Summary
In the ClpXP proteolytic machine, ClpX uses the energy of ATP hydrolysis to unfold protein substrates and translocate them through a central pore and into the degradation chamber of ClpP. Here, we demonstrate a bipartite system of ClpX-ClpP interactions that serves multiple functional roles. High-affinity contacts between six loops near the periphery of the hexameric ClpX ring and a ClpP ring establish correct positioning and increase degradation activity but are insensitive to nucleotide state. These static peripheral interactions maintain a stable ClpXP complex, while other parts of this machine change conformation hundreds of times per minute. By contrast, relatively weak axial contacts between loops at the bottom of the ClpX central channel and N-terminal loops of ClpP vary dynamically with the nucleotide state of individual ClpX subunits, control ATP-hydrolysis rates, and facilitate efficient protein unfolding. Thus, discrete static and dynamic interactions mediate binding and communication between ClpX and ClpP.
Introduction
Collaboration between AAA+ ATPases and compartmental peptidases is essential for energy-dependent protein degradation, which is carried out by large multi-subunit complexes in all organisms (for review, see Gottesman et al., 1997; Neuwald et al., 1999; Sauer et al., 2004). The AAA+ ATPases in these proteolytic complexes recognize, unfold, and translocate protein substrates into an internal degradation chamber of the peptidase. The eukaryotic 26S proteasome and the bacterial ClpXP, ClpAP, HslUV, Lon, and FtsH proteases are examples of ATP-dependent degradation machines.
Distinct proteins often perform the ATPase and peptidase functions of ATP-dependent proteases. In ClpXP, ClpAP, and HslUV, for example, protein degradation occurs in the ClpP or HslV enzymes, whereas protein recognition, unfolding, and translocation is carried out by hexameric rings of the ClpX, ClpA, or HslU ATPases. Similarly, the 26S proteasome consists of the 20S peptidase and a 19S complex that contains multiple ATPases. In the peptidases, the active sites are sequestered in an aqueous chamber formed by stacks of heptameric or hexameric rings (Loewe et al., 1995; Groll et al., 1997; Bochtler et al., 1997; Wang et al., 1997). Coaxial stacking of the ATPase and peptidase rings creates a continuous central channel, allowing protein substrates in denatured and extended conformations to be translocated through a narrow pore and into the peptidase chamber (Beuron et al., 1998; Bochtler et al., 2000; Sousa et al., 2000; Ortega et al., 2000; Ortega et al., 2002). A key question is how the ATPase and peptidase components of these proteolytic machines coordinate activities during degradation, which typically requires hundreds of repetitive mechanical cycles driven by chemical cycles of ATP binding, hydrolysis, and nucleotide release (for review, see Sauer et al., 2004).
We have been studying Escherichia coli ClpXP as a paradigm of an ATP-dependent protease. Electron-microscopy studies show that a ClpX hexamer contacts a heptameric ring of ClpP14 (Grimaud et al., 1998). Formation of this complex is essential for protein degradation and has several additional consequences. For example, ClpXP is more active than ClpX in disassembly of macromolecular complexes, ClpX activates ClpP peptidase activity, ClpP represses ClpX ATPase activity, and ClpX-ClpP affinity correlates with rates of substrate degradation and the state of the ClpP active sites (Jones et al., 1998; Grimaud et al., 1998; Kim et al., 2001; Joshi et al., 2004). Currently, the mechanisms that mediate ClpX-ClpP communication are largely unknown.
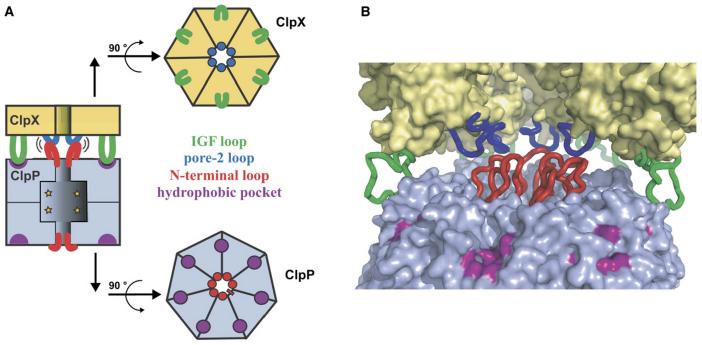
Some determinants of ClpX-ClpP binding are established. For example, IGF tripeptides, present in loops that project from the ClpX hexamer, are required for ClpP recognition (Kim et al. 2001; Singh et al., 2001). Modeling and mutagenesis based on crystal structures suggest that these IGF sequences dock in pockets located near the outer edge of the ClpP ring (Fig. 1; Wang et al., 1997; Kim et al. 2001; Kim and Kim, 2003; Bewley et al., 2006). It has been proposed that only a subset of the six IGF loops in ClpX contact ClpP, with nucleotide-dependent changes in these interactions coordinating functional communication (Joshi et al., 2004). The N-terminal region of ClpP is also important for complex formation (Kang et al., 2004; Gribun et al., 2005; Bewley et al., 2006). These ClpP residues surround the entry pore and, in one of two observed conformations, form a loop that protrudes towards the central channel of ClpX (Fig. 1; Bewley et al., 2006). However, the ClpX sites contacted by these N-terminal ClpP loops have not been identified.
