SUMMARY
Peptidoglycan recognition proteins (PGRPs) are structurally conserved through evolution, but their functions in innate immunity are different in invertebrates and vertebrates. We asked what the functions of PGRPs in fish are and whether they are indispensable for defense against infection, because fish are the first vertebrates that developed adaptive immunity, but still rely solely on innate immunity during early development of embryos. We identified and cloned three zebrafish PGRPs, and showed that they are highly expressed in eggs, developing embryos, and in adult tissues that contact external environment. Zebrafish PGRPs have both peptidoglycan-lytic amidase activity and broad-spectrum bactericidal activity, which is a unique feature. Furthermore, we demonstrated that in the developing zebrafish embryo one of these PGRPs is essential for defense and survival during bacterial infections. This is the first demonstration of an absolute requirement for innate immunity in defense against infections in fish embryos and for a PGRP protein for survival in vertebrates.
INTRODUCTION
Innate immunity arose in the early multicellular organisms and has remained an essential component of defense mechanisms in all metazoans. Although all vertebrates developed acquired immunity, early vertebrates' eggs and embryos, which are laid and develop in water, must solely rely on innate immunity in defense against infections, until their acquired immune system develops.
Peptidoglycan Recognition Proteins (PGRPs or PGLYRPs) is a family of innate immunity molecules that were first identified in insects (Yoshida et al., 1996; Kang et al., 1998) and then in mammals (Kang et al., 1998; Liu et al., 2001), other vertebrates, mollusks, and echinoderms (Dziarski and Gupta, 2006b). The invertebrate and vertebrate PGRP proteins are highly conserved in their structure and recognize bacteria through their cell wall component, peptidoglycan.
Peptidoglycan is a major component of bacterial cell wall and is a polymer of β(1-4)-linked N-acetylglucosamine (GlcNAc) and N-acetylmuramic acid (MurNAc). MurNAc has short peptides, typically containing four alternating L- and D-amino acids. The third amino acid in Gram-negative bacteria and Gram-positive bacilli is m-diaminopimelic acid (DAP) and in most other Gram-positive bacteria (including Gram-positive cocci) it is L-lysine (Lys) (Dziarski and Gupta, 2006a, b).
Despite the structural similarities, invertebrate and vertebrate PGRPs have acquired different functions. Insects have many PGRPs that function in cell activation, phagocytosis, and/or hydrolysis of peptidoglycan (Werner et al., 2000). The cell activating PGRPs stimulate either the Toll (Drosophila PGRP-SA, PGRP-SD, and PGRP-SC1) (Michel et al., 2001; Bischoff et al., 2004; Garver et al., 2006) or the Imd (Drosophila PGRP-LC) (Choe et al., 2002; Gottar et al., 2002; Ramet et al., 2002) signal transduction pathways, which induce transcription of antimicrobial peptides active against Gram-positive bacteria and fungi (Lemaitre et al., 1996) or Gram-negative bacteria (Lemaitre et al., 1995), respectively. The Toll and Imd pathways are preferentially triggered by either Lys-type or DAP-type peptidoglycan, respectively (Leulier et al., 2003; Kaneko and Silverman, 2005). Other PGRPs, such as silkworm PGRP-S, activate the prophenol-oxidase cascade, which also generates antimicrobial products (Yoshida et al., 1996; Park et al., 2006). Finally, some PGRPs, such as Drosophila PGRP-SC1, PGRP-LB, and PGRP-SB1 are N-acetylmuramoyl-L-alanine amidases (Kim et al., 2003; Mellroth et al., 2003; Mellroth and Steiner 2006), which hydrolyze the lactyl-amide bond between MurNAc and L-Ala. The function of these insect amidases is to prevent excessive activation of the immune system by bacteria (Mellroth et al., 2003; Zaidman-Remy et al. 2006; Bischoff et al., 2006).
Mammals have a family of four PGRPs: PGLYRP-1, PGLYRP-2, PGLYRP-3, and PGLYRP-4 (Liu et al., 2001; Dziarski and Gupta, 2006b). These proteins were initially hypothesized to function as pattern recognition receptors similar to insect PGRPs (Liu et al., 2001). However, we have demonstrated that human PGRPs have two major functions: amidase and antibacterial activities. PGLYRP-2, is an N-acetylmuramoyl-L-alanine amidase and is constitutively expressed in the liver and secreted into the bloodstream (Gelius et al., 2003; Wang et al., 2003; Zhang et al., 2005). Its expression is also induced in keratinocytes upon bacterial stimulation (Wang et al., 2005; Li et al., 2006). PGLYRP-1 is present in the granules of polymorphonuclear leukocytes (Tydell et al., 2002; Dziarski et al., 2003). PGLYRP-3 and PGLYRP-4 are expressed in tissues that come in contact with the environment (Lu et al., 2006). PGLYRP-1, PGLYRP-3, and PGLYRP-4 are bactericidal and kill a wide range of Gram-positive and Gram-negative bacteria (Lu et al., 2006; Wang et al. 2007). These PGLYRPs do not have amidase activity, but their bactericidal activity relies on targeting bacterial cell wall and does not involve peptidoglycan hydrolysis or permeabilization of the cytoplasmic membrane (Lu et al., 2006; Wang et al. 2007).
Insect PGRPs are not known to be bactericidal, except for Drosophila PGRP-SB1, which has both N-acetylmuramoyl-L-alanine amidase and bactericidal activities (Mellroth and Steiner, 2006). However, PGRP-SB1 is bactericidal for only one bacterial species, Bacillus megaterium, which is likely due to its amidase activity (Mellroth and Steiner, 2006). By contrast, mammalian PGLYRPs have broad-spectrum bactericidal activity that does not rely on amidase activity (Tydell et al., 2002; Lu et al., 2006; Wang et al. 2007).
To gain insight into the evolution of the function of PGRPs in innate immunity, we have identified a family of PGLYRPs in an earlier vertebrate, the zebrafish, and we studied their functions and role in defense against bacterial infections. Zebrafish and other teleosts have a well-developed immune system that is similar to the mammalian immune system, and unlike invertebrates, includes both innate and adaptive immunity (Yoder et al., 2002; Lam et al., 2004; Trede et al., 2004;). However, fish were the first vertebrates to develop adaptive immunity, and it is not known whether their innate immune system has greater similarities to the invertebrate or to the higher vertebrate systems. Zebrafish, thus, serves as an important model for the evolutionary analysis of immune mechanisms and the role of innate immunity in lower vertebrates (Trede et al., 2001; Yoder et al.; 2002; Lam et al., 2004; DeVries et al., 2006; Trede et al., 2004).
Results
Cloning and Sequence Analysis of Zebrafish PGLYRPs
In order to identify pglyrp genes in the zebrafish we searched GenBank databases for genes homologous to human PGLYRPs. We identified four pglyrp genes in the zebrafish genome (about 70% of the genome is sequenced). We have cloned three of these genes and have named them pglyrp-2, pglyrp-5, and pglyrp-6. Zebrafish PGLYRP-2 was designated 2 because it had the highest homology to mammalian PGLYRP-2 compared to other zebrafish PGLYRPs. The other two zebrafish PGLYRPs were not considered orthologs of the remaining three mammalian PGLYRPs, because unlike these mammalian PGLYRPs, they are predicted amidases, they are larger than PGLYRP-1, and they do not have two PGRP domains, which are present in mammalian PGLYRP-3 and PGLYRP-4. Thus, we named them PGLYRP-5 and PGLYRP-6. We were unable to clone the full length cDNA for the fourth zebrafish PGLYRP which was identified from an EST clone. The genome organization (Fig. S1) and sequence analysis (Fig. S2) of zebrafish PGLYRPs is presented in the Supplementary Results.
Zebrafish PGLYRP proteins have no predicted signal peptides. However, our immunohistochemistry data demonstrate that all three proteins are present in the lumen of blood vessels, which indicates that they are secreted. Zebrafish PGLYRPs are not glycosylated (data not shown), unlike human PGLYRPs. Zebrafish PGLYRPs, similar to most vertebrate and invertebrate PGRPs, all have one C-terminal PGRP domain, which have 43%, 36%, and 42%, conserved identities and 59%, 54%, and 57% conserved similarities, respectively, with PGRP domains in human PGLYRPs (Fig. S2). All three zebrafish PGLYRPs are predicted to have amidase activity, because they all have the conserved four Zn2+-binding amino acids, corresponding to His411, Tyr447, His522, and Cys530 in human PGLYRP-2, which is an amidase (Wang et al., 2003). These four amino acids are conserved in all amidase-active PGRPs and in bacteriophage and bacterial type-2 amidases, such as bacteriophage T7 lysozyme. By contrast, human PGLYRP-1, PGLYRP-3, and PGLYRP-4, which are not amidases, do not have the last Cys conserved (corresponding to Cys530 in human PGLYRP-2).
Thus, we have identified four pglyrp genes in zebrafish and we have cloned full-length cDNA for three of these genes. Zebrafish PGLYRP-2, PGLYRP-5, and PGLYRP-6, each have one C-terminal PGRP domain that is conserved through evolution and these proteins are predicted to be amidases.
Differential Expression of Zebrafish PGLYR-2, PGLYR-5, and PGLYR-6
We performed both RNA and protein analysis to identify tissues that express the different PGLYRP proteins. Total RNA isolated from different tissues from adult zebrafish was analyzed by Northern blots. Both PGLYRP-2 and PGLYRP-5 mRNA were strongly expressed in the egg and had lower levels of expression in the liver (Fig. 1). PGLYRP-5 mRNA was also expressed in the intestine and gill (Fig. 1). In contrast, PGLYRP-6 was strongly expressed in the intestine and weakly expressed in the liver (Fig. 1).
Figure 1. Zebrafish PGLYRP-2, PGLYRP-5, and PGLYRP-6 mRNAs are differentially expressed in various tissues.
