Abstract
The proliferating cell nuclear antigen (PCNA), a crucial component of eukaryotic cell cycle and DNA replication complexes, is induced by the adenovirus E1A 243R oncoprotein through a cis-acting element termed the PERE (PCNA-E1A responsive element). The PERE contains a sequence homologous to an activating transcription factor (ATF) motif, and ATF-1 is a major component of PERE-protein complexes. We have identified a second PERE-binding protein, the cAMP response element binding protein (CREB) transcription factor, which forms heterodimers with ATF-1 at this site. CREB, but not ATF-1, is able to mediate transactivation of a minimal PCNA-chloramphenicol acetyltransferase reporter by E1A 243R. Further analysis revealed that the transcriptional coactivator, the CREB-binding protein (CBP), associates with PERE-related complexes, and that CBP is able to mediate a strong transactivation response to E1A 243R at the PCNA promoter. Experiments conducted with mutants in the E1A or CREB components support a model whereby E1A 243R transactivates the PCNA promoter via a CBP-CREB-PERE pathway. These findings delineate a paradigm by which E1A 243R can target and transactivate specific DNA promoter sequences.
Keywords: p300, activating transcription factor
The proliferating cell nuclear antigen (PCNA) is a highly conserved protein and a requisite component of cellular complexes essential for eukaryotic DNA replication and repair. PCNA is involved in both leading- and lagging-strand DNA synthesis during DNA replication (1, 2) as an auxiliary factor of DNA polymerase δ (3, 4). It is trimeric in structure (5, 6) and is postulated to function in a manner akin to a sliding clamp to increase the processivity of DNA synthesis (7). Consistent with its role in DNA replication, PCNA is induced in response to serum (8) and mitogenic growth factors (9–11) and is required for growth and cell cycle progression in both yeast (12) and mammalian cells (11, 13). It is found in quaternary complexes containing cyclins, cyclin-dependent kinases (CDK), and the CDK-inhibitor p21 (14–17). The function of PCNA in DNA replication can be inhibited by direct interaction with p21 (18). PCNA also is required for DNA repair (19–22) and is coinduced with cellular p53 levels in cells exposed to UV radiation (23). Compatible with the roles of both proteins in DNA damage repair function, p53 can transactivate the human PCNA promoter through a p53-binding site in a concentration-dependent manner (24).
Stimulation of the DNA replication machinery is a key feature in the process of neoplastic cellular transformation. From this aspect, the regulation of PCNA expression by the adenovirus E1A oncogene products represents an instructive model system for study of the molecular mechanisms underlying transformation. The adenovirus E1A oncoproteins possess both transcriptional and transforming properties that allow them to perturb normal host-cell growth control through interactions with a number of cellular targets. The major adenovirus E1A gene products, E1A 243R and E1A 289R, are identical in composition with the exception of conserved region three (CR3), a promiscuous and potent transactivation domain (25–28) unique to the latter species. Although the smaller E1A 243R protein lacks CR3, it is able to induce PCNA expression in both viral infection and transient expression assays conducted in HeLa cells (29, 30).
E1A induces PCNA expression through an increase in promoter activity (31), and mutational analysis has defined a novel cis-acting PCNA E1A-responsive element, termed the PERE (32). The PERE resides between nucleotides −59 and −45 relative to the transcription start site in the PCNA promoter and contains a sequence from −52 to −45 which is homologous to an activating transcription factor (ATF) motif (32). Both basal and E1A 243R-induced PCNA expression require this element in transient expression assays (32) and moreover, the PERE can confer E1A-responsiveness to a heterologous promoter (33).
To understand how E1A 243R transactivates PCNA expression we have sought to identify cellular factors that bind to the PERE. Previously, data indicated that the transcription factor ATF-1 is a major component of PERE-binding protein complexes (34). In this report, we demonstrate that the cAMP response element binding protein (CREB) transcription factor also binds to the PERE and heterodimerizes with ATF-1 at this site. Results of transfection experiments show that CREB is able to mediate a response to E1A 243R in vivo, whereas ATF-1 does not. Furthermore, the transcriptional coactivator, the CREB-binding protein (CBP), appears to associate with PERE-protein complexes and like CREB, can mediate induction of the PCNA promoter by E1A 243R. Taken together, our data support a model by which the E1A 243R oncoprotein targets and transactivates the PCNA promoter via CBP and the sequence-specific CREB transcription factor.
MATERIALS AND METHODS
Plasmids.
ATF-1 and CREB cDNA sequences were inserted in-frame into the unique 5′ XbaI and 3′ BamHI sites of the cytomegalovirus (CMV) promoter-driven GAL4 DNA-binding domain (DBD) expression construct pCGGAL4 (1 to 94) vector (35) by conventional subcloning techniques and PCR. The pGAL4-CBP effector construct was created by subcloning the XbaI and BamHI CBP cDNA inserts from pGAL4CBP 8.0 (36) into the unique 5′ XbaI–3′ BamHI sites of pCGGAL4 (35). Reporter plasmids G5PCNA-CAT, PCNA-87 CAT, and ATF-BAM CAT have been described previously (30, 33). Vectors expressing the E1A 243R oncoprotein (pCMV 12S), a truncated E1A 12S product (pCMV12S.FS), E1A mutants: pCMV Δ 2–36, pCMV Δ 30–85, pCMV Δ 2–85, pCMV Δ 86–120, pCMV Δ 120–140, the E1B 19-kDa protein (pCMV19K), and pON260, a plasmid expressing the β-galactosidase gene under the control of the CMV promoter have been described previously (30, 37–39). Rous sarcoma virus-ATF-1 (40) was provided by S. Wagner (University of Massachusetts Medical Center), and Rous sarcoma virus-CREB (41) and mutant GAL4-CREB plasmid (GAL4-CREB pm) were provided by M. Gilman (Ariad Pharmaceuticals).