Figure 1. Model for ClpX-ClpP interactions.
(A) Cartoons showing stacking of ClpX hexamer and ClpP heptamer with interactions between ClpX IGF loops (green) and hydrophobic ClpP pockets (purple) and between ClpX pore-2 loops (blue) and N-terminal ClpP loops (red), which are shown with pseudo 6-fold symmetry (Bewley et al., 2006).
(B) Interaction model based on the ClpP structure (Bewley et al., 2006) and a ClpX model based on the subunit structure of H. pylori ClpX (Kim and Kim, 2003). The ClpX hexamer and pore-2 loops were modeled from the HslU structure (Bochtler et al., 2000). Two ClpX subunits were removed to improve the view of the pore-2 loops. Color scheme as in panel A.
Here, we probe the function of specific ClpX structural elements and the nucleotide states of subunits in supporting ClpP binding and communication. Using covalently linked ClpX variants to allow alterations of individual subunits (Martin et al., 2005), we find that each of the six IGF loops in a ClpX hexamer is required for strong ClpP binding and full degradation activity but in a static or nucleotide-independent fashion. We also identify important interactions between loops at the bottom of the central channel of ClpX (pore-2 loops) and the N-terminal loops of ClpP (Fig. 1). These axial ClpX-ClpP interactions stabilize ClpXP, depend on the highly dynamic nucleotide states of individual ClpX subunits, control the rate of ATP hydrolysis, and assist in unfolding of native protein substrates.
Results
Nomenclature and interaction assays
Many of our experiments probe ClpP interactions with ClpX hexamers composed of covalently linked subunits. Single-chain constructs of full-length ClpX are insoluble but constructs with subunits lacking the N-domain (ClpX-ΔN) are well behaved and fully active in degradation of substrates with ssrA degradation tags (Martin et al., 2005). In these linked ClpX variants, W designates a wild-type ClpX-ΔN subunit; WW, WWW, and WWWWWW indicate two, three, and six linked “subunits”, respectively; and WW/WW/WW and WWW/WWW refer to pseudo-hexamers formed by noncovalent association of linked dimers or trimers.
ClpXP complex formation was tested by qualitative pull-downs and three assays to determine apparent binding affinities: (i) monitoring ClpX ATP hydrolysis as a function of ClpP concentration (Kim et al., 2001, Joshi et al., 2004); (ii) titrating ClpP against ClpX and assaying degradation of saturating concentrations of GFP-ssrA; and (iii) measuring ClpP cleavage of a decapeptide as a function of ClpX concentration. In each assay, the ClpP-binding properties of a WWW/WWW or WWWWWW hexamer of ClpX were similar to those of wild-type ClpX (not shown), demonstrating that covalent linkage of subunits or deletion of the N-domain does not perturb ClpP binding. For WWW/WWW, the ClpP affinities measured by the ATPase, GFP-ssrA degradation, and peptidase assays were 50 nM, 70 nM, and 170 nM, respectively (Table 1). The slightly weaker interaction measured by the peptidase assay probably results from using ATPγS rather than ATP and/or using excess ClpX, which allows two ClpX hexamers to bind one ClpP tetradecamer (Grimaud et al., 1998).
Table 1.
Effects of ClpX mutations on ATP-hydrolysis and ClpP interactions.