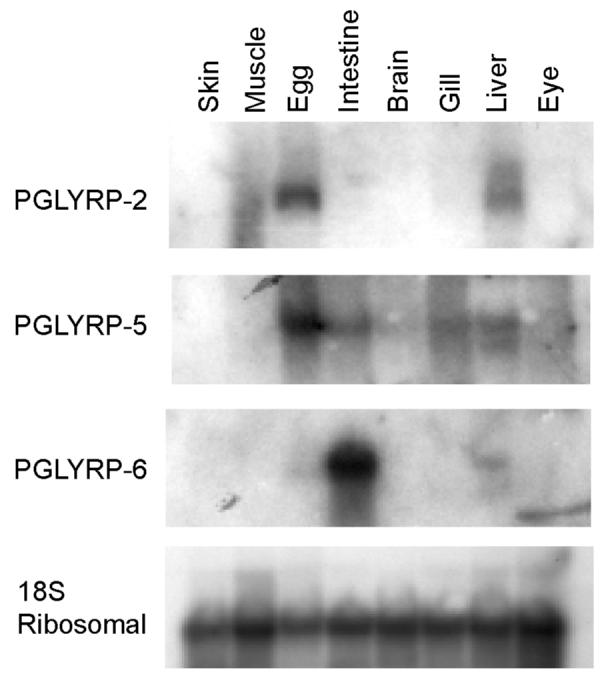
Northern blots with RNA isolated from different adult zebrafish tissues were hybridized with the indicated probes and exposed to X-ray film. Hybridization with the 18S ribosomal probe demonstrates equal loading of the RNA samples. The results are from one of three similar experiments.
In situ hybridization of whole fish sections using RNA probes for the three PGLYRPs demonstrated a similar pattern of differential expression. PGLYRP-2 and PGLYRP-5 transcripts were strongly expressed in the developing oocytes (Stages II, III, and IV shown for PGLYRP-2, Fig. 2A; stage V shown for PGLYRP-5, Fig. 2E). PGLYRP-2 and PGLYRP-5 mRNAs were also expressed in hepatocytes in the liver (Fig. 2, C and G). PGLYRP-6 was strongly expressed in the absorptive cells of the intestine with no expression in the goblet cells (Fig. 2I). The staining was specific for all PGLYRPs as only the antisense probes (Fig. 2, A, C, E, G, and I) and not the sense probes (Fig. 2, B, D, F, H, and J) hybridized.
Figure 2. PGLYRP-2, PGLYRP-5, and PGLYRP-6 transcripts are differentially expressed in developing oocytes, liver, and intestine of adult zebrafish.
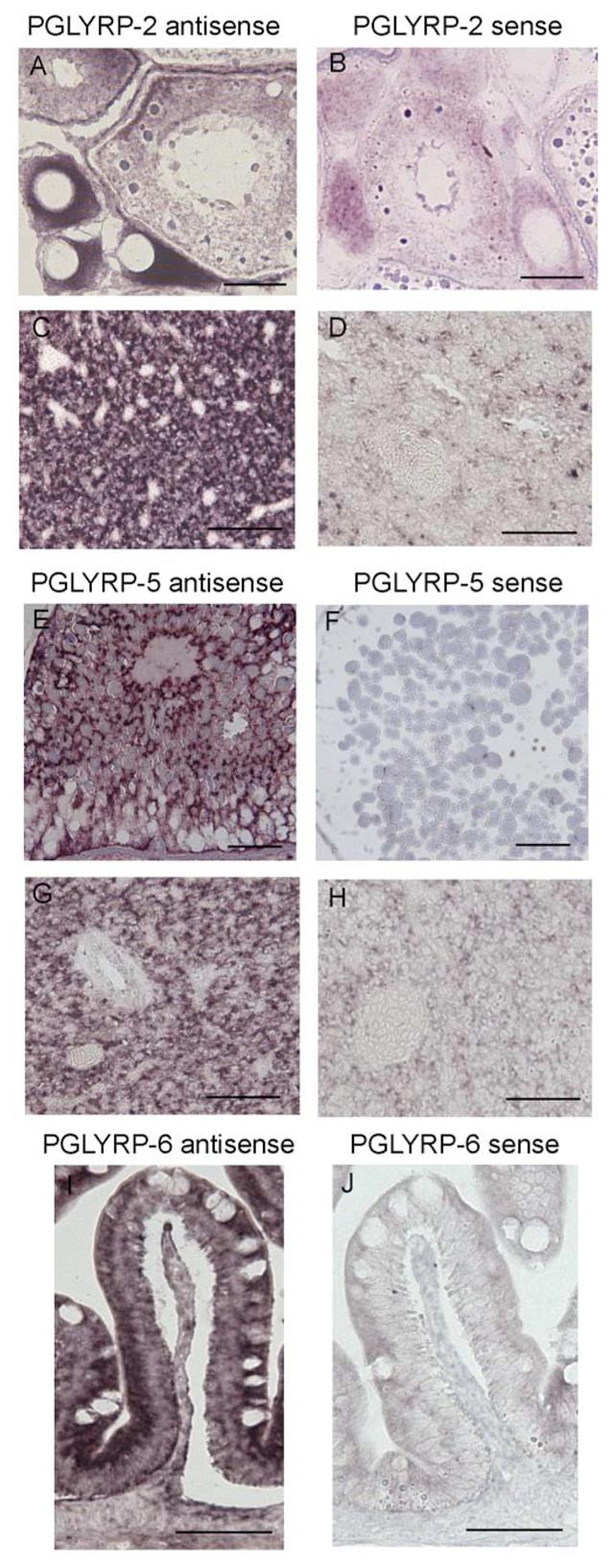
Sections of zebrafish developing oocytes (A and B) and liver (C and D) were hybridized with PGLYRP-2 antisense (A and C) or sense (B and D) RNA probes. Sections of developing oocytes (E and F) and liver (G and H) were hybridized with PGLYRP-5 antisense (E and G) or sense (F and H) probes. Sections of intestine (I and J) were hybridized with PGLYRP-6 antisense (I) or sense (J) probes. Bar = 50 μm. The results are from one of three similar experiments.
We next identified tissues that express PGLYRP proteins in sections of whole adult fish using antibodies specific to each of the zebrafish proteins. PGLYRP-2, PGLYRP-5, and PGLYRP-6 proteins were expressed in hepatocytes and in the lumen of sinusoids in the liver (Fig. 3, A-C). PGLYRP-2 and PGLYRP-5 proteins were highly expressed in the exocrine acinar cells of the pancreas, whereas PGLYRP-6 had lower levels of expression (Fig. 3, E-G). In contrast, PGLYRP-6 was highly expressed in the absorptive cells of the intestine, whereas PGLYRP-2 and PGLYRP-5 had lower levels of expression (Fig. 3, I-K) and no expression of either PGLYRP proteins in the goblet cells. PGLYRP-2 was strongly expressed in Stages III to V of the developing oocyte (Fig. 3M), whereas PGLYRP-5 had somewhat lower levels of expression (Fig. 3N) and PGLYRP-6 had the lowest levels of expression (Fig. 3O). The three PGLYRPs were detected in the gills of the zebrafish and were present in a population of epithelial cells lining the gill lamella and in the vascular lumen of the gills (Fig. 3, Q-S). PGLYRP-2, PGLYRP-5, and PGLYRP-6 were expressed in the alarm cells, but not in the mucous cells and not in the taste buds of the integumentary system (Fig. 3, U-W). PGLYRP-2, PGLYRP-5, and PGLYRP-6 were also expressed in the epithelial cells of the kidney tubules (data not shown). There was no staining with the control antibody (Fig. 6, D, H, L, P, T, and X) indicating specific detection of PGLYRP proteins in these tissues.
Figure 3. Zebrafish PGLYRP-2, PGLYRP-5, and PGLYRP-6 proteins are expressed in the liver, pancreas, intestine, developing oocytes, gills, and skin of adult zebrafish.
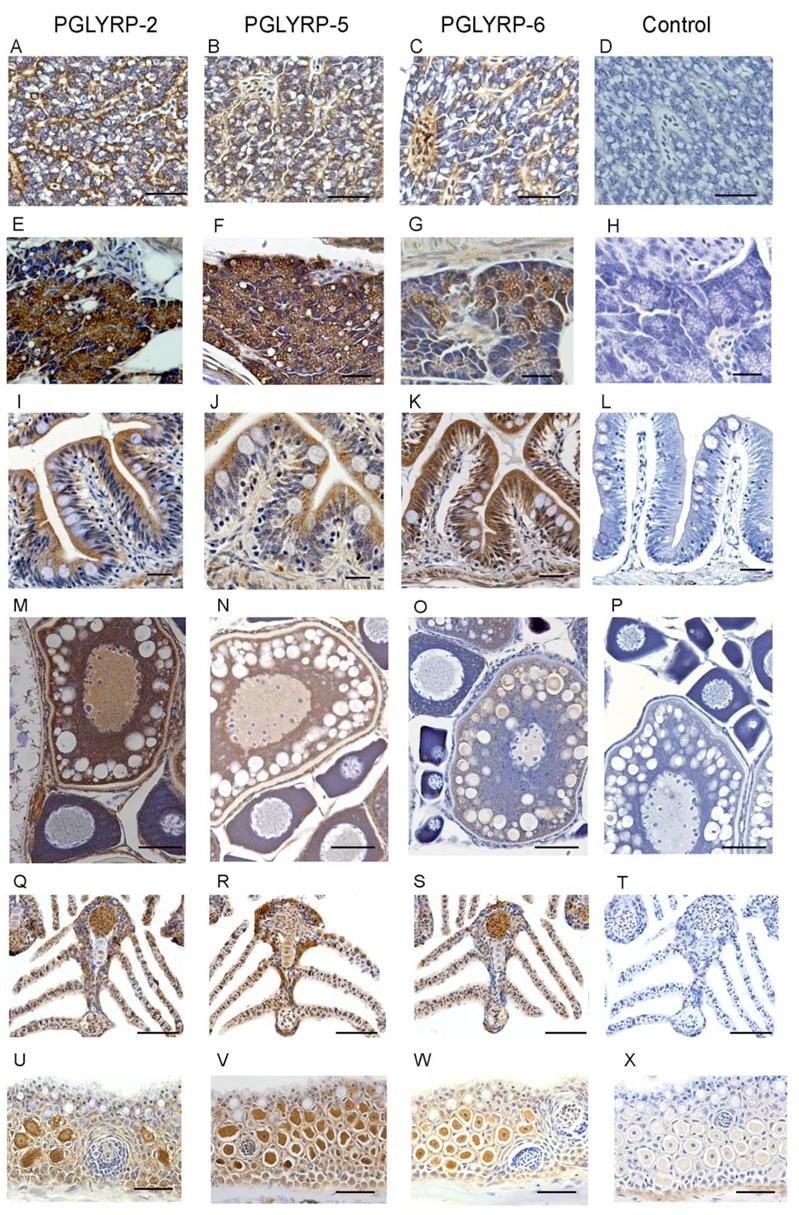
Sections of zebrafish liver (A-D), pancreas (E-H), intestine (I-L), developing oocytes (M-P), gills (Q-T), and skin (U-X) were reacted with anti-PGLYRP-2 (A, E, I, M, Q, U), anti-PGLYRP-5 (B, F, J, N, R, V), anti-PGLYRP-6 (C, G, K, O, S, W), or control (D, H, L, P, T, X) antibody, stained with peroxidase and counterstained with hematoxylin. Bar for A to L = 25 μm and M to X = 50 μm. The results are from one of three similar experiments.