Cell Culture.
Monolayer cultures of HeLa cells (ATCC CCL2) were grown under 5% CO2 in DMEM supplemented with 10% fetal calf serum and 100 μg each of penicillin and streptomycin per ml. Monolayer cultures of human 293 cells were grown in the same medium containing 10% calf serum instead of 10% fetal calf serum.
Transfections.
Transient expression assays were performed by calcium phosphate precipitation as previously described (31). HeLa cells at 50% confluence in 60-mm plates were transfected with 5 μg of reporter construct, 5 μg of effector construct, 1 μg pCMVβGAL (pON260), 0.5 μg of pCMV19K, 0.5 μg of E1A expression plasmid (pCMV12S.FS, pCMV12S, or pCMV E1A mutants), and salmon sperm DNA to 20 μg total DNA per plate. Human 293 cells were transfected with 5 μg of G5PCNA-CAT reporter construct, 5 μg of effector construct, 1 μg of pCMVβGAL (pON260), and salmon sperm DNA to a total of 20 μg per 60-mm plate. Cells were washed twice with PBS 16 hr after transfection, and fresh medium was added to the plates. Cells were harvested 45–48 hr after transfection.
Enzyme Assays.
Extracts from cells were prepared by freezing and thawing cells in 0.25 M Tris⋅HCl (pH 8.0). Assays for chloramphenicol acetyltransferase (CAT) activity were performed to yield values within the linear range of this assay and were quantified with a Betascan System (AMBIS). CAT activity was expressed as a percentage of chloramphenicol acetylated by 50 μl of extract incubated at 37°C for 1 hr. β-Galactosidase activity was expressed as the optical density at 420 nm obtained with 20 μl of extract incubated at 37°C. CAT activity results were normalized to β-galactosidase activity to adjust for transfection efficiency and expressed relative to the level of CAT activity obtained by cotransfection of reporter plasmid with pCMV12S.FS.
Electrophoretic Mobility Shift Assays (EMSA).
Double-stranded oligonucleotide probes in EMSAs were end-labeled with [α-32P]dATP (3,000 Ci/mmol) using DNA polymerase I (Klenow fragment) and purified by PAGE. DNA binding assays were performed essentially as described in ref. 34. In brief, HeLa cell nuclear extracts (5 μg) were incubated with 1× EMSA buffer (12 mM Hepes, pH 7.6, 50 mM NaCl, 1 mM DTT, and 5% (vol/vol) glycerol, 2 μg poly(dI-dC)⋅poly(dI-dC), and oligonucleotide probes (20,000 cpm) in a final volume of 20 μl. DNA binding reaction mixtures were resolved on a 5% polyacrylamide (29:1 Bis)-0.5× 89 mM Tris/89 mM boric acid/2 mM EDTA, pH 8.0 gel, after a 15-min incubation at 37°C. In EMSA experiments with in vitro translated proteins, 5 μg of BSA (Boehringer Manheim) was included in the DNA binding mixtures. Gels were subject to electrophoresis at 4°C (10 V/cm), and dried for autoradiography.
In Vitro Transcription and Translation.
Plasmids pT7CREB (42) and pGEM-ATF-1 (40) were used to synthesize full-length CREB and ATF-1 proteins, respectively, using the ProMega TNT Kit or homemade T7 RNA polymerase and rabbit reticulocyte lysate. EMSAs were performed with unlabeled proteins, and [35S]methionine-labeled proteins were synthesized in parallel reactions. Translation products were analyzed in SDS/12% PAGE.
RESULTS
CREB-PERE Complexes.
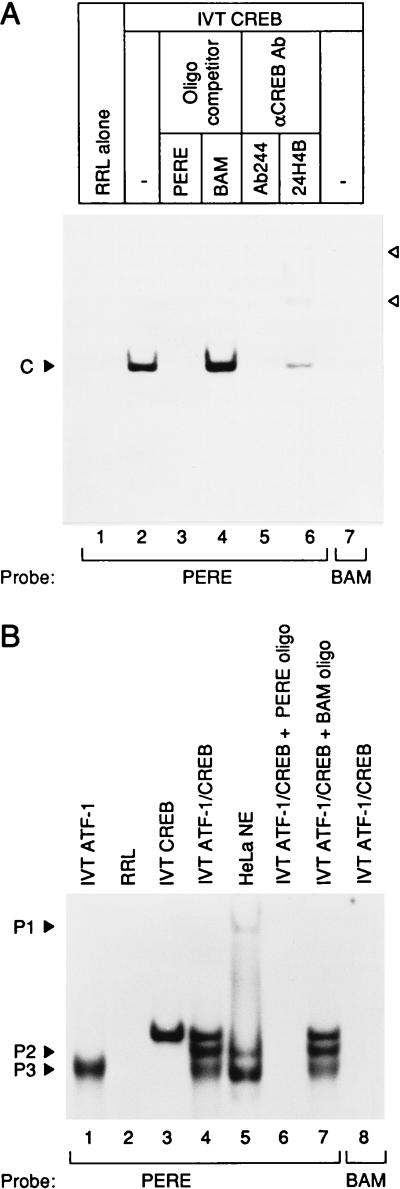
The transcription factor ATF-1 binds to the PERE (34), yet ATF-1 is reportedly unresponsive to E1A (42, 43). ATF-1 binds to DNA as either a homodimer or a heterodimer with the CREB transcription factor (40, 44, 45). Previous antibody-interference assays did not reveal any CREB binding at the PERE (34), although protein purification and UV-crosslinking studies demonstrated that a protein of ≈45 kDa, similar to the molecular mass of CREB (41), associates with the PERE (data not shown). To test directly whether CREB can bind to the PERE, we performed EMSAs with CREB protein synthesized in a cell-free translation system (IVT). With the radiolabeled PERE probe IVT CREB formed a specific complex (Fig. 1A, lane 2), not observed with unprogrammed reticulocyte lysate (lane 1). The addition of excess unlabeled wild-type PERE competitor oligonucleotide abrogated the CREB complex (lane 3), whereas complex formation was unaffected by the addition of excess cold BAM oligonucleotide (lane 4), which contains a 4-bp mutation within the core of the ATF-consensus binding site. Moreover, IVT CREB failed to form a complex when the BAM oligonucleotide was used as a probe (lane 7). Finally, anti-CREB antibody (Ab244) was able to inhibit formation of the IVT CREB complex with a faint indication of a supershifted band (lane 5) and CREB antibody (24H4B) could supershift the IVT CREB complex bound to the DNA probe (lane 6), confirming CREB binding to the PERE site.