| ATP hydrolysis (min-1 enz-1) | apparent ClpP affinity (nM) | ||||
|---|---|---|---|---|---|
| ClpX variant | basal ratea | % change with ClpP | ATPase assay | protease assay | peptidase assay |
| WWW/WWW | 113 | -49 | 50 | 70 | 170 |
| WWWWWW | 107 | -47 | / | 81 | / |
| WWWWWWL | 112 | -30 | / | 2800 | / |
| WWWWWLWL | 135 | n.b. | n.b. | n.b. | |
| WWLW/WWLW | 141 | n.b. | n.b. | n.b. | |
| WE/WE/WE | 118 | -38 | / | / | / |
| EWL/EWL/EWL | 131 | n.b. | n.b. | n.b. | |
| WEL/WEL/WEL | 130 | n.b. | n.b. | n.b. | |
| WWWWWR | 209 | -41 | 40 | / | / |
| WWWWRR | 363 | -44 | 53 | / | / |
| RWW/RWW | 260 | -53 | 55 | 75 | / |
| WWR/WWR | 245 | -51 | / | / | / |
| WWWRRR | 390 | -24 | 160 | / | / |
| WR/WR/WR | 135 | -36 | 170 | / | / |
| RWWRRW | 377 | -34 | 150 | / | / |
| WRR/WRR | 100 | n.b. | n.b. | ||
| WRRRRR | 22 | n.b. | n.b. | ||
| ClpX-ΔN | 105 | -48 | 50 | / | / |
| ClpX-ΔN Δpore-2 | 550 | +5 | ‡ | ‡ | 2300 |
| ClpX-ΔN R191Q | 210 | -44 | 40 | / | / |
| ClpX-ΔN D194N | 170 | -12 | 210 | / | / |
| ClpX-ΔN I198A | 125 | -42 | 82 | / | / |
| ClpX-ΔN R200Q | 317 | -20 | 330 | / | 660 |
| ClpX-ΔN D201N | 710 | +3 | ‡ | ‡ | 2600 |
| RWE/RWE | 380 | -63 | 63 | / | / |
| RWEDN/RWEDN | 697 | -29 | 640 | / | / |
| RWDNE/RWDNE | 1090 | -30 | 1400 | / | 2900 |
| RDNWE/RDNWE | 928 | -40 | 430 | / | / |
| RDNWEDN/RDNWEDN | 940 | +5 | ‡ | / | 2900 |
| RWDNEDN/RWDNEDN | 929 | +17 | / | / | 2400 |
| values below for binding of ClpX variants to ClpP RA12 | |||||
| WWW/WWW | 113 | -30 | / | 320 | 930 |
| ClpX-ΔN D201N | / | ‡ | 1900 | ||
| RWDNE/RWDNE | / | / | 2400 | ||
| WWWWWWL | / | 9500 | / | ||
ATPase rates were determined in the absence of ClpP and protein substrate, unlike the values reported by Martin et al. (2005)
value not determined
no binding detected in pull-down assays or in ATPase or protease assays
assay precluded because mutant ClpXP complex did not degrade GFP-ssrA or showed a small change in ATPase activity
D201N.
Errors were estimated to be ±5% for ATPase rates and ±20% for apparent affinities based upon replicate measurements for WWW/WWW (± stdev for n ≥ 3). All ClpX variants in this table lack the N-terminal domain.
Six IGF loops are required for strong ClpP binding and efficient degradation
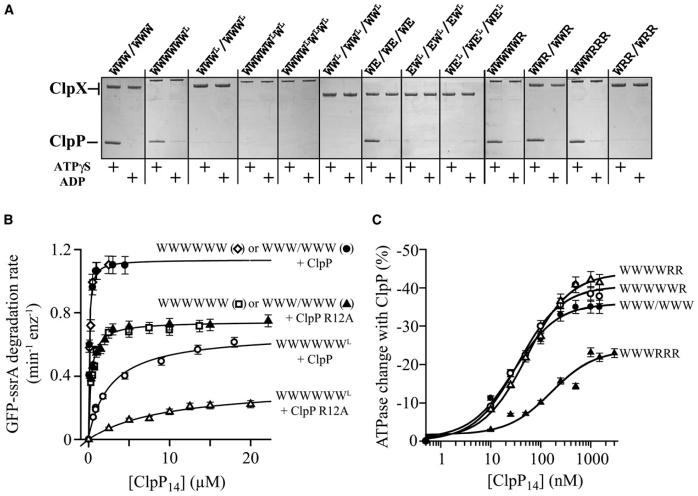
To determine how many of the six IGF loops in a ClpX hexamer are required for ClpP binding, we constructed and purified covalent ClpX hexamers containing mixtures of normal W subunits and WL subunits, where the L superscript (for loopless) signifies replacing residues 264-278 of the IGF loop with a shorter GSGSG sequence. A variant missing one IGF loop (WWWWWWL) bound ClpP weakly in the pull-down assay (Fig. 2A) and had an apparent ClpP affinity reduced about 50-fold (Kapp = 2.8 μM; Fig. 2B). Variants missing two or more IGF loops showed no detectable binding to ClpP in pull-down assays (Fig. 2A) or in GFP-ssrA degradation assays (not shown), irrespective of the positions of the mutant subunits in the hexamer. We conclude that all six IGF loops in the ClpX hexamer contribute to ClpP binding.
Figure 2. Role of ClpX IGF loops and nucleotide state in ClpP binding.
(A) SDS PAGE of pull-down assays containing ClpP and His6-tagged single-chain ClpX or variants with subunits missing the IGF loop (WL, EL) or constrained to an ATP-free state (R). Binding observed with ADP is non-specific.
(B) Deletion of one ClpX IGF loop reduced affinity for ClpP and the ClpP R12A mutant. Binding was assayed by changes in the initial rate of GFP-ssrA degradation. Data in this panel and panel C were fit to the Michaelis-Menten equation; Table 1 lists fitted Kapp values. Bars represent the error of linear fits of kinetic data used to determine degradation rates.