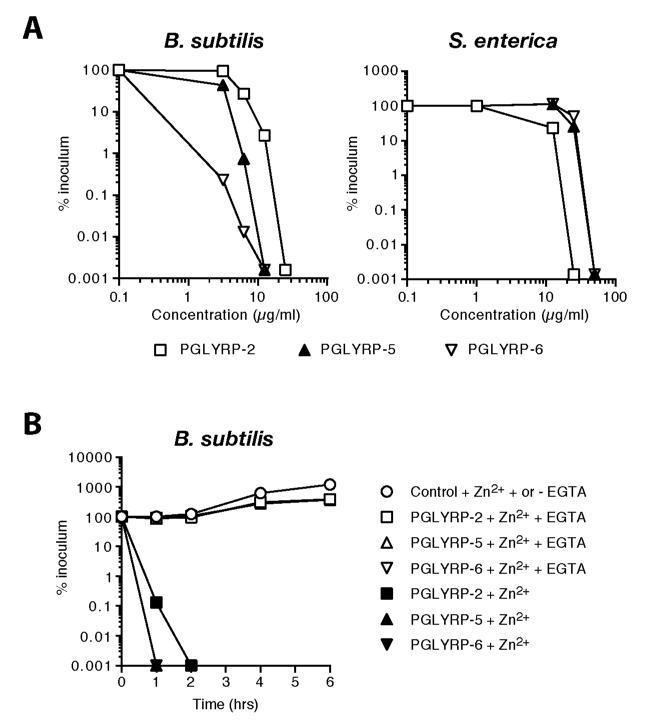
Figure 6. Zebrafish PGLYRPs are bactericidal (kill 99% of bacteria) at ∼1.3-13 μg/ml (0.02-0.2 μM) for B. subtilis and at ∼15-33 μg/ml (0.2-1.0 μM) for S. enterica, and their bactericidal activity requires Zn2+.
A, Bacteria were incubated for 1 to 2 hrs with the indicated concentrations of the indicated PGLYRPs. B, PGLYRPs or BSA as a control, purified in the presence of Zn2+, were treated or not treated with EGTA, as indicated. The numbers of bacteria were determined by colony counts, and the results are means of two to three experiments.
These data demonstrate that zebrafish PGLYRPs are selectively expressed in a wide range of tissues including those that are exposed to bacteria in the surrounding water such as gills, skin, and eggs, and also in tissues that would be exposed to bacteria during a systemic infection, such as liver.
Amidase Activity of Zebrafish PGLYRP-2, PGLYRP-5, and PGLYRP-6
Based on the presence of all four conserved amino acids needed for the amidase activity of insect and mammalian PGLYRPs and bacteriophage type-2 amidases (Fig. S2), we hypothesized that zebrafish PGLYRPs also have amidase activity for bacterial peptidoglycan. To test this hypothesis, we determined whether purified zebrafish PGLYRP proteins hydrolyze peptidoglycan.
We first used soluble polymeric S. aureus peptidoglycan labeled with biotin on the N-terminal glycine in its peptide as a substrate. All three zebrafish PGLYRPs hydrolyzed S. aureus peptidoglycan (Fig. 4A). Amidase-active PGRPs and other type-2 amidases hydrolyze the lactyl-amide bond between the MurNAc and L-Ala, which is the first amino acid in the stem peptide. To determine which bond in peptidoglycan is hydrolyzed by zebrafish PGLYRPs, we used synthetic peptidoglycan fragments (MurNAc-pentapeptides) and identified the digestion products using mass spectrometry. Furthermore, based on our sequence analysis zebrafish PGLYRP-2 and PGLYRP-6 were predicted to be DAP-specific (Supplementary Results). We tested whether zebrafish PGLYRPs hydrolyze both DAP- and Lys-containing muramyl peptides (Fig. 4B). We also used a muramyl-peptide with N-acetylated Lys, MurNAc-L-Ala-D-isoGln-LLys(NHAc)-D-Ala-D-Ala (which mimics Gly that is often bound to the γ-amino group of Lys in Gram-positive peptidoglycans), to determine whether zebrafish PGLYRPs have a preference for hydrolyzing such a substituted peptide. The results demonstrated that all three zebrafish PGLYRPs are N-acetylmuramoyl-L-alanine amidases (Fig. 4C). PGLYRP-6 hydrolyzed both Lys- (non-acetylated and N-acetylated) and DAP-containing muropeptides equally well. PGLYRP-2 hydrolyzed DAP-containing and N-acetylated-Lys-containing muropeptides equally well, but was less effective on non-acetylated Lys-containing muropeptide. PGLYRP-5 had lower activity than PGLYRP-2 and PGLYRP-6 and showed preference for DAP-containing muropeptide (Fig. 4).
Figure 4. Zebrafish PGLYRP proteins are enzymes with N-acetylmuramoyl-L-alanine amidase activity.
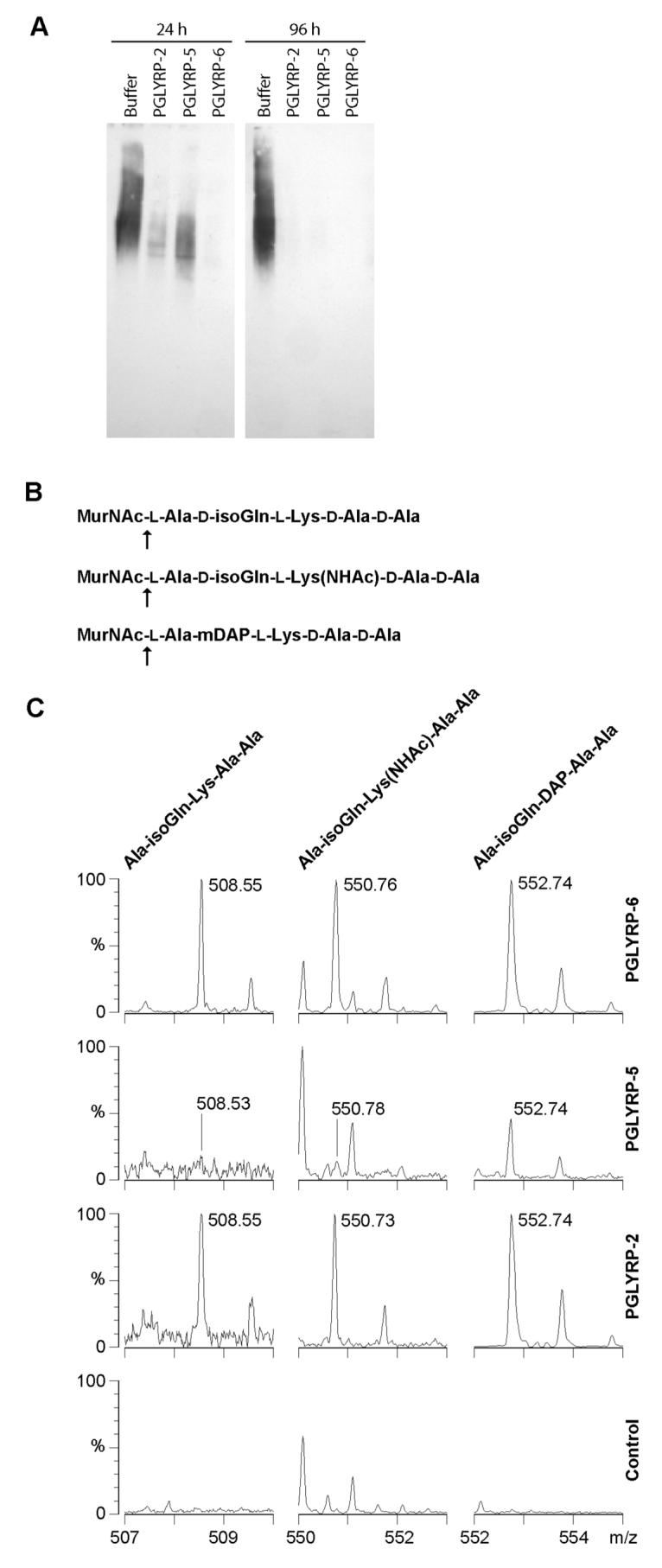
A, S. aureus soluble polymeric peptidoglycan, labeled with biotin on terminal Gly, was incubated in buffer alone or with the indicated PGLYRPs for 24 or 96 hrs. High molecular weight polymeric biotin-peptidoglycan was detected on a Western blot with streptavidin-peroxidase. Hydrolysis of the peptide from the glycan chain removes the biotin-labeled peptide. The results are from one of two similar experiments. B and C, The muropeptides indicated in (B) were incubated in buffer alone (control) or in the presence of the indicated PGLYRPs, and the hydrolysis products were identified using MALDI-TOF MS; peaks for the indicated peptides and observed masses ([M + Na]+) are shown in (C). The vertical scale in the left-hand PGLYRP-5 and PGLYRP-2 panels was expanded to reveal minor peaks. The unlabeled peaks are either matrix or buffer adduct peaks that are the same in both digested and undigested samples, or reflect natural isotopic distribution of the digestion products. K+ adducts of the same peptides were also detected (not shown). The hydrolyzed bond is indicated in (B) by an arrow. Identical hydrolysis products were generated by incubation of the same substrates with human PGLYRP-2, a proven N-acetyl-muramoyl-L-alanine amidase (Wang et al., 2003), as a positive control (not shown). The results are from one of two similar experiments.