Figure 1.
CREB transcription factor binds to the PERE. (A) Full-length CREB was synthesized in vitro and incubated with α-32P-labeled PERE or BAM probe. IVT-CREB formed a specific complex (C) on the PERE probe not present with the mutant BAM probe. PERE probe was also incubated with unprogrammed reticulocyte lysate (RRL) as a control. Supershifted CREB complexes produced by anti-CREB antibodies are denoted by arrowheads. (B) CREB forms a heterodimer with ATF-1 on the PERE and forms complexes comigrating with complexes P2 and P3. Full-length ATF-1 and CREB were synthesized in vitro and bound to the PERE probe individually or mixed together in DNA binding reactions. The positions of P1–3, DNA-protein complexes formed from HeLa cell nuclear extract (5 μg), are indicated. The PERE oligonucleotide (5′-CAGCGTGGTGACGTCGCAACG-3′) contains sequences from −60 to −40 relative to the transcription start site of the human PCNA promoter. The BAM oligonucleotide (5′-CAGCGTGGTGGATCCGCAACG-3′) harbors a 4-bp mutation within the ATF consensus binding site that abrogates PCNA promoter transactivation by E1A in vivo (30, 32). Anti-CREB antibodies (Ab244, M. Montminy, Harvard Medical School; 24HB4, Santa Cruz Biotechnology) were preincubated in the DNA binding mixture for at least 1 hr at 4°C before the addition of radiolabeled probe.
As shown previously (34), the PERE gives a single band with cell-free synthesized ATF-1 (Fig. 1B, lane 1). This band has a greater mobility than the band formed with IVT CREB (lane 3). Significantly, when IVT CREB and IVT ATF-1 were incubated together, three complexes were formed (lane 4). Their specificity for the PERE was demonstrated by competition experiments in which all three bands were abrogated by excess unlabeled PERE oligonucleotide but unaffected by the unlabeled BAM oligonucleotide (lanes 6 and 7). No complexes were formed with the BAM oligonucleotide probe (lane 8). The two faster-migrating bands comigrated with P2 and P3 (compare lanes 4 and 5), two PERE-related complexes that form with HeLa cell nuclear extracts and correlate with PCNA promoter activity in vivo (34). The middle band, which comigrates with complex P2, appeared only when both CREB and ATF-1 were present together, implying that it is a heterodimer of CREB and ATF-1. Complex P3 presumably contains a homodimer of ATF-1 because it comigrates with the band derived from IVT ATF-1 (compare lanes 1 and 5). We deduce that the stoichiometric ratios of CREB and ATF-1 in the HeLa cell nuclear extract are such that the majority of CREB is heterodimerized with ATF-1 and the CREB homodimer is undetectable in the conditions used in our PERE EMSAs.
E1A Transactivation via CREB.
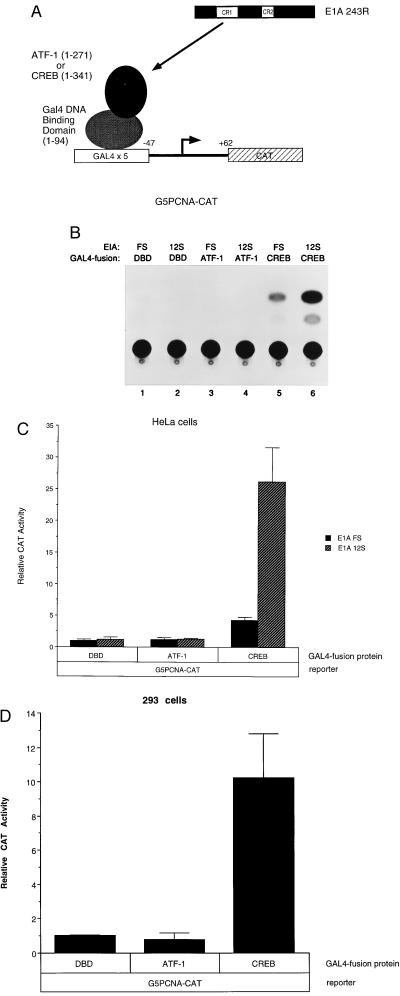
To determine whether ATF-1 or CREB can mediate a response to E1A 243R at the PCNA promoter, we tested each of these proteins in transient expression assays conducted in HeLa cells. As diagrammed in Fig. 2A, GAL4-fusion constructs encoding either ATF-1 or CREB were cotransfected with the G5PCNA-CAT reporter, in which the PERE and PCNA gene sequences upstream of it are replaced by five tandem GAL4 DNA-binding sites (33). When brought to the GAL4 DNA-binding site, GAL4-CREB increased basal transcription levels from the G5PCNA-CAT reporter (Fig. 2B, lane 5; Fig. 2C). Moreover, GAL4-CREB was able to mediate a response to E1A 243R when cotransfected with the wild-type E1A 12S expression plasmid (≈5- to 6-fold over basal levels, obtained with the 12S.FS control; compare lanes 5 and 6). The ability of E1A 243R to transactivate the minimal PCNA promoter via CREB was a specific effect in that E1A 243R was unable to transactivate the promoter when either GAL4-ATF-1 (lane 4) or the GAL4 DBD alone (lane 2) was bound to the promoter. We carried out similar transfections in adenovirus-transformed 293 cells, which express endogenous E1A protein. Again, the GAL4-CREB effector plasmid increased G5PCNA-CAT expression whereas neither GAL4-ATF-1 or the GAL4 DBD alone had any detectable effect on CAT activity (Fig. 2D).