(C) ClpP affinity of ClpX with different numbers of subunits constrained to the ATP-free state (R) was measured by changes in ATPase activity. Bars represent the error of linear fits of kinetic data used to determine ATPase rates.
At saturating ClpP, the rate of WWWWWWL-mediated degradation of GFP-ssrA was only about half of the WWWWWW or WWW/WWW values (Fig. 2B). Several lines of evidence indicate that this difference does not reflect a substrate-recognition or protein-unfolding defect. WWWWWWL and WWWWWW had the same KM for ClpP degradation of ssrA-tagged substrates and, as reported for loopless ClpX (Joshi et al., 2004), mediated the same rate of substrate unfolding/translocation in the absence of ClpP (not shown). Moreover, WWWWWWL/ClpP also degraded an unfolded model substrate, CM-titin-ssrA, at roughly 60% of the WWWWWW/ClpP rate. The presence of a GroEL mutant that traps unfolded GFP-ssrA (Weber-Ban et al., 1999) made no significant difference in the apparent rate of degradation by WWWWWWL/ClpP (not shown), showing that the denatured polypeptide emerging from the pore of the WWWWWWL ClpX ring is almost always threaded directly into ClpP. Together, these results suggest that removing a single IGF loop from the ClpX hexamer decreases the rate of protein degradation by slowing protein translocation into ClpP.
We compared the consequences of the single IGF-loop deletion in ClpX with those caused by a mutation in the N-terminal loop of ClpP (R12A), which weakens but does not prevent ClpX binding (Bewley et al., 2006). Complexes of R12A ClpP with WWWWWW or WWWWWWL had lower maximal rates of GFP-ssrA degradation than complexes of wild-type ClpP with these ClpX variants (Fig. 2B). The effects of the IGF-loop and R12A mutations on degradation activity under saturating conditions were approximately additive, as were the energetic effects of these mutations on ClpX-ClpP binding (Fig. 2B; Table 1). Thus, interactions mediated by the ClpX IGF-loops and ClpP N-terminal loops independently influence substrate degradation rates and ClpX-ClpP binding affinity.
IGF interactions and nucleotide state
ClpX saturated with ATP has two empty nucleotide sites, and subunits in the hexamer can be ATP bound, ADP bound, or nucleotide free depending upon the stage in ATP hydrolysis (Hersch et al., 2005). Because ClpX subunits cycle through different nucleotide-dependent conformations, our finding that six IGF loops are required for strong ClpP binding does not rule out the possibility that only the IGF loops of ClpX subunits in specific states contact ClpP at any given time (Joshi et al., 2004). Mutations can prevent ATP hydrolysis and limit the nucleotide states accessible to ClpX subunits. ATP-bound E185Q (E) subunits are trapped in an ATP-state conformation, whereas R370K (R) subunits behave like ATP-free subunits (Joshi et al., 2004, Hersch et al., 2005). A single-chain variant with two R, two W, and two E subunits (RWE/RWE) is active in ClpP-mediated degradation, demonstrating that ClpX can function with a few subunits capable of hydrolyzing ATP and the rest trapped in fixed nucleotide states (Martin et al., 2005).
We asked if a subset of IGF loops in ClpX subunits constrained to specific nucleotide states or neighboring such subunits supported ClpP binding. Linked ClpX variants containing 3-4 IGF loops in wild-type (W), ATP-state (E), or ATP-free (R) subunits were constructed with IGF-loop deletions in other subunits (WL, EL, or RL). Variants with IGF loops in three W subunits adjacent to loopless E subunits (WEL/WEL/WEL) or in three E subunits (EWL/EWL/EWL) did not bind ClpP (Fig. 2A). Similarly, no binding was detected for variants with IGF-loops in four W subunits next to loopless R subunits (WRLW/WRLW) or in four W and R subunits neighboring loopless W subunits (RWLW/RWLW) (not shown). The corresponding ClpX variants with six IGF loops (e.g., RWW/RWW) bind ClpP normally as judged by degradation activity (Martin et al., 2005), confirming the importance of six IGF-loop contacts.
ATP or ATPγS support ClpX binding to and activation of ClpP peptidase activity, whereas ADP does not (Grimaud et al., 1998; Joshi et al., 2004). Thus, some aspect of nucleotide state must affect ClpP recognition. To address this issue, we assayed the ClpP affinity of ClpX variants with different numbers of R subunits to mimic ATP-free subunits. ClpP bound ClpX hexamers containing one or two R subunits with wild-type affinity and bound about 3-fold more weakly to hexamers with three R subunits (Fig. 2C; Table 1). By contrast, hexamers with four or more R subunits did not bind ClpP (Fig. 2A), but such molecules are functionally irrelevant because working ClpX has three or four ATP-bound subunits (Hersch et al., 2005; Martin et al., 2005). The important result, however, is that the nucleotide state of an individual subunit in an active ClpX hexamer has little impact on ClpP affinity. Because all six IGF loops in a ClpX hexamer are required for strong ClpP binding, we conclude that the IGF-loops in both ATP-bound and ATP-free ClpX subunits contact ClpP.