These data prove that all three zebrafish PGRPs are type 2 amidases. The proven amidase activity is consistent with the earlier discussed prediction of amidase activity of all three zebrafish PGLYRPs based on the presence of all four conserved Zn2+-binding amino acids. Our results are consistent with the idea that peptidoglycan-binding groove specific for DAP-containing peptide can also accommodate Lys-containing peptide, which is less bulky, i.e., contains H found in Lys instead of COOH found in DAP (which is the only difference between Lys and DAP).
Bactericidal Activity of Zebrafish PGLYRP-2, PGLYRP-5, and PGLYRP-6
We have recently demonstrated that three human PGLYRPs (PGLYRP-1, PGLYRP-3, and PGLYRP-4) are bactericidal (Lu et al., 2006; Wang et al., 2007). Therefore, here we tested the hypothesis that zebrafish PGLYRPs may be also bactericidal. Moreover, because the antibacterial activity of amidase-active Drosophila PGRP-SB1 is limited to B. megaterium (Mellroth and Steiner, 2006), we tested whether zebrafish PGLYRPs have a broad or a narrow spectrum bactericidal activity.
All three zebrafish PGLYRPs were strongly bactericidal for Gram-positive bacteria: they reduced the numbers of Bacillus subtilis, Staphylococcus aureus, and Listeria monocytogenes by 4 to 5 logs in 1, 2, or 4 hrs, respectively (Fig. 5). These PGLYRPs also had similar bactericidal effect on Gram-negative bacteria, although the sensitivity of Gram-negative bacteria to various proteins somewhat varied: Salmonella enterica was highly sensitive to all three proteins, whereas Escherichia coli, Proteus vulgaris, and Pseudomonas aeruginosa were less sensitive to PGLYRP-6, and P. vulgaris and P. aeruginosa were also less sensitive to PGLYRP-5 or PGLYRP-2, respectively (Fig. 5).
Figure 5. Zebrafish PGLYRP proteins are bactericidal for several Gram-positive and Gram-negative bacteria.
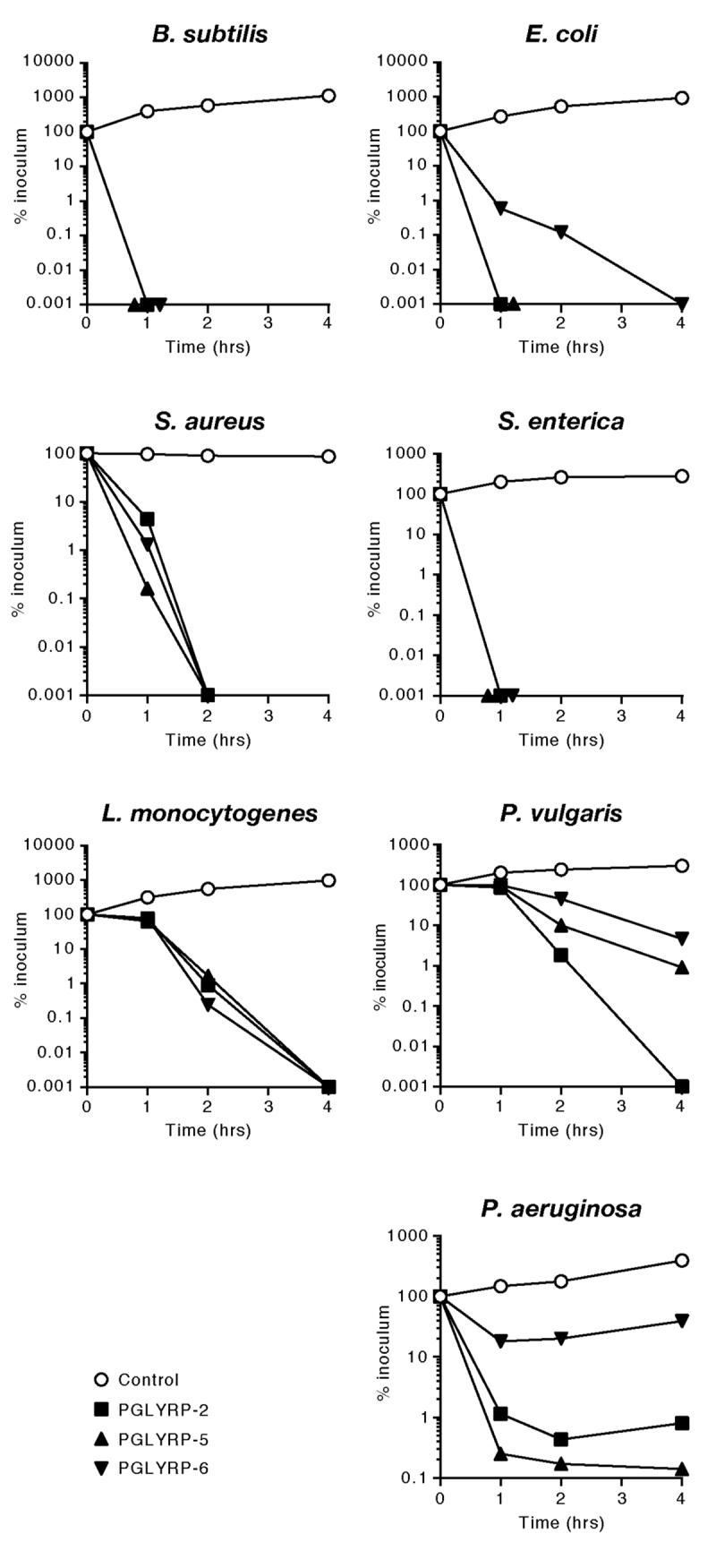
The indicated bacteria were incubated with 100 μg/ml of the indicated PGLYRPs or BSA as a control, and the numbers of bacteria were determined by colony counts. The results are means of two to three experiments.
The LD99 (the concentration that kills 99% of bacteria) of PGLYRP-2, PGLYRP-5, and PGLYRP-6 for B. subtilis were 13 μg/ml (0.2 μM), 6 μg/ml (0.2 μM), and 1.3 μg/ml (0.02 μM), respectively; and for S. enterica 15 μg/ml (0.2 μM), 33 μg/ml (1.0 μM), and 33 μg/ml (0.6 μM), respectively (Fig. 6A). Thus, Gram-positive B. subtilis is most sensitive to PGLYRP-6 and Gram-negative S. enterica is most sensitive to PGLYRP-2.
Because the bactericidal activity of human PGLYRPs required Zn2+, we then tested whether bactericidal activity of zebrafish PGLYRPs also required Zn2+. All three zebrafish PGLYRPs, similar to human PGLYRPs, required 5 μM Zn2+ for their bactericidal activity. They were not bactericidal when purified and assayed without Zn2+, and removal of Zn2+ with EGTA, which has a high affinity for Zn2+ (the log stability constant for Zn2+ is 12.9, compared to 11.0 for Ca2+) abolished the bactericidal activities of all three PGLYRPs (Fig. 6B).
These results demonstrate that zebrafish PGLYRPs, in addition to their amidase activity and unlike other vertebrate amidases, are also bactericidal. Zebrafish PGLYRPs, similar to human bactericidal PGLYRPs, have broad-spectrum Zn2+-dependent bactericidal activity, and at least one zebrafish PGLYRP (PGLYRP-6) has higher bactericidal activity than human PGLYRPs.