Figure 2.
CREB mediates transactivation by E1A 243R of the PCNA promoter. (A) Experimental design and schematic diagram of the G5PCNA-CAT reporter plasmid and GAL4-fusion protein constructs. G5PCNA-CAT (33) has five tandem GAL4 DNA-binding sites located upstream of the PCNA basal promoter (sequences from −47 to +62) linked to a CAT reporter. GAL4-ATF-1, GAL4-CREB, or GAL4-DBD were cotransfected into HeLa cells with either pCMV12S.FS or pCMV12S. A representative experiment shown in B and C displays an average of three independent transfections performed in duplicate, with SD indicated. Transient expression assays with human 293 cells were carried out with GAL4-ATF-1, GAL4-CREB, or GAL4-DBD and the G5PCNA-CAT reporter. (D) An average of two independent experiments carried out in duplicate, with SD noted.
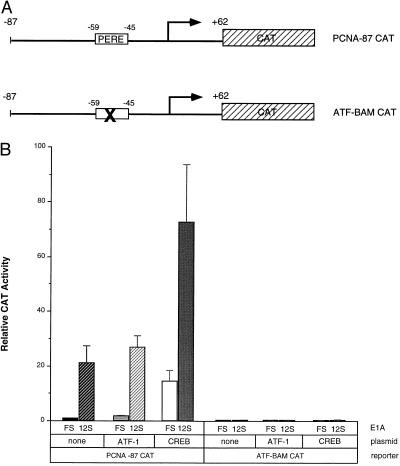
We then examined the effects of CREB and ATF-1 overexpression in the context of the wild-type PCNA-87 CAT and mutant ATF-BAM CAT reporter constructs (Fig. 3A). Consistent with the results obtained with the G5PCNA-CAT reporter, transient expression assays indicated that ATF-1 overexpression had no effect upon PCNA-CAT basal or E1A 243R-induced transcription, whereas cotransfection of a CREB expression plasmid increased both basal and E1A 243R-induced transcription from PCNA-87 CAT (Fig. 3B). The increase brought about by CREB overexpression was dose-dependent (data not shown) and dependent upon the PERE site. CREB’s effect was eliminated by the ATF-BAM mutation (Fig. 3B), which also abrogates PCNA transactivation by E1A 243R (30) and abolishes CREB and ATF-1 binding to the PERE in vitro (Fig. 1 A and B; ref. 34). Taken together, these data point to CREB as a limiting factor for PCNA transcription, and to its role in regulation by the E1A 243R oncoprotein.
Figure 3.
CREB enhances both basal and E1A-transactivated transcription from the wild-type PCNA promoter. (A) Schematic diagram of wild-type PCNA-87 CAT and ATF-BAM CAT reporters (30). Both constructs contain PCNA promoter sequences from −87 to +62 linked to the CAT reporter. The X marked within the PERE box of the ATF-BAM CAT construct denotes a 4-bp mutation that abolishes E1A 243R transactivation of the PCNA promoter in vivo and CREB and ATF-1 binding in vitro. (B) Rous sarcoma virus-ATF-1 (40) (5 μg) or 5 μg of Rous sarcoma virus-CREB (41) were cotransfected into HeLa cells with 5 μg of PCNA-87 CAT or ATF-BAM CAT and 0.5 μg or pCMV12S.FS or pCMV12S as indicated. Results represent three independent experiments performed in duplicate with SD indicated.
Involvement of CBP.