ClpX pore-2 loops mediate ClpP binding and communication
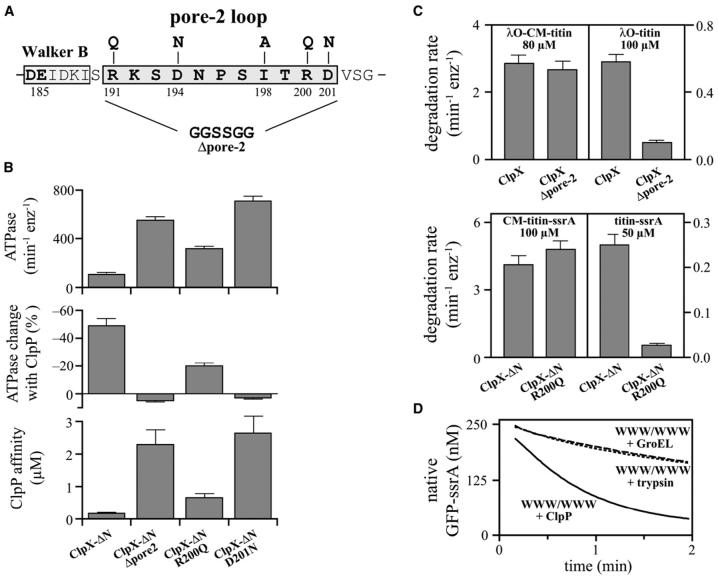
Because ClpX-ClpP interactions vary as a function of nucleotide (Joshi et al., 2004), we searched for regions of ClpX other than the IGF-loops that might contact ClpP and mediate nucleotide dependence. In models of the complex, the ClpX pore-2 loops (residues 191-201) can be positioned near the N-terminal ClpP loops, which are known to be important for ClpX binding (Fig. 1). To test the role of the pore-2 region, we constructed and purified unlinked ClpX mutants containing a deletion/substitution mutation (Δpore-2) or single-residue substitutions at five highly conserved sequence positions (Fig. 3A). For consistency with single-chain variants, these mutants were made in ClpX-ΔN.
Figure 3. Effects of pore-2 loop mutations in ClpX.
(A) Sequence of the pore-2 loop of E. coli ClpX and positions of mutations.
(B) Bar graphs show effects of selected pore-2 loop mutations in a ClpX-ΔN background on basal ATPase activity, ClpP repression of ATPase activity, and apparent ClpP affinity (determined by the decapeptide cleavage assay). Error bars (1 s.d.; n=3).
(C) Degradation rates of denatured or native substrates in the presence of ClpP and full-length ClpX, ClpX-ΔN, or variants with pore-2 mutations. Error bars (1 s.d.; n=3).
(D) ClpX (1 μM WWW/WWW) denatures GFP-ssrA (250 nM) more efficiently in the presence of ClpP (2 μM) as assayed by loss of native fluorescence. In the experiment without ClpP, the D87K GroEL mutant (2.5 μM; dashed line) or trypsin (1 μM; dotted line) were present to trap or degrade unfolded GFP-ssrA.
Several pore-2 mutations weakened ClpP binding, with roughly 15-fold decreases in affinity for D201N and Δpore-2, and smaller effects for D194N and R200Q (Table 1; Fig. 3B). Notably, the conservative Asp201→Asn mutation had functional consequences as severe as replacing the entire pore-2 loop. In addition, each pore-2 mutation increased the basal rate of ClpX ATP hydrolysis, with Δpore-2 and D201N again having the largest effects (Table 1). ClpP binding repressed the ATPase activity of “parental” ClpX-ΔN about 50%, as reported for wild-type ClpX (Kim et al., 2001), but did not reduce ATP hydrolysis of the D201N and Δpore-2 mutants, and only modestly repressed the D194N and R200Q mutants (Fig. 3B; Table 1). Overall, these results indicate that pore-2 loop residues play roles in controlling the ATPase rate of ClpX, in determining ClpP affinity, and in mediating ClpP repression of ClpX ATPase activity.
Pore-2 mutations affect ssrA-tag recognition and substrate unfolding
In addition to their effects on ClpXP affinity and communication, we found that severe pore-2 mutations altered ClpX’s ability to recognize ssrA-tagged substrates and to unfold native proteins. Specifically, the D201N and Δpore-2 mutations prevented degradation of folded or unfolded ssrA-tagged substrates in the presence of saturating ClpP and eliminated binding to a fluorescein-labeled ssrA peptide in anisotropy experiments (not shown). Next, we tested a substrate with a different degradation signal, the λO tag (Flynn et al., 2003). Because the N-domain is required for recognition of λO-tagged substrates (Singh et al., 2001), we constructed a Δpore-2 mutant in otherwise full-length ClpX for these studies. In complex with ClpP, this ClpX mutant and wild-type ClpX degraded denatured λO-CM-titin at similar rates, but the mutant degraded native λO-titin at less than 20% of the wild-type rate (Fig. 3C). Hence, the pore-2 loop appears to play a role in protein unfolding but not in translocation of λO-tagged substrates.