PGLYRP-2 and PGLYRP-5 are expressed in the developing embryos and PGLYRP-5 is required for in vivo protection against infection
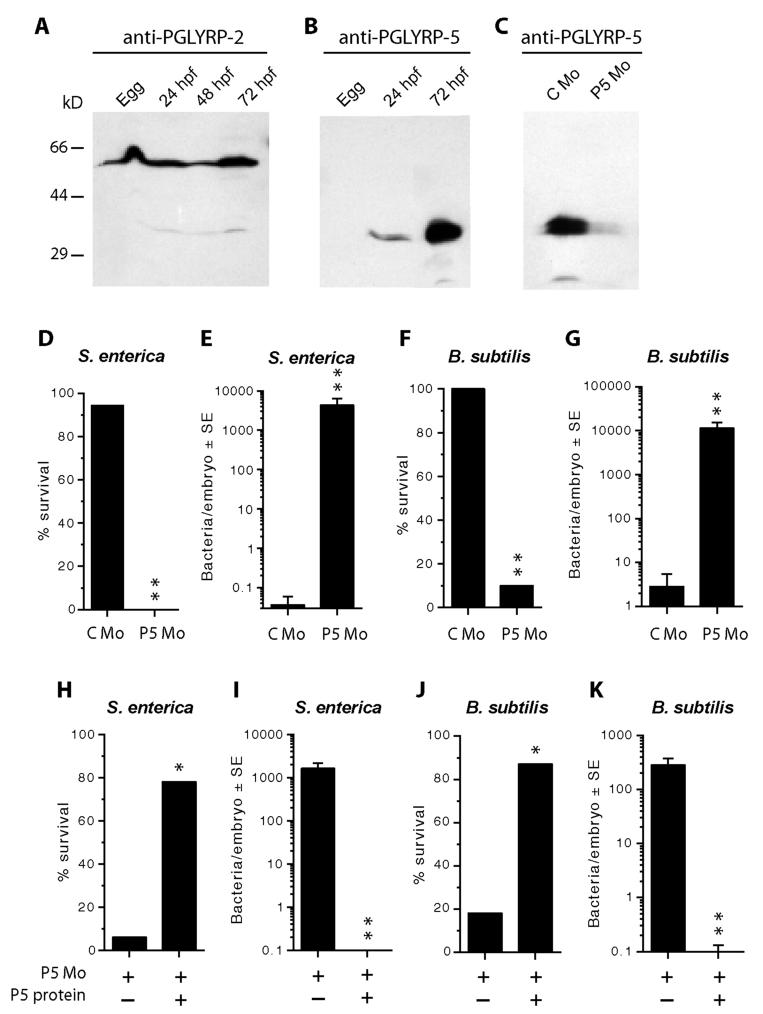
Lower vertebrates such as fish depend on innate immunity, particularly during the early stages of development as adaptive immunity matures later. In order to identify the role of PGLYRPs in the embryo during bacterial infection, we first analyzed the expression of these proteins in zebrafish embryos at different stages of development. PGLYRP-2 protein is expressed in eggs and in embryos (Fig. 7A), whereas PGLYRP-5 protein is not expressed in the eggs, weakly expressed in the 24 hpf embryo, and strongly expressed in the 72 hpf embryo (Fig. 7B). PGLYRP-6 protein could not be detected in the eggs or in the early stages of development.
Figure 7. PGLYRP-5 and PGLYRP-2 are expressed in the developing embryos and PGLYRP-5 is essential for in vivo protection against infection.
A-C, Cell lysates from eggs or from embryos at the indicated developmental stage were analyzed by Western blots using anti-PGLYRP-2 (A) or anti-PGLYRP-5 (B and C) antibody. Size markers are shown on the left. The results are from one of three similar experiments. D-K, Eggs at 2-8 cell stage were injected with 1 nl of 0.5 mM PGLYRP-5 morpholino (P5 Mo) or a control morpholino (C Mo). At the onset of circulation (∼30 hpf), embryos were injected with S. enterica (D, E) or B. subtilis (F, G) or with S. enterica (H, I) or B. subtilis (J, K) and PGLYRP-5 protein (P5 protein) or buffer. Embryos were assayed for survival at 4 hpi or lysed for determining numbers of recovered bacteria/embryo at 2-3 hpi. The data in each panel are from 2-3 experiments with the following total numbers of embryos with C Mo and P5 Mo or − and + P5 protein, respectively: D, 18 and 24; E, 35 and 43; F, 24 and 19; G, 24 and 21; H, 16 and 9; I, 16 and 12; J, 16 and 15; K, 20 and 28. The survival and the numbers of recovered bacteria were significantly different at P=0.0002 (*) or P<0.0001 (**) between PGLYRP-5 morpholino- and control morpholino-injected groups, or PGLYRP-5 protein-injected and buffer-injected groups.
We next determined the role of PGLYRP proteins in resistance to bacterial infection in the developing embryo. We initially selected PGLYRP-2 and PGLYRP-5 for these experiments, because both proteins are expressed in the developing embryos (Fig. 7A and B). Morpholinos that bind mRNA and block translation were designed for both PGLYRP-2 and PGLYRP-5. The morpholino designed to block PGLYRP-5 specifically inhibited translation of this protein and there was no inhibition with the control morpholino (Fig. 7C). PGLYRP-2 protein, however, was already synthesized in high amounts in the developing oocytes and its concentration could not be reduced by injection of morpholinos (data not shown).
Embryos, in which the expression of the endogenous PGLYRP-5 protein was inhibited by an injection of a specific morpholino, were then tested for resistance to infections with the Gram-negative (S. enterica) and the Gram-positive (B. subtilis) bacteria. Embryos injected with either a control (C Mo) or the PGLYRP-5 (P5 Mo) morpholino were subsequently challenged with S. enterica and assayed for survival at 4 hrs post-challenge. Embryos injected with control morpholino had 94% survival, whereas embryos injected with PGLYRP-5 morpholino had 0% survival (Fig. 7D, P<0.0001, control vs. PGLYRP-5 morpholino). To confirm that the increased mortality of the embryos was due to the developing infection (manifested by increasing numbers of bacteria in the embryos), the numbers of S. enterica recovered from live embryos, which were lysed at 2-3 hpi, were determined. Embryos injected with PGLYRP-5 morpholino had significantly (P<0.0001) higher numbers of bacteria than the embryos injected with a control morpholino (Fig. 7E). The latter embryos were able to eliminate virtually all injected bacteria within 4 hrs (Fig. 7E).
Similar results were obtained in infection experiments with B. subtilis. Embryos injected with a control morpholino or with PGLYRP-5 morpholino followed by B. subtilis infection had 100% and 10% survival respectively (Fig. 7F, P<0.0001, control vs. PGLYRP-5 morpholino). To determine the numbers of B. subtilis recovered, live embryos were lysed at 2-3 hpi. Embryos injected with PGLYRP-5 morpholino had significantly (P<0.0001) higher numbers of bacteria than the embryos injected with a control morpholino (Fig. 7G). The latter embryos were able to eliminate virtually all injected bacteria within 4 hrs (Fig. 7G).
To confirm the in vivo role of PGLYRP-5 in eliminating bacteria and in the survival of the infected embryos, rescue experiments were then performed. Embryos were first injected with PGLYRP-5 morpholino to suppress the production of endogenous PGLYRP-5 protein. These embryos were then injected with purified PGLYRP-5 protein or buffer alone at the time of S. enterica or B. subtilis challenge. Injection of PGLYRP-5 protein, but not buffer, significantly (P=0.0002) increased the survival of the embryos infected with S. enterica (Fig. 7H) and B. subtilis (Fig. 7J). Similarly, injection of PGLYRP-5 protein, but not buffer, enabled the embryos to kill the infecting bacteria (S. enterica, Fig. 7E; and B. subtilis, Fig. 7K, P<0.0001). Thus, these results confirm that PGLYRP-5 protein is responsible for eliminating both Salmonella and Bacillus from the embryos and for the survival of the embryos during bacterial infections.
Discussion
Our results demonstrate the essential role of innate immunity in defense against infections in fish and identify the component of their innate immune system responsible for this effect (bactericidal PGRPs). Because fish eggs and developing embryos have to survive in bacteria-containing water before adaptive immunity develops, during that time they have to rely solely on innate immunity for defense against infections and survival. Up until now this requirement was assumed, but, to the best of our knowledge, there was no experimental evidence for it. Moreover, our results demonstrate an absolute requirement for a PGRP molecule in defense and survival against bacterial infections in vertebrates.
Zebrafish PGLYRP-2, PGLYRP-5, and PGLYRP-6 proteins (which we identified and cloned) are highly conserved and have one PGRP domain, which is homologous to the PGRP domains of other invertebrate and vertebrate PGRPs. Zebrafish PGLYRPs, similar to human PGLYRPs, are selectively expressed in a wide range of tissues including the liver, intestine, pancreas, maturing oocytes, gills, and skin and are secreted in the lumen of the sinusoids in the liver and in the blood vessels of the gills. All three proteins are expressed in the developing oocytes, although there may be quantitative differences in their expression. PGLYRP-2 is also strongly expressed in the eggs and both PGLYRP-2 and PGLYRP-5 are expressed in the developing embryo. This expression of PGLYRPs in a wide range of tissues suggests an important role for this family of proteins in defense of adult zebrafish against bacterial infections. Furthermore, the expression profile of PGLYRPs would allow the fish to combat bacteria both in the surrounding water and internalized bacteria during a systemic infection. Expression of PGLYRPs in the eggs also equips the developing embryo with innate immune mechanisms to fight bacterial infections before the development of adaptive immunity.
We have demonstrated that PGLYRP-2, PGLYRP-5, and PGLYRP-6 digest polymeric peptidoglycan. Furthermore, the three proteins specifically hydrolyze the lactyl-amide bond between the MurNAc and L-Ala, which confirms that PGLYRP-2, PGLYRP-5, and PGLYRP-6 are N-acetylmuramoyl-L-alanine amidases, similar to mammalian PGLYRP-2 and Drosophila PGRP-SC1, PGRP-LB, and PGRP-SB1 (Wang et al., 2003; Gelius et al., 2003; Kim et al., 2003; Mellroth et al., 2003; Sang et al., 2005; Mellroth and Steiner, 2006).
In addition to the amidase activity, zebrafish PGLYRP-2, PGLYRP-5, and PGLYRP-6 are also bactericidal. The three proteins kill several different Gram-positive bacteria equally efficiently, but show somewhat differential killing of several Gram-negative bacteria. Some of these bacteria are human pathogens and are also pathogenic for zebrafish, producing symptoms similar to those in humans (van der Sar et al., 2003; van der Sar et al., 2004). However, zebrafish PGLYRPs are different from human bactericidal PGLYRP-1, PGLYRP-3, and PGLYRP-4, as the human proteins have no amidase activity. Human PGLYRP-2 is an amidase, however it is not known if it is also bactericidal. One insect PGRP, Drosophila PGRP-SB1, has both amidase and antibacterial activities (Mellroth and Steiner, 2006). However, the spectrum of antibacterial activity of PGRP-SB1 was very narrow and limited to only B. megaterium, unlike the broad spectrum of antibacterial activity by human (Lu et al., 2006; Wang et al., 2007) and zebrafish PGLYRPs (this study). The bactericidal activity of PGLYRP-2, PGLYRP-5, and PGLYRP-6 requires zinc, similar to human PGLYRPs (Lu et al., 2006; Wang et al., 2007).