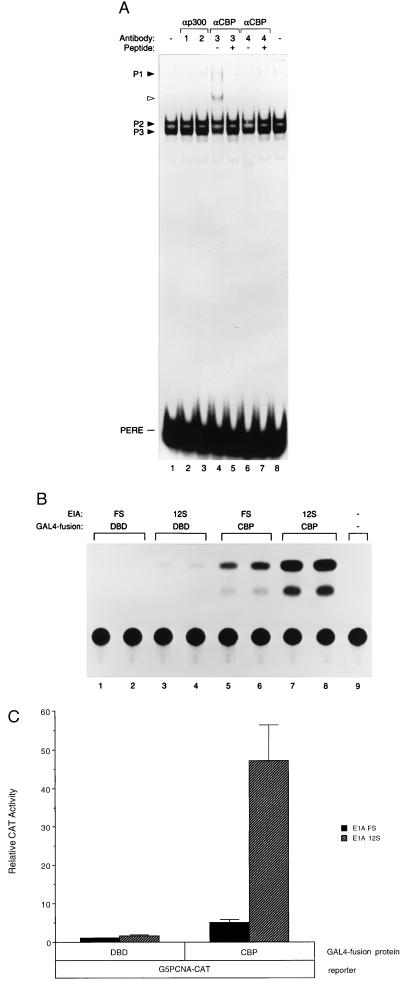
The discovery of CREB binding at the PERE prompted us to examine the possible involvement of the CBP and/or its functional homologue p300 in mediating PCNA’s response to E1A. Although CBP is distinct from p300, the two proteins share many structural and functional features as transcriptional coactivators that bind to similar regions of the E1A oncoprotein (46–48). Furthermore, previous mutational analyses of the E1A 243R protein suggested that those regions of E1A that bind to CBP/p300 are required for optimal PCNA induction by E1A 243R (39). In mobility shift experiments, an antibody specific for CBP (A-22) gave rise to a supershifted PERE-associated complex and concomitantly diminished the intensity of complexes P2 and P3 (Fig. 4A; lane 4). A second anti-CBP antibody (C-20) also reduced the intensity of the P2 and P3 complexes, but no supershifted complex was observed (lane 6). Although these effects were not drastic, they were reproducible, and several observations argue for their significance. Preincubation of the anti-CBP antibodies with their respective antigenic peptides blocked the supershift and/or diminution of P2 and P3 complexes, demonstrating specificity of the effects by these antibodies (lanes 5 and 7). Furthermore, these anti-CBP antibodies had no effect on IVT ATF-1 or CREB in EMSAs (data not shown). In contrast, antibodies directed against the cellular factor p300 had no detectable effect on complexes P2 and P3 (lanes 2 and 3). While these data are inconclusive with respect to p300, they implicated CBP in PERE-bound complexes.
Figure 4.
CBP is a component of PERE-associated complexes. (A) Effects of antibodies to CBP or p300 in EMSAs. HeLa cell nuclear extract (5 μg) was preincubated for 1 hr at 4°C with antibody (1 μg) to p300 (lanes 2 and 3) or CBP (lanes 4–7) before incubation with radiolabeled PERE probe for 15 min at 37°C. Antibodies were as follows: 1, anti-p300 (C-20; Santa Cruz Biotechnology); 2, anti-p300 (Upstate Biotechnology, Lake Placid, NY; catalog no. 05-222); 3, anti-CBP (A-22; Santa Cruz Biotechnology); 4, anti-CBP (C-20; Santa Cruz Biotechnology). For lanes 6 and 8, the antibody was first preincubated with its antigenic peptide (A-22 P or C-20 P; Santa Cruz Biotechnology) for 2 hr at 4°C before mixing with nuclear extract. No antibodies were added in lanes 1 and 8. Complexes P1–3 are noted, and the complex supershifted by anti-CBP antibody in lane 4 is marked by an open arrowhead. (B and C) CBP mediates transactivation by E1A 243R when bound to the PCNA promoter. Five micrograms of pGAL4-CBP or GAL4 DBD alone was cotransfected into HeLa cells with 5 μg of G5PCNA-CAT reporter and 0.5 μg of either pCMV12S.FS or pCMV12S where indicated. A representative experiment is shown in B, and an average of three independent experiments performed in duplicate is shown in C with SD indicated.
We therefore examined whether CBP, like CREB, can mediate a response to E1A 243R when bound to the GAL4 DNA-binding sites of the G5PCNA-CAT reporter. Previous reports indicated that E1A 243R is able to repress CREB-mediated and p300/CBP-mediated transcription at certain promoters (47, 48). As expected, when cotransfected with this reporter plasmid, GAL4-CBP increased basal transcription activity (Fig. 4B, lanes 5 and 6; Fig. 4C). In marked contrast to what has previously been observed at other promoters (47, 48), but consistent with E1A 243R transactivation of the PCNA promoter, cotransfection of E1A 12S dramatically transactivated GAL4-CBP when bound to the G5PCNA-CAT reporter (Fig. 4B, lanes 7 and 8; Fig. 4C).
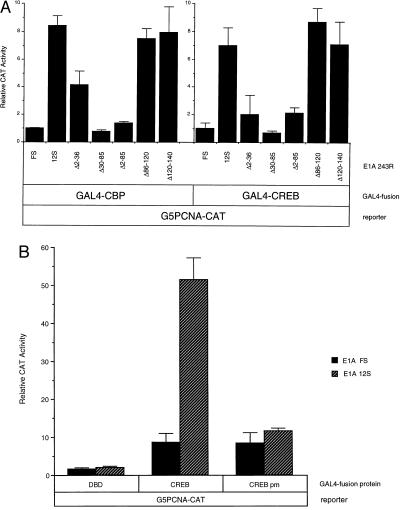
To determine if the p300/CBP binding domain of E1A 243R was necessary to transactivate both CBP and CREB, we used a panel of E1A mutants that differ in their ability to interact with p300/CBP. Similar to results shown in Figs. 2 and 4, wild-type E1A 243R (E1A 12S) transactivated GAL4-CREB and GAL4-CBP ≈6- and ≈8-fold, respectively, in transient expression experiments with the G5PCNA-CAT reporter (Fig. 5A). However, E1A proteins harboring deletions of amino acids from 2–36 (E1A Δ2–36), 30–85 (E1A Δ30–85), or 2–85 (E1A Δ2–85), which are unable to interact with p300/CBP (49, 50), are impaired for activation of G5PCNA-CAT through GAL4-CREB or GAL4-CBP (Fig. 5A). In contrast, deletions of E1A that do not affect p300/CBP binding did not adversely affect E1A’s ability to transactivate the G5PCNA-CAT reporter via GAL4-CREB or GAL4-CBP (Fig. 5A), E1A Δ86–120, and E1A Δ120–140). The pattern of stimulation by the various E1A mutants was similar when either GAL4-CREB and GAL4-CBP was used.