The milder R200Q pore-2 mutation in ClpX-ΔN also weakened binding to ssrA-tagged substrates, increasing KM for degradation of CM-titin-ssrA about 20-fold (not shown). At substrate concentrations well above KM, this mutant degraded unfolded CM-titin-ssrA normally but degraded native titin-ssrA and GFP-ssrA very slowly (Fig. 3C; not shown). Thus, several different pore-2 mutations prevent or weaken ClpX recognition of the ssrA degradation signal and compromise the ability of ClpXP to denature native proteins but not to translocate unfolded proteins.
Part of the unfolding defect of pore-2 mutants could be explained if ClpP helps ClpX denature native proteins. To test this model, we assayed the rate of denaturation of native GFP-ssrA by excess ClpX or ClpXP (Fig. 3D). In these single-turnover experiments, GFP fluorescence is lost concomitantly with denaturation and is a direct measure of the rate of enzymatic unfolding as long as denatured GFP-ssrA cannot refold. To prevent refolding when ClpP was absent, we used a GroEL mutant to trap denatured GFP-ssrA or trypsin to degrade this unfolded substrate. Under these conditions, GFP-ssrA unfolding by the WWW/WWW ClpX variant was about 3-fold slower than in the presence of ClpP (Fig. 3D). Importantly, the kinetics were the same with both trapping procedures, and doubling the GroEL trap or trypsin concentrations did not alter the results (not shown). Thus, both trapping procedures efficiently prevent GFP-ssrA refolding. Additional controls showed that trypsin did not degrade native GFP-ssrA or change the ClpXP-mediated denaturation rate and that differences in denaturation rates did not arise from differences in GFP-ssrA binding (not shown). Taken together, these results demonstrate that ClpXP is a more powerful protein unfoldase than ClpX alone.
The pore-2 loop of ClpX interacts with the N-terminal loop of ClpP
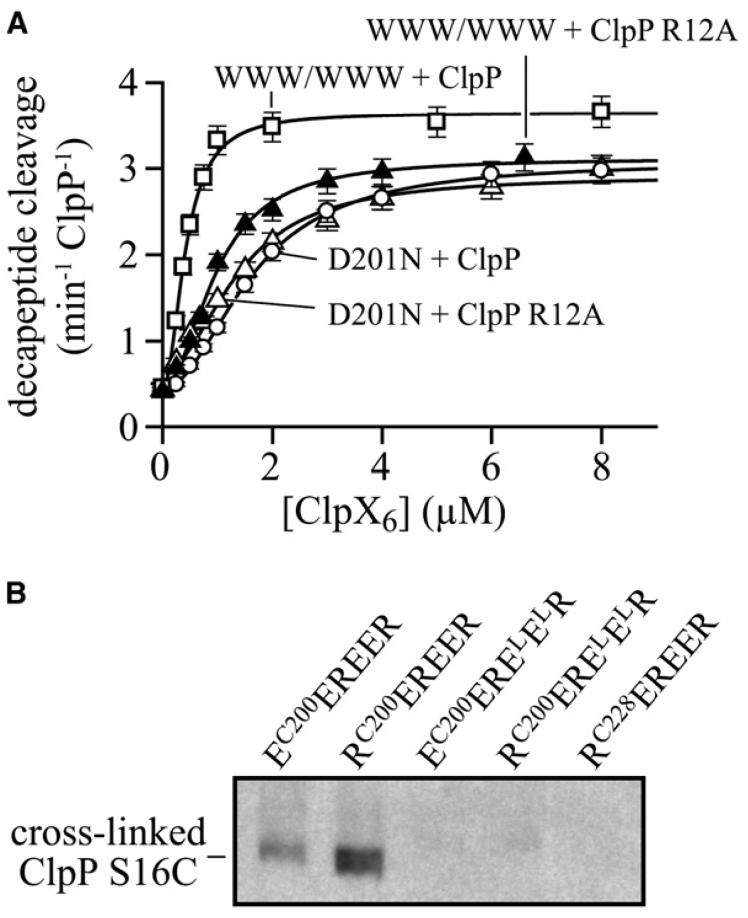
As a test for interactions, we compared the effects of the R12A N-terminal ClpP loop mutation on binding to ClpX with or without the D201N pore-2 mutation. The R12A mutation reduced binding to a ClpX variant with wild-type pore-2 loops but did not decrease binding to the D201N mutant (Fig. 4A). The non-additivity of these mutant effects is consistent with contacts between the N-terminal loops of ClpP and the pore-2 loops of ClpX.