The killing mechanism by human bactericidal PGRPs most likely targets the bacterial cell wall and does not involve peptidoglycan hydrolysis or permeabilization of the cytoplasmic membrane (Lu et al., 2006; Wang et al. 2007). Zebrafish PGRPs, unlike human PGRPs, have both amidase and bactericidal activities. Thus, their mechanism of action may be different.
We have further demonstrated that PGLYRP-5 is required in the zebrafish embryo for eliminating bacteria and for survival of the embryo during bacterial infections. We selected S. enterica and B. subtilis for these experiments, because both bacteria are highly sensitive to the bactericidal activity of zebrafish PGLYRPs; are free-living and found in water and thus infect aquatic life such as fish and turtles; and are not highly pathogenic for zebrafish and are rapidly eliminated in the normal developing embryo. The ability of the host to eliminate bacteria using innate immune mechanisms may be an important property that differentiates less pathogenic from more pathogenic bacteria. PGLYRP-5 is expressed in the developing embryo at an estimated concentration of 100 μg/ml at 24 hpf. This concentration of PGLYRP-5 protein is based on an estimated total volume of 0.3 μl/embryo (24 hpf) and the amount of endogenous PGLYRP-5 protein detected on the Western blot (30 ng per 24 hpf embryo). This concentration of PGLYRP-5 is more than sufficient to kill 1-5 × 107 bacteria/ml (100-500 bacteria/embryo) – the numbers of bacteria that were injected into the embryo. This calculation is based on our in vitro bactericidal assays, in which LD99 of PGLYRP-5 is 6 μg/ml for B. subtilis and 33 μg/ml for S. enterica at 1-5 × 106 bacteria/ml, and was verified in our rescue experiments with recombinant PGLYRP-5 injected into PGLYRP-5-depleted embryos (Fig. 7H-K).
This is the first demonstration of an essential role for a PGLYRP protein in the survival of vertebrates, which contrasts the results of two previous studies: one study showed partial requirement for PGLYRP-1 in resistance to infection in mice (Dziarski et al., 2003), and the second study concluded that PGLYRP-2 was not essential for immunity in mice (Xu et al., 2004). Our results also demonstrate the importance of innate immunity molecules in the defense against bacterial pathogens, especially during the early stages of development when there is no adaptive immunity.
In conclusion, we have cloned a family of PGLYRP proteins from an earlier vertebrate, the zebrafish, and we have identified the function of PGLYRP-2, PGLYRP-5, and PGLYRP-6 proteins. Zebrafish PGLYRP proteins are expressed in several tissues in adult zebrafish and are differentially expressed in the developing embryos. These proteins are unique compared to other vertebrate PGLYRPs, because they have both amidase activity and a broad-spectrum bactericidal activity. Furthermore, zebrafish PGLYRP-5 is essential for defense against bacterial infections in the developing embryos. Thus, our results demonstrate that in zebrafish innate immunity (and PGLYRP proteins in particular) is essential for defense against infections and survival of developing embryos.
EXPERIMENTAL PROCEDURES
Maintenance of zebrafish
Adult zebrafish, Danio rerio, were obtained from the Zebrafish International Resource Center (supported by grant #RR12546 from NIH-NCRR), University of Oregon. Zebrafish were housed and maintained in our facilities according to the established protocols described in the “The Zebrafish Book”, University of Oregon (www.zfin.org). All experiments with fish were approved by the Institutional Animal Care and Use Committee at Indiana University School of Medicine Northwest.
Cloning of zebrafish pglyrp genes
We identified four zebrafish PGLYRP genes in the GenBank databases and have cloned cDNAs for three PGLYRPs. All cloning was done by RACE reactions and by RT-PCR using total RNA isolated from adult zebrafish tissues. The cDNAs for the three PGLYRPs were initially cloned using RACE nested primers (see Table S1) based on the following EST clones: CF999082 and CK029852 (PGLYRP-2); CO92224 (PGLYRP-5); and AL922460 (PGLYRP-6). RACE reactions were done using the First-Choice RLM-RACE kit (Ambion). The 5' RACE products for the three PGLYRPs were extended at the 5' end using primers based on the GenBank predicted protein sequences XM691972 (PGLYRP-2), XM697301 (PGLYRP-5), and XM695448 (PGLYRP-6) to give full length cDNAs. The 3' RACE reactions resulted in clones with poly(A) tails. All clones were analyzed by restriction digestion and sequencing. Final sequences were analyzed for signal peptides using the SPScan program (Genetic Computer Group, Madison, WI) and for transmembrane domains using the Swiss TMpred program. Multiple sequence alignments were performed with the ClustalW program using MacVector (Genetic Computer Group, Madison, WI). The nucleotide sequences reported in this paper have been submitted to the GenBank with the accession numbers DQ447202, DQ447203 and DQ447204.
In-vitro transcription and digoxigenin labeled RNA probes
Zebrafish PGLYRP cDNAs were amplified with primers (Table S1) and subcloned into the pGEM-T vector (Promega). This vector has dual opposed promoters that allow in vitro transcription of both strands of the cloned DNA. For generating RNA probes the clones were digested with Nco I or Nde I, the fragments were purified on an agarose gel, and used for separate in vitro transcription reactions using the DIG RNA Labeling Kit (Roche). Transcription was performed with SP6 RNA polymerase and T7 RNA polymerase to generate antisense and sense RNA probes.
Expression and purification of PGLRP proteins
The coding regions for zebrafish PGLYRPs were amplified (Table S1) and subcloned into the expression vector pMT/BiP/V5-His vector (Invitrogen). Stable S2 cell lines were generated, expression of PGLYRPs was induced with 500 μM CuSO4, and proteins were purified as described before (Lu et al., 2006). All purification buffers included 40 μM ZnSO4; elution buffer also included 10% glycerol; and dialysis buffer included 10 μM ZnSO4 and 10% glycerol (Wang et al., 2007). The purity of the proteins was detected on Coomassie blue-stained blots (Fig. S3). To remove divalent cations proteins were treated with 100 μM EGTA for 10 min at room temperature and then used for bactericidal assays. To test whether zebrafish PGLYRPs are glycosylated, proteins were treated with the N-glycosidase, PNGase, and analyzed by Western blots as described (Lu et al., 2006).
RNA and Northern blots
Total RNA from eggs and 24 hours post fertilization (hpf), 48 hpf, and 72 hpf embryos or adult zebrafish skin, muscle, liver, eggs, gills, intestine, brain, liver, and eyes was purified using Trizol reagent (Invitrogen). 30 μg of RNA from each sample was analyzed by Northern blots. PGLYRP cDNA clones were amplified with primers (Table S1) and used as a probe. The 18S ribosomal clone, IMAGE ID 5604426, was used as a control. DNA was labeled with 32P using RadPrime DNA Labeling System (Invitrogen) and hybridizations were done in ExpressHyb solution at 68°C (Wang et al., 2005). All probes were highly specific, as they did not cross-hybridize with other members of the PGLYRP family (data not shown).
In-Situ Hybridization
Adult zebrafish were fixed in Bouin's Reagent for 3 hrs, rinsed in 70% ethanol, embedded in paraffin and 5 μm thick, longitudinal sections, were prepared by Transgenic Pathology Laboratory (University of California, Davis). The sections were incubated in prehybridization buffer for 4 hrs at 60°C and an overnight incubation at 55°C in hybridization buffer containing 10 ng of digoxigenin labeled RNA probe. The sections were washed at 55°C in the following series of wash solutions: 2x SSC, 2x SSC with 50% formamide, 2x SSC with 25% formamide, 1x SSC, and 0.1x SSC and then blocked with 2% normal sheep serum. The sections were incubated with sheep anti-DIG-alkaline phosphatase antibody overnight and followed by color development with nitroblue tetrazolium and 5-bromo-chloro-3-indolyl phosphate. The sections were counterstained with fast red.
Antibodies and Western Blots
Anti-PGLYRP antibodies to the following peptides: CETETHPKIKERLN for PGLYRP-2, CSVLPKLRDRLQNNE for PGLYRP-5, and CVRRNLNQDERKKLD for PGLYRP-6 were made and affinity purified (Lu et al., 2006). These Abs were specific for each PGLYRP, as demonstrated by their reactivity on Western blots with only one PGLYRP from which the peptide was derived, and no reactivity with the other PGLYRPs (Fig. S3). An Ab to an unrelated peptide, which did not react with zebrafish PGLYRPs, prepared and purified by the same method, was used as a negative control. V5 mouse monoclonal Ab was obtained from Invitrogen.
Total cell protein was prepared from eggs and embryos in SDS sample buffer (The Zebrafish Book, www.zfin.org). Lysates from 20 eggs or embryos were analyzed by SDS-gels and Western blots, using antibodies specific for zebrafish PGLYRPs.
Immunohistochemistry
Adult zebrafish were fixed in Bouin's Reagent and sectioned as described above for In-Situ Hybridization. Following standard deparaffinization, re-hydration, and quenching of endogenous peroxidase by 30 min incubation in 0.3% H2O2, the slides were incubated with 1 μg/ml of anti-PGLYRP-2, anti-PGLYRP-5, anti-PGLYRP-6 or control Ab (obtained and tested for specificity as described above) overnight, followed by biotinylated second Ab and Vectastain Elite ABC kit (Vector) with 3,3' diaminobenzidene as a substrate (this generates a brown reaction product) and counterstaining with hematoxylin (blue).