Figure 5.
(A) Transactivation of PCNA through CREB and CBP by E1A requires p300/CBP binding regions of E1A 243R. The G5PCNA-CAT reporter plasmid was cotransfected into HeLa cells with either GAL4-CREB or GAL4-CBP and wild-type, or pCMV12S.FS, or mutant E1A expression plasmids (pCMVΔ2–36, pCMVΔ30–85, pCMVΔ2–85, pCMVΔ86–120, and pCMVΔ120–140) as indicated. CAT activity values represent an average of three independent experiments performed in duplicate with SD noted. (B) Transactivation of CREB by E1A requires CBP. HeLa cells were transiently transfected with the G5PCNA-CAT reporter plasmid and either GAL4-CREB or GAL4-CREB pm and wild-type pCMV12S or the control pCMV12S.FS plasmid. Relative CAT activity values represent an average of two independent experiments performed in duplicate with SD indicated.
As a further test of CBP’s involvement, we used a mutant GAL4-CREB fusion protein deficient in its ability to interact with CBP and assessed its ability to mediate transactivation of the G5PCNA-CAT reporter by E1A 243R. CBP has been shown to bind specifically to CREB protein phosphorylated at a conserved serine residue via a direct mechanism (36, 51). Mutation of this serine residue abolishes the cAMP-dependent transactivation of CREB and its ability to bind to CBP (36). Consistent with our hypothesis that E1A 243R is transactivating CREB via CBP, a mutant GAL4-CREB plasmid containing a serine to alanine substitution at residue 133 (GAL4-CREB pm) could not mediate transactivation of the G5PCNA-CAT reporter by E1A 243R as compared with wild-type GAL4-CREB protein (Fig. 5B). Taken together, the observations presented in Fig. 5 indicate that transactivation of PCNA expression by E1A occurs through direct interactions between CREB, CBP, and E1A 243R.
DISCUSSION
The E1A 243R protein generally represses transcription by inhibiting enhancer function (50, 52), a property that correlates with its ability to bind to p300/CBP (49, 53). In spite of this effect, E1A 243R is capable of transactivating a number of viral and cellular promoters (29, 31, 54–57). E2F-dependent transactivation and the relief of YY1 transcriptional repression by E1A 243R are two mechanisms by which this oncoprotein has been shown to transactivate promoters (54, 58–60). However, the human PCNA promoter lacks a clearly defined E2F-binding site (31), and although we have demonstrated the binding of YY1 to the initiator sequence of the promoter (34), no functional significance has been attributed to YY1-binding at the PCNA promoter in the context of E1A transactivation (32).
Because E1A is not a sequence-specific DNA binding protein (61, 62) its ability to transactivate DNA sequences presumably involves protein-protein interactions. Our data suggest a model in which E1A 243R is targeted to the PCNA promoter at the PERE site via a CREB-CBP-DNA pathway and may provide an example of an alternative mechanism by which E1A 243R can activate transcription at certain promoters in a sequence-specific fashion. Once localized at the PCNA promoter, how does E1A 243R up-regulate gene transcription? The presence of CBP at the PERE provides a physical bridge between E1A and the general transcription machinery, as CBP has been demonstrated to interact with TFIIB (63). One can envisage that E1A 243R alters interactions between CBP and general transcription factors, thereby promoting the assembly of initiation complexes on the PCNA basal promoter and increasing transcription. Alternatively, because those regions of E1A 243R that bind to the retinoblastoma-related p107 protein also are required for optimal transactivation by E1A (39), additional interactions between E1A 243R and p107 may be required to induce PCNA expression. It is possible that E1A mediates the relief of transcriptional inhibition of cellular factors, including p107, which associate with the PCNA promoter.
The binding of CBP/p300 to E1A is well established as an important interaction for the transforming and growth-inducing abilities of this viral oncoprotein. As PCNA is tightly associated with cellular growth activities, its transactivation by E1A may represent one link between the processes of transcription and neoplastic transformation. Furthermore, the association of CREB and CBP with the PCNA promoter has implications for growth control in normal cells. The involvement of CREB and CBP in the regulation of PCNA gene expression provides new insights into additional mechanisms by which the cell cycle and normal cellular growth may be influenced via these two proteins.
Acknowledgments
We thank P. Wendel for expert tissue culture, M. Gilman and N. Jones for CREB expression plasmids, S. Wagner and M. Green for pGEM-ATF-1, M. Montminy for CREB antibody, R. Goodman for CBP reagents, W. Herr for pCGGAL4, and C. Labrie for helpful discussions. This work was supported by National Institutes of Health Grant CA 13106.