Figure 4. Interaction of the ClpX pore-2 loop with the ClpP N-terminal loop.
(A) The ClpP R12A mutation reduced the affinity for single-chain ClpX-ΔN with wild-type pore-2 loops (WWW/WWW) but not for ClpX-ΔN with the D201N pore-2 mutation. Binding was measured by changes in decapeptide cleavage, and data were fit to the equation a + b*(1/(1+(Kapp/[ClpX])2) to account for binding cooperativity. Bars represent the error of linear fits of kinetic data used to determine peptide-cleavage rates.
(B) Western blot, probed with anti-ClpP antibody, assaying disulfide crosslinking between the ClpP with the N-terminal loop S16C mutation and single-chain ClpX hexamers with the pore-2-loop R200C mutation in one subunit. Crosslinking occurred for the EC200EREER and RC200EREER ClpX variants, but not for control variants that do not bind ClpP or have the cysteine mutation on the other side of ClpX.
To establish that a pore-2 loop of ClpX directly contacts an N-terminal loop of ClpP, we used disulfide crosslinking. In the N-terminal loop of ClpP, we introduced the S16C mutation. In ClpX, we introduced an R200C mutation into the pore-2 loop of either an E or an R subunit in single-chain hexamers that were ATPase deficient to prevent cycling and conformational changes (EC200EREER and RC200EREER). To form disulfide crosslinks, we activated these hexamers by formation of a mixed disulfide with dithionitrobenzoic acid, added ClpP S16C and ATPγS for 3 min, and then quenched the reaction by alkylating free cysteines with iodoacetic acid. Following non-reducing SDS PAGE, disulfide-linked ClpXP complexes were detected by western blotting (Fig. 4B). Both the EC200 and RC200 ClpX hexamers crosslinked to ClpP S16C, although the RC200 variant showed a higher yield of product. No crosslinking was observed for otherwise isogenic control hexamers lacking two IGF loops or with the cysteine on the opposite face of ClpX from the pore-2 loop (Fig. 4B). We conclude that residues in the pore-2 loop of ClpX and the N-terminal loop of ClpP are sufficiently close in complexes to allow efficient disulfide crosslinking, with nucleotide state playing a role in determining proximity.
Nucleotide state influences interaction of the ClpX pore-2 loop with ClpP
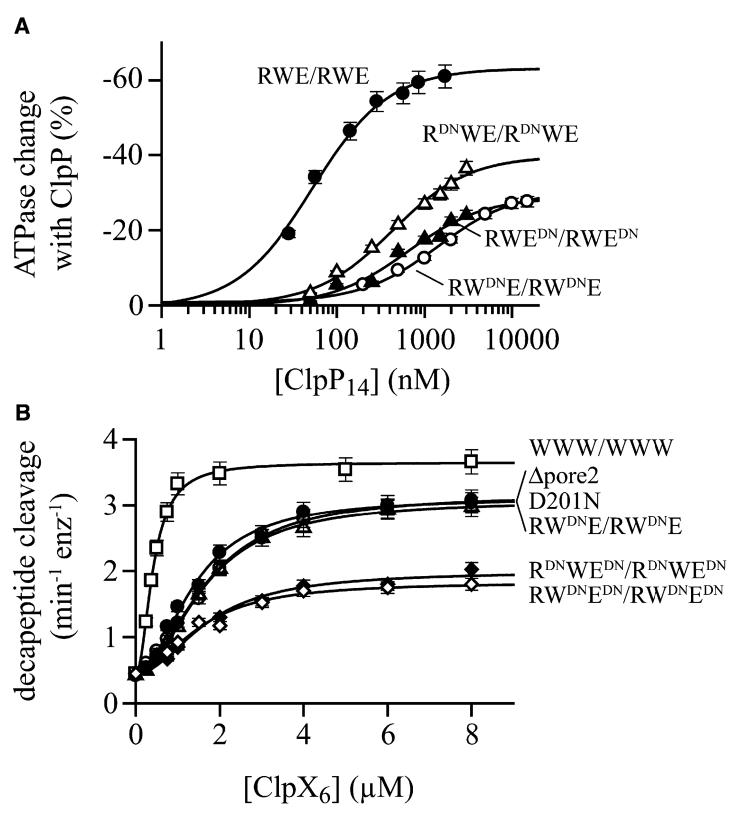
To determine if pore-2-loop interactions with ClpP are nucleotide dependent, we mimicked the varied nucleotide states of individual subunits in wild-type ClpX by introducing D201N mutations into just the R, the W, or the E subunits of RWE/RWE. These mutations reduced ClpP affinity from 63 nM to 430 nM (R), 640 nM (E), and 1.4 μM (W) (Fig. 5A; Table 1). Thus, the pore-2 loops of different subunits in the RWE/RWE hexamer contribute unequally to ClpP binding. The positions of the mutant D201N subunits in RWE/RWE also affected maximal ClpP repression of ATPase activity (Fig. 5A; Table 1). Thus, the phenotypes of pore-2 mutations depend on the nucleotide state of ClpX subunits.