Peptidoglycan digestion and amidase assays
Soluble uncrosslinked peptidoglycan, purified by vancomycin-affinity chromatography from S. aureus Rb, was labeled with biotin on the N-terminal glycine of its interpeptide bridge, and its hydrolysis was measured as described previously (Wang et al., 2003). Peptidoglycan-biotin (500 ng) was incubated for 24 or 96 hrs at 37°C with 500 ng of recombinant PGLYRP-2, PGLYRP-5, PGLYRP-6 or buffer alone. Enzyme-digested peptidoglycan-biotin was subjected to SDS-PAGE, blotted on Immobilon-P, and high Mr peptidoglycan-biotin was detected with streptavidin-peroxidase and enhanced chemiluminescence as described previously (Wang et al., 2003).
To determine which bond in peptidoglycan is hydrolyzed by zebrafish PGLYRPs, 10 μg of synthetic peptidoglycan fragments (Kumar et al., 2005) were incubated for 72 hrs in 4 mM TRIS pH 7.6, with 60 mM NaCl, 4 μM ZnSO4, and 4% glycerol, with 2 μg zebrafish PGLYRPs, or human PGLYRP-2 as a positive control, or in buffer alone as a negative control. The samples were analyzed by MALDI-TOF mass spectrometry and digestion products were identified as described previously (Wang et al., 2003).
Antimicrobial assay
Bactericidal activity of zebrafish PGLYRP proteins was assayed as described (Lu et al., 2006; Wang et al., 2007). Briefly, PGLYRP proteins purified from S2 cell supernatants, or BSA as a control, were incubated with bacteria in a buffer containing 5 mM TRIS, pH 7.6, 150 mM NaCl, 5 μM ZnSO4, 5% glycerol, and 1% LB (50% Schaeffer's medium was used for B. subtilis instead of LB). The following bacteria were assayed for sensitivity: B. subtilis, L. monocytogenes, S. aureus (clinical isolate Rb), E. coli K12, P. vulgaris, P. aeruginosa, and S. enterica. The number of viable bacteria was determined by colony counts. All other controls were performed as described previously (Lu et al., 2006). Bactericidal activity is defined as an at least 2 log10 decrease in the number of inoculated bacteria in 6 hrs; bacteriostatic activity is defined as an inhibition of growth of bacteria, or a decrease in the number of inoculated bacteria of less than 2 log10 in 6 hrs. The results are means of 3 (mostly) or 2 experiments; the average standard errors were 15% of the mean (ranged from 8% to 24%) and are not shown, because the error bars were mostly smaller than the size of the symbols in the figures. The significance of differences was calculated using the Student's t-test, and all differences in bacterial numbers greater than 1 log10 were statistically significant at P<0.05.
Infection of zebrafish embryos
Zebrafish eggs were rinsed with 0.003% sodium hypochlorite, suspended in embryo medium and transferred into an agarose injection plate (Grabher et al., 2004). PGLYRP-5 (AAGATGAAGAAACTGTGCTGCATGG) and control (CCTCTTACCTCAGTTACAATTTATA) morpholinos (Gene Tools) were at 0.5 mM concentration. A PV830 Pneumatic PicoPump (World Precision Instrument, Sarasota, FL) was used to pressure inject 1 nl of morpholino solution into the yolk of the 2-8 cell stage developing embryos. The embryos were placed at 28°C in embryo medium and observed under the microscope to ensure normal development. In initial experiments embryos were observed for ∼60 hpf (this is ∼26 hrs longer than the infection experiments) and there were no obvious morphological differences between the PGLYRP-5 and control morpholino-injected embryos. At 28-32 hpf the embryo heart was beating and the circulation had commenced. Embryos similar in development were selected from PGLYRP-5 and control morpholino-injected groups and the embryo yolk was injected with 1 nl of an exponentially growing culture of S. enterica or B. subtilis, diluted to produce 100-500 bacteria per injection. In initial experiments the entry of the bacteria into the circulation was monitored using phenol red mixed with bacteria. The injected embryos were observed under the microscope and the sample was seen rapidly entering the circulation. The dose of bacteria injected into each embryo was verified by dispensing an identical volume onto LB plates just before and after each injection group. Subsequent culture of these plates showed the same numbers of bacteria injected into each embryo treatment group. Following bacterial injections the embryos were scored for survival after 4 hours or were individually lysed and plated on LB agar to determine the numbers of recovered bacteria at 2-3 hours. Longer time points were not used because all PGLYRP-5 morpholino-injected embryos challenged with bacteria were dead. For rescue experiments PGLYRP-5 purified protein (0.4 ng/embryo) was injected into embryos with S. enterica or B. subtilis and individual embryos were assayed as described above. We used two types of control injections: (i) control morpholinos, which had no effect on PGRP expression, embryo development, and the course of infection; and (ii) vehicle or buffer injections instead of morpholinos, bacteria, or PGRP protein, which had no harmful effects on the embryos (buffer-injected embryos developed normally or had normal course of infections if subsequently challenged with bacteria). The survival results were analyzed for significance of differences by the Chi-square test, and the numbers of recovered bacteria are presented as geometric means and the significance of differences was determined by the Mann-Whitney U-test.
Supplementary Material
Acknowledgements
We are grateful to Patrick Bankston and William Hamlett for help in interpreting the immunohistochemistry slides and David Lains, Christen Mumaw, and Nancy Mangini for advice on maintenance of zebrafish. This work was supported by USPHS Grants AI28797 and AI56395 from NIH to R.D. and D.G. and 1R01GM065248 to G-J. B.
None of the authors of this manuscript have a financial interest related to this work.
Abbreviations
- DAP
diaminopimelic acid
- hpf
hours post fertilization
- Lys
lysine
- MurNAc
N-acetylmuramic acid
- PGLYRP or PGRP
peptidoglycan recognition proteins
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bischoff V, Vignal C, Boneca IG, Michel T, Hoffmann JA, Royet J. Function of the drosophila pattern-recognition receptor PGRP-SD in the detection of Gram-positive bacteria. Nat. Immunol. 2004;5:1175–1180. doi: 10.1038/ni1123. [DOI] [PubMed] [Google Scholar]
- Bischoff V, Vignal C, Duvic B, Boneca IG, Hoffmann JA, Royet J. Down regulation of the Drosophila immune response by peptidoglycan-recognition proteins SC1 and SC2. PLoS Pathog. 2006;2:e14. doi: 10.1371/journal.ppat.0020014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe K-M, Werner T, Stoven S, Hultmark D, Anderson KV. Requirement for a peptidoglycan recognition protein (PGRP) in Relish activation and antibacterial immune responses in Drosophila. Science. 2002;296:359–362. doi: 10.1126/science.1070216. [DOI] [PubMed] [Google Scholar]
- DeVries ME, Kelvin AA, Xu L, Ran L, Robinson J, Kelvin DJ. Defining the origins and evolution of the chemokine/chemokine receptor system. J. Immunol. 2006;176:401–415. doi: 10.4049/jimmunol.176.1.401. [DOI] [PubMed] [Google Scholar]
- Dziarski R, Gupta D. Mammalian PGRPs: novel antibacterial proteins. Cell. Microb. 2006a;8:1059–1069. doi: 10.1111/j.1462-5822.2006.00726.x. [DOI] [PubMed] [Google Scholar]
- Dziarski R, Gupta D. The peptidoglycan recognition proteins (PGRPs) Genome Bio. 2006b;7:232. doi: 10.1186/gb-2006-7-8-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dziarski R, Platt KA, Gelius E, Steiner H, Gupta D. Defect in neutrophil killing and increased susceptibility to infection with nonpathogenic gram-positive bacteria in peptidoglycan recognition protein-S (PGRP-S)-deficient mice. Blood. 2003;102:689–697. doi: 10.1182/blood-2002-12-3853. [DOI] [PubMed] [Google Scholar]
- Garver LS, Wu J, Wu LP. The peptidoglycan recognition protein PGRPSC1a is essential for Toll signaling and phagocytosis of Staphylococcus aureus in Drosophila. Proc. Natl. Acad. Sci. U S A. 2006;103:660–665. doi: 10.1073/pnas.0506182103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelius E, Persson C, Karlsson J, Steiner H. A mammalian peptidoglycan recognition protein with N-acetylmuramoyl-L-alanine amidase activity. Biochem. Biophys. Res. Commun. 2003;306:988–994. doi: 10.1016/s0006-291x(03)01096-9. [DOI] [PubMed] [Google Scholar]
- Gottar M, Gobert V, Michel T, Belvin M, Duyk G, Hoffmann JA, Ferrandon D, Royet J. The Drosophila immune response against Gram-negative bacteria is mediated by a peptidoglycan recognition protein. Nature. 2002;416:640–644. doi: 10.1038/nature734. [DOI] [PubMed] [Google Scholar]
- Grabher C, Joly J-S, Wittbrodt J. Highly efficient zebrafish transgenesis mediated by the meganuclease I-SceI. Methods Cell Biol. 2004;77:381–401. doi: 10.1016/s0091-679x(04)77021-1. [DOI] [PubMed] [Google Scholar]
