas 88% of the methyl-tetrahydromethanopterin:coenzyme M methyltransferase (methyltransferase) was associated with the vesicles. The activity of the methyltransferase was stimulated 4.6-fold by ATP and 10-fold by ATP plus a reducing agent [e.g., Ti(III)]. In addition, methyltransferase activity depended on the presence of Na+ (apparent Km = 0.7 mM) and Na+ was pumped into the lumen of the vesicles in the course of methyl transfer from methyl-tetrahydromethanopterin not only to coenzyme M but also to hydroxycobalamin. Both methyl transfer reactions were inhibited by 1-iodopropane and reconstituted by illumination. A model for the methyl transfer reactions is presented.
Full text
PDF

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- BROT N., WEISSBACH H. ENZYMATIC SYNTHESIS OF METHIONINE. CHEMICAL ALKYLATION OF THE ENZYME-BOUND COBAMIDE. J Biol Chem. 1965 Jul;240:3064–3070. [PubMed] [Google Scholar]
- Banerjee R. V., Matthews R. G. Cobalamin-dependent methionine synthase. FASEB J. 1990 Mar;4(5):1450–1459. doi: 10.1096/fasebj.4.5.2407589. [DOI] [PubMed] [Google Scholar]
- Deppenmeier U., Blaut M., Jussofie A., Gottschalk G. A methyl-CoM methylreductase system from methanogenic bacterium strain Gö 1 not requiring ATP for activity. FEBS Lett. 1988 Dec 5;241(1-2):60–64. doi: 10.1016/0014-5793(88)81031-7. [DOI] [PubMed] [Google Scholar]
- Deppenmeier U., Blaut M., Mahlmann A., Gottschalk G. Reduced coenzyme F420: heterodisulfide oxidoreductase, a proton- translocating redox system in methanogenic bacteria. Proc Natl Acad Sci U S A. 1990 Dec 1;87(23):9449–9453. doi: 10.1073/pnas.87.23.9449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ELLMAN G. L. A colorimetric method for determining low concentrations of mercaptans. Arch Biochem Biophys. 1958 Apr;74(2):443–450. doi: 10.1016/0003-9861(58)90014-6. [DOI] [PubMed] [Google Scholar]
- Hippe H., Caspari D., Fiebig K., Gottschalk G. Utilization of trimethylamine and other N-methyl compounds for growth and methane formation by Methanosarcina barkeri. Proc Natl Acad Sci U S A. 1979 Jan;76(1):494–498. doi: 10.1073/pnas.76.1.494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaesler B., Schönheit P. The sodium cycle in methanogenesis. CO2 reduction to the formaldehyde level in methanogenic bacteria is driven by a primary electrochemical potential of Na+ generated by formaldehyde reduction to CH4. Eur J Biochem. 1989 Dec 8;186(1-2):309–316. doi: 10.1111/j.1432-1033.1989.tb15210.x. [DOI] [PubMed] [Google Scholar]
- Kengen S. W., Daas P. J., Duits E. F., Keltjens J. T., van der Drift C., Vogels G. D. Isolation of a 5-hydroxybenzimidazolyl cobamide-containing enzyme involved in the methyltetrahydromethanopterin: coenzyme M methyltransferase reaction in Methanobacterium thermoautotrophicum. Biochim Biophys Acta. 1992 Feb 1;1118(3):249–260. doi: 10.1016/0167-4838(92)90282-i. [DOI] [PubMed] [Google Scholar]
- Ma K., Linder D., Stetter K. O., Thauer R. K. Purification and properties of N5,N10-methylenetetrahydromethanopterin reductase (coenzyme F420-dependent) from the extreme thermophile Methanopyrus kandleri. Arch Microbiol. 1991;155(6):593–600. doi: 10.1007/BF00245355. [DOI] [PubMed] [Google Scholar]
- Müller V., Winner C., Gottschalk G. Electron-transport-driven sodium extrusion during methanogenesis from formaldehyde and molecular hydrogen by Methanosarcina barkeri. Eur J Biochem. 1988 Dec 15;178(2):519–525. doi: 10.1111/j.1432-1033.1988.tb14478.x. [DOI] [PubMed] [Google Scholar]
- Pol A., van der Drift C., Vogels G. D. Corrinoids from Methanosarcina barkeri: structure of the alpha-ligand. Biochem Biophys Res Commun. 1982 Sep 30;108(2):731–737. doi: 10.1016/0006-291x(82)90890-7. [DOI] [PubMed] [Google Scholar]
- Sauer F. D. Tetrahydromethanopterin methyltransferase, a component of the methane synthesizing complex of Methanobacterium thermoautotrophicum. Biochem Biophys Res Commun. 1986 Apr 29;136(2):542–547. doi: 10.1016/0006-291x(86)90474-2. [DOI] [PubMed] [Google Scholar]
- Stupperich E., Steiner I., Rühlemann M. Isolation and analysis of bacterial cobamides by high-performance liquid chromatography. Anal Biochem. 1986 Jun;155(2):365–370. doi: 10.1016/0003-2697(86)90447-1. [DOI] [PubMed] [Google Scholar]
- te Brömmelstroet B. W., Geerts W. J., Keltjens J. T., van der Drift C., Vogels G. D. Purification and properties of 5,10-methylenetetrahydromethanopterin dehydrogenase and 5,10-methylenetetrahydromethanopterin reductase, two coenzyme F420-dependent enzymes, from Methanosarcina barkeri. Biochim Biophys Acta. 1991 Sep 20;1079(3):293–302. doi: 10.1016/0167-4838(91)90072-8. [DOI] [PubMed] [Google Scholar]
- van der Meijden P., Heythuysen H. J., Pouwels A., Houwen F., van der Drift C., Vogels G. D. Methyltransferases involved in methanol conversion by Methanosarcina barkeri. Arch Microbiol. 1983 Jun;134(3):238–242. doi: 10.1007/BF00407765. [DOI] [PubMed] [Google Scholar]