ABBREVIATIONS
- PCNA
proliferating cell nuclear antigen
- CREB
cAMP response element binding protein
- PERE
PCNA-E1A responsive element
- ATF
activating transcription factor
- CBP
CREB-binding protein
- CAT
chloramphenicol acetyltransferase
- EMSA
electrophoretic mobility shift assay
- DBD
DNA-binding domain
- CMV
cytomegalovirus
References
- 1.Prelich G, Tan C-K, Kostura M, Mathews M B, So A G, Downey K M, Stillman B. Nature (London) 1987;326:517–520. doi: 10.1038/326517a0. [DOI] [PubMed] [Google Scholar]
- 2.Waga S, Stillman B. Nature (London) 1994;369:207–212. doi: 10.1038/369207a0. [DOI] [PubMed] [Google Scholar]
- 3.Bravo R, Frank R, Blundell P A, Macdonald-Bravo H. Nature (London) 1987;326:515–517. doi: 10.1038/326515a0. [DOI] [PubMed] [Google Scholar]
- 4.Prelich G, Kostura M, Marshak D R, Mathews M B, Stillman B. Nature (London) 1987;326:471–475. doi: 10.1038/326471a0. [DOI] [PubMed] [Google Scholar]
- 5.Kong X-P, Onrust R, O’Donnell M, Kuriyan J. Cell. 1992;69:425–437. doi: 10.1016/0092-8674(92)90445-i. [DOI] [PubMed] [Google Scholar]
- 6.Brand S R, Bernstein R M, Mathews M B. J Immunol. 1994;153:3070–3078. [PubMed] [Google Scholar]
- 7.Tan C-K, Castillo C, So A G, Downey K M. J Biol Chem. 1986;261:12310–12316. [PubMed] [Google Scholar]
- 8.Almendral J M, Huebsch D, Blundell P A, MacDonald-Bravo H, Bravo R. Proc Natl Acad Sci USA. 1987;84:1575–1579. doi: 10.1073/pnas.84.6.1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bravo R, MacDonald-Bravo H. EMBO J. 1984;3:3177–3181. doi: 10.1002/j.1460-2075.1984.tb02276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Matsumoto K, Moriuchi T, Koji T, Nakane P. EMBO J. 1987;6:637–642. doi: 10.1002/j.1460-2075.1987.tb04802.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jaskulski D, Gatti C, Travali S, Calabretta B, Baserga R. J Biol Chem. 1988;263:10175–10179. [PubMed] [Google Scholar]
- 12.Bauer G A, Burgers P M J. Nucleic Acids Res. 1990;18:261–265. doi: 10.1093/nar/18.2.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu Y-C, Marraccino R L, Keng P C, Bambara R A, Lord E M, Chou W-G, Zain S B. Biochemistry. 1989;28:2967–2974. doi: 10.1021/bi00433a034. [DOI] [PubMed] [Google Scholar]
- 14.Xiong Y, Zhang H, Beach D. Cell. 1992;71:505–514. doi: 10.1016/0092-8674(92)90518-h. [DOI] [PubMed] [Google Scholar]
- 15.Xiong Y, Hannon G J, Zhang H, Casso D, Kobayashi R, Beach D. Nature (London) 1993;366:701–704. doi: 10.1038/366701a0. [DOI] [PubMed] [Google Scholar]
- 16.Xiong Y, Zhang H, Beach D. Genes Dev. 1993;7:1572–1583. doi: 10.1101/gad.7.8.1572. [DOI] [PubMed] [Google Scholar]
- 17.Zhang H, Xiong Y, Beach D. Mol Biol Cell. 1993;4:897–906. doi: 10.1091/mbc.4.9.897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Waga S, Hannon G J, Beach D, Stillman B. Nature (London) 1994;369:574–578. doi: 10.1038/369574a0. [DOI] [PubMed] [Google Scholar]
- 19.Nichols A F, Sancar A. Nucleic Acids Res. 1992;20:2441–2446. doi: 10.1093/nar/20.10.2441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shivji M K K, Kenny M K, Wood R D. Cell. 1992;69:367–374. doi: 10.1016/0092-8674(92)90416-a. [DOI] [PubMed] [Google Scholar]
- 21.Li R, Waga S, Hannon G, Beach D, Stillman B. Nature (London) 1994;371:534–537. doi: 10.1038/371534a0. [DOI] [PubMed] [Google Scholar]
- 22.Smith M L, Chen I T, Zhan Q, Bae I, Chen C Y, Gilmer T M, Kastan M B, O’Connor P M, Fornace A J. Science. 1994;266:1376–1380. doi: 10.1126/science.7973727. [DOI] [PubMed] [Google Scholar]
- 23.Hall P A, McKee P H, Menage H D, Dover R, Lane D P. Oncogene. 1993;8:203–207. [PubMed] [Google Scholar]
- 24.Morris G F, Bischoff J R, Mathews M B. Proc Natl Acad Sci USA. 1996;93:895–899. doi: 10.1073/pnas.93.2.895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Berk A J. Annu Rev Genet. 1986;20:45–79. doi: 10.1146/annurev.ge.20.120186.000401. [DOI] [PubMed] [Google Scholar]
- 26.Moran E, Mathews M B. Cell. 1987;48:177–188. doi: 10.1016/0092-8674(87)90418-1. [DOI] [PubMed] [Google Scholar]
- 27.Nevins J R. Adv Vir Res. 1989;37:35–83. doi: 10.1016/s0065-3527(08)60832-5. [DOI] [PubMed] [Google Scholar]
- 28.Shenk T, Flint T. Adv Canc Res. 1991;57:47–85. doi: 10.1016/s0065-230x(08)60995-1. [DOI] [PubMed] [Google Scholar]
- 29.Zerler B, Roberts R J, Mathews M B, Moran E. Mol Cell Biol. 1987;7:821–829. doi: 10.1128/mcb.7.2.821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Morris G F, Mathews M B. J Virol. 1991;65:6397–6406. doi: 10.1128/jvi.65.12.6397-6406.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Morris G F, Mathews M B. J Biol Chem. 1990;265:16116–16125. [PubMed] [Google Scholar]