- Kaneko T, Silverman N. Bacterial recognition and signalling by the Drosophila IMD pathway. Cell. Microbiol. 2005;7:461–469. doi: 10.1111/j.1462-5822.2005.00504.x. [DOI] [PubMed] [Google Scholar]
- Kang D, Liu G, Lundstrom A, Gelius E, Steiner H. A peptidoglycan recognition protein in innate immunity conserved from insects to humans. Proc. Natl. Acad. Sci. U S A. 1998;95:10078–10082. doi: 10.1073/pnas.95.17.10078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M-S, Byun M, Oh B-H. Crystal structure of peptidoglycan recognition protein LB from Drosophila melanogaster. Nat. Immunol. 2003;4:787–793. doi: 10.1038/ni952. [DOI] [PubMed] [Google Scholar]
- Kumar S, Roychowdhury A, Ember B, Wang Q, Guan R, Mariuzza RA, Boons GJ. Selective recognition of synthetic lysine and meso diaminopimelic acid-type peptidoglycan fragments by human peptidoglycan recognition protein-Ialpha and -S. J. Biol. Chem. 2005;280:37005–37012. doi: 10.1074/jbc.M506385200. [DOI] [PubMed] [Google Scholar]
- Lam SH, Chua HL, Gong Z, Lam TJ, Sin YM. Development and maturation of the immune system in zebrafish, Danio rerio: a gene expression profiling, in situ hybridization and immunological study. Dev. Comp. Immunol. 2004;28:9–28. doi: 10.1016/s0145-305x(03)00103-4. [DOI] [PubMed] [Google Scholar]
- Lemaitre B, Kromer-Metzger E, Michaut L, Nicolas E, Meister M, Georgel P, Reichhart JM, Hoffmann JA. A recessive mutation, immune deficiency (imd), defines two distinct control pathways in the Drosophila host defense. Proc. Natl. Acad. Sci. U S A. 1995;92:9465–9469. doi: 10.1073/pnas.92.21.9465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemaitre B, Nicolas E, Michaut L, Reichhart JM, Hoffmann JA. The dorsoventral regulatory gene cassette spatzle/Toll/cactus controls the potent antifungal response in Drosophila adults. Cell. 1996;86:973–983. doi: 10.1016/s0092-8674(00)80172-5. [DOI] [PubMed] [Google Scholar]
- Leulier F, Parquet C, Pili-Floury S, Ryu J-H, Caroff M, Lee W-J, Mengin-Lecreulx D, Lemaitre B. The Drosophila immune system detects bacteria through specific peptidoglycan recognition. Nat. Immunol. 2003;4:478–484. doi: 10.1038/ni922. [DOI] [PubMed] [Google Scholar]
- Li X, Wang S, Wang H, Gupta D. Differential expression of peptidoglycan recognition protein 2 in the skin and liver requires different transcription factors. J. Biol. Chem. 2006;281:20738–20748. doi: 10.1074/jbc.M601017200. [DOI] [PubMed] [Google Scholar]
- Lim J-H, Kim M-S, Kim H-E, Yano T, Oshima Y, Aggarwal K, Goldman WE, Silverman N, Kurata S, Oh B-H. Structural basis for preferential recognition of diaminopimelic acid-type peptidoglycan by a subset of peptidoglycan recognition proteins. J. Biol. Chem. 2006;281:8286–8295. doi: 10.1074/jbc.M513030200. [DOI] [PubMed] [Google Scholar]
- Liu C, Xu Z, Gupta D, Dziarski R. Peptidoglycan recognition proteins: a novel family of four human innate immunity pattern recognition molecules. J. Biol. Chem. 2001;276:34686–34694. doi: 10.1074/jbc.M105566200. [DOI] [PubMed] [Google Scholar]
- Lu X, Wang M, Qi J, Wang H, Li X, Gupta D, Dziarski R. Peptidoglycan recognition proteins are a new class of human bactericidal proteins. J. Biol. Chem. 2006;281:5895–5907. doi: 10.1074/jbc.M511631200. [DOI] [PubMed] [Google Scholar]
- Mellroth P, Karlsson J, Steiner H. A scavenger function for a Drosophila peptidoglycan recognition protein. J. Biol. Chem. 2003;278:7059–7064. doi: 10.1074/jbc.M208900200. [DOI] [PubMed] [Google Scholar]
- Mellroth P, Steiner H. PGRP-SB1: An N-acetylmuramoyl l-alanine amidase with antibacterial activity. Biochem. Biophys. Res. Comm. 2006;350:994–999. doi: 10.1016/j.bbrc.2006.09.139. [DOI] [PubMed] [Google Scholar]
- Michel T, Reichhart JM, Hoffmann JA, Royet J. Drosophila Toll is activated by Gram-positive bacteria through a circulating peptidoglycan recognition protein. Nature. 2001;414:756–759. doi: 10.1038/414756a. [DOI] [PubMed] [Google Scholar]
- Park JW, Je B-R, Piao S, Inamura S, Fujimoto Y, Fukase K, Kusumoto S, Soderhall K, Ha N-C, Lee BL. A synthetic peptidoglycan fragment as a competitive inhibitor of the melanization cascade. J. Biol. Chem. 2006;281:7747–7755. doi: 10.1074/jbc.M510058200. [DOI] [PubMed] [Google Scholar]
- Ramet M, Manfruelli P, Pearson A, Mathey-Prevot B, Ezekowitz RAB. Functional genomic analysis of phagocytosis and identification of a Drosophila receptor for E. coli. Nature. 2002;416:644–648. doi: 10.1038/nature735. [DOI] [PubMed] [Google Scholar]
- Sang Y, Ramanathan B, Ross CR, Blecha F. Gene silencing and overexpression of porcine peptidoglycan recognition protein long isoforms: involvement in beta-defensin-1 expression. Infect. Immun. 2005;73:7133–7141. doi: 10.1128/IAI.73.11.7133-7141.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swaminathan CP, Brown PH, Roychowdhury A, Wang Q, Guan R, Silverman N, Goldman WE, Boons G-J, Mariuzza RA. Dual strategies for peptidoglycan discrimination by peptidoglycan recognition proteins (PGRPs) Proc. Natl. Acad. Sci. U S A. 2006;103:684–689. doi: 10.1073/pnas.0507656103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trede NS, Langenau DM, Traver D, Look AT, Zon LI. The use of zebrafish to understand immunity. Immunity. 2004;20:367–379. doi: 10.1016/s1074-7613(04)00084-6. [DOI] [PubMed] [Google Scholar]
- Trede NS, Zapata A, Zon LI. Fishing for lymphoid genes. Trends Immunol. 2001;22:302–307. doi: 10.1016/s1471-4906(01)01939-1. [DOI] [PubMed] [Google Scholar]
- Tydell CC, Yount N, Tran D, Yuan J, Selsted ME. Isolation, characterization, and antimicrobial properties of bovine oligosaccharide-binding protein. A microbicidal granule protein of eosinophils and neutrophils. J. Biol. Chem. 2002;277:19658–19664. doi: 10.1074/jbc.M200659200. [DOI] [PubMed] [Google Scholar]
- van der Sar AM, Appelmelk BJ, Vandenbroucke-Grauls CMJE, Bitter W. A star with stripes: zebrafish as an infection model. Trends Microbiol. 2004;12:451–457. doi: 10.1016/j.tim.2004.08.001. [DOI] [PubMed] [Google Scholar]
- van der Sar AM, Musters RJP, van Eeden FJM, Appelmelk BJ, Vandenbroucke-Grauls CMJE, Bitter W. Zebrafish embryos as a model host for the real time analysis of Salmonella typhimurium infections. Cell. Microbiol. 2003;5:601–611. doi: 10.1046/j.1462-5822.2003.00303.x. [DOI] [PubMed] [Google Scholar]
- Wang H, Gupta D, Li X, Dziarski R. Peptidoglycan recognition protein 2 (N-acetylmuramoyl-L-Ala amidase) is induced in keratinocytes by bacteria through the p38 kinase pathway. Infect. Immun. 2005;73:7216–7225. doi: 10.1128/IAI.73.11.7216-7225.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Liu L-H, Wang S, Li X, Lu X, Gupta D, Dziarski R. Human Peptidoglycan Recognition Proteins Require Zinc to Kill Both Gram-Positive and Gram-Negative Bacteria and Are Synergistic with Antibacterial Peptides. J. Immunol. 2007;178:3116–3125. doi: 10.4049/jimmunol.178.5.3116. [DOI] [PubMed] [Google Scholar]
- Wang Z-M, Li X, Cocklin RR, Wang M, Wang M, Fukase K, Inamura S, Kusumoto S, Gupta D, Dziarski R. Human peptidoglycan recognition protein-L is an N-acetylmuramoyl-L-alanine amidase. J. Biol. Chem. 2003;278:49044–49052. doi: 10.1074/jbc.M307758200. [DOI] [PubMed] [Google Scholar]
- Werner T, Liu G, Kang D, Ekengren S, Steiner H, Hultmark D. A family of peptidoglycan recognition proteins in the fruit fly Drosophila melanogaster. Proc. Natl. Acad. Sci. USA. 2000;97:13772–13777. doi: 10.1073/pnas.97.25.13772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M, Wang Z, Locksley RM. Innate immune responses in peptidoglycan recognition protein L-deficient mice. Mol. Cell. Biol. 2004;24:7949–7957. doi: 10.1128/MCB.24.18.7949-7957.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoder JA, Nielsen ME, Amemiya CT, Litman GW. Zebrafish as an immunological model system. Microbes Infect. 2002;4:1469–1478. doi: 10.1016/s1286-4579(02)00029-1. [DOI] [PubMed] [Google Scholar]
- Yoshida H, Kinoshita K, Ashida M. Purification of a peptidoglycan recognition protein from hemolymph of the silkworm, Bombyx mori. J. Biol. Chem. 1996;271:13854–13860. doi: 10.1074/jbc.271.23.13854. [DOI] [PubMed] [Google Scholar]
- Zhang Y, van der Fits L, Voerman JS, Melief M-J, Laman JD, Wang M, Wang H, Wang M, Li X, Walls CD, Gupta D, Dziarski R. Identification of serum N-acetylmuramoyl-l-alanine amidase as liver peptidoglycan recognition protein 2. Biochem. Biophys. Acta. 2005;1752:34–46. doi: 10.1016/j.bbapap.2005.07.001. [DOI] [PubMed] [Google Scholar]
- Zaidman-Remy A, Herve M, Poidevin M, Pili-Floury S, Kim MS, Blanot D, Oh BH, Ueda R, Mengin-Lecreulx D, Lemaitre B. The Drosophila amidase PGRP-LB modulates the immune response to bacterial infection. Immunity. 2006;24:463–473. doi: 10.1016/j.immuni.2006.02.012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.