- 32.Labrie C, Morris G F, Mathews M B. Mol Cell Biol. 1993;13:1697–1707. doi: 10.1128/mcb.13.3.1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Morris G F, Labrie C, Mathews M B. Mol Cell Biol. 1994;14:543–553. doi: 10.1128/mcb.14.1.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Labrie C, Lee B H, Mathews M B. Nucleic Acids Res. 1995;23:3732–3741. doi: 10.1093/nar/23.18.3732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tanaka M, Clouston W M, Herr W. Mol Cell Biol. 1994;14:6046–6055. doi: 10.1128/mcb.14.9.6046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chrivia J C, Kwok R P S, Lamb N, Hagiwara M, Montminy M R, Goodman R H. Nature (London) 1993;365:855–859. doi: 10.1038/365855a0. [DOI] [PubMed] [Google Scholar]
- 37.Spaete R R, Mocarski E S. J Virol. 1985;56:135–143. doi: 10.1128/jvi.56.1.135-143.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.White E, Cipriani R. Mol Cell Biol. 1990;10:120–130. doi: 10.1128/mcb.10.1.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kannabiran C, Morris G F, Labrie C, Mathews M B. J Virol. 1993;67:507–515. doi: 10.1128/jvi.67.1.507-515.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu F, Thompson M A, Wagner S, Greenberg M E, Green M R. J Biol Chem. 1993;268:6714–6720. [PubMed] [Google Scholar]
- 41.Berkowitz L A, Gilman M Z. Proc Natl Acad Sci USA. 1990;87:5258–5262. doi: 10.1073/pnas.87.14.5258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Flint K J, Jones N C. Oncogene. 1991;6:2019–2026. [PubMed] [Google Scholar]
- 43.Liu F, Green M R. Cell. 1990;61:1217–1224. doi: 10.1016/0092-8674(90)90686-9. [DOI] [PubMed] [Google Scholar]
- 44.Hurst H C, Masson N, Jones N C, Lee K A W. Mol Cell Biol. 1990;10:6192–6203. doi: 10.1128/mcb.10.12.6192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rehfuss R P, Walton K M, Loriaux M M, Goodman R H. J Biol Chem. 1991;266:18431–18434. [PubMed] [Google Scholar]
- 46.Arany Z, Sellers W R, Livingston D M, Eckner R. Cell. 1994;77:799–800. doi: 10.1016/0092-8674(94)90127-9. [DOI] [PubMed] [Google Scholar]
- 47.Arany Z, Newsome D, Oldread E, Livingston D M, Eckner R. Nature (London) 1995;374:81–84. doi: 10.1038/374081a0. [DOI] [PubMed] [Google Scholar]
- 48.Lundblad J R, Kwok R P, Laurance M E, Harter M L, Goodman R H. Nature (London) 1995;374:85–88. doi: 10.1038/374085a0. [DOI] [PubMed] [Google Scholar]
- 49.Whyte P, Williamson N M, Harlow E. Cell. 1989;56:67–75. doi: 10.1016/0092-8674(89)90984-7. [DOI] [PubMed] [Google Scholar]
- 50.Stein R W, Corrigan M, Yaciuk P, Whelan J, Moran E. J Virol. 1990;64:4421–4427. doi: 10.1128/jvi.64.9.4421-4427.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Parker D, Ferreri K, Nakajima T, LaMorte V J, Evans R, Koerber S C, Hoeger C, Montminy M R. Mol Cell Biol. 1996;16:694–703. doi: 10.1128/mcb.16.2.694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rochette-Egly L, Fromental C, Chambon P. Genes Dev. 1990;4:137–150. doi: 10.1101/gad.4.1.137. [DOI] [PubMed] [Google Scholar]
- 53.Jelmsa T N, Howe J A, Mymryk J S, Evelegh C M, Cunnif N F A, Bayley S T. Virology. 1989;170:120–130. doi: 10.1016/0042-6822(89)90518-7. [DOI] [PubMed] [Google Scholar]
- 54.Bagchi S, Raychaudhuri P, Nevins J R. Cell. 1990;62:659–669. doi: 10.1016/0092-8674(90)90112-r. [DOI] [PubMed] [Google Scholar]
- 55.Leff T, Elkaim R, Goding C R, Jalinot P, Sassone-Corsi P, Perricaudet M, Kédinger C, Chambon P. Proc Natl Acad Sci USA. 1984;81:4381–4385. doi: 10.1073/pnas.81.14.4381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mudryj M, Hiebert S W, Nevins J R. EMBO J. 1990;7:2179–2184. doi: 10.1002/j.1460-2075.1990.tb07387.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.van Dam H, Duyndam M, Rottier R, Bosch A, Vries-Smits L D, Herrlich P, Zantema A, Angel P, van der Eb A J. EMBO J. 1993;12:479–487. doi: 10.1002/j.1460-2075.1993.tb05680.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bandara L R, La Thangue N B. Nature (London) 1991;351:494–497. doi: 10.1038/351494a0. [DOI] [PubMed] [Google Scholar]
- 59.Shi Y, Seto E, Chang L-S, Shenk T. Cell. 1991;67:377–388. doi: 10.1016/0092-8674(91)90189-6. [DOI] [PubMed] [Google Scholar]
- 60.Lee J-S, Galvin K M, See R H, Eckner R, Livingston D, Moran E, Shi Y. Genes Dev. 1995;9:1188–1198. doi: 10.1101/gad.9.10.1188. [DOI] [PubMed] [Google Scholar]
- 61.Chatterjee P K, Bruner M, Flint S J, Harter M L. EMBO J. 1988;7:835–841. doi: 10.1002/j.1460-2075.1988.tb02882.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ferguson B, Krippl B, Andrisani O, Jones N, Westphal H, Rosenberg M. Mol Cell Biol. 1985;5:2653–2661. doi: 10.1128/mcb.5.10.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kwok R P S, Lundblad J R, Chrivia J C, Richards J P, Bachinger H P, Brennan R G, Roberts S G R, Green M R, Goodman R H. Nature (London) 1994;370:223–226. doi: 10.1038/370223a0. [DOI] [PubMed] [Google Scholar]