Figure 5. Nucleotide state and pore-2 interactions.
(A) The affinity of ClpP for the single-chain RWE/RWE ClpX variant was reduced to different extents by pore-2 D201N mutations in just the R subunits, just the W subunits, and just the E subunits. Binding was assayed by changes in ATPase activity and data were fit to the Michaelis-Menten equation; apparent affinities are listed in Table 1. Bars represent the error of linear fits of kinetic data used to determine ATPase rates.
(B) ClpX variants with two, four, or six D201N mutations per hexamer bind ClpP with similar affinities. Binding was assayed by changes in decapeptide cleavage and data were fit as described in panel A of Fig. 4. Bars represent the error of linear fits of kinetic data used to determine peptide-cleavage rates.
In the decapeptide assay, the maximum ClpP cleavage rate stimulated by RWE/RWE ClpX was reduced by 18%, 13%, and 32% for D201N mutations in just the R, the W, or the E subunits, and by almost 60% when both the R and E subunits or the W and E subunits were mutant (Fig. 5B). Surprisingly, D201N mutations in all six subunits of unlinked ClpX only reduced maximum cleavage by 18%. Thus, ClpX pore-2 mutations reduce ClpP activity in a manner that depends on nucleotide state and the number of mutations. It is possible that the pore-2 loops of wild-type ClpX stabilize an open-pore conformation of ClpP and/or that mutant loops occlude the pore.
In some ClpX variants, the introduction of two, four, or six D201N substitutions had roughly comparable effects on ClpP affinity (Fig. 5B; Table 1). This non-additivity suggests that pore-2 loops interact cooperatively, possibly supporting a subset of pore-2 loops that directly contact ClpP. Moreover, the ATPase activities of RWE/RWE variants with D201N mutations just in the E subunits or just in the R subunits were 2- to 3-fold higher than those of the parent enzyme (Table 1). Because ATP hydrolysis is catalyzed only by the W subunits in these variants, ATPase activity must depend on the pore-2 loops of neighboring subunits. Thus, the pore-2 loops in a ClpX hexamer appear to interact with each other and with the N-terminal loops of ClpP.
Discussion
ClpX and other proteolytic ATPases change structure as they transit through the ATP-fueled mechanical cycles that drive protein unfolding and translocation. This structural diversity raises the question of how these machines dock with their partner peptidases. Is docking relatively static or does the peptidase adapt to each structural change in the ATPase? Both mechanisms appear to contribute to ClpXP stabilization and communication, with one set of structural elements involved in strong but relatively static interactions and another set responsible for weak but dynamic interactions.
Static contacts mediated by IGF-loop interactions
It is known that the IGF loops of ClpX play important roles in ClpP binding (Kim et al., 2001; Singh et al., 2001; Joshi et al., 2004). Our present results add three important new facts. First, each of the six IGF loops in a ClpX hexamer contributes to tight ClpP binding. Deleting one IGF loop decreases ClpX-ClpP affinity markedly; deleting more eliminates binding. Second, each IGF loop is necessary for efficient ClpXP proteolysis. Removing even one loop reduces maximal degradation by 35-45%, probably as a consequence of slower substrate translocation caused by misalignment of the ClpX WWWWWWL and ClpP rings and/or by incomplete opening of the ClpP pore. Third, IGF-loop interactions with ClpP are essentially independent of the nucleotide state of individual ClpX subunits. Under standard conditions of ATP excess, at least two subunits in the ClpX hexamer are nucleotide free and the rest, on average, are in the ATP-state (Hersch et al., 2005; Martin et al., 2005). Single-chain ClpX hexamers with two ATP-free subunits (mimicked by the R370K mutation) bind ClpP with wild-type affinity, and three of these mutant subunits reduce ClpP affinity only 3-fold. Because six IGF loops are needed for strong ClpP binding, however, the IGF loops in both ATP-bound and ATP-free ClpX subunits must make important contributions to ClpP affinity.
The “static” nature of the contacts between the IGF loops of ClpX and ClpP could arise because the IGF loops are flexible, and thus conformational changes in any given ClpX subunit do not propagate through these loops to the binding sites on ClpP. This mechanism is analogous to the use of springs or shock absorbers to damp vibrations. Alternatively, the IGF loops could be rather rigid and facilitate static contacts with ClpP because their anchoring points in ClpX remain comparatively motionless relative to the parts of the ClpX machine that change conformation during the ATPase cycle.
Dynamic contacts between ClpX pore-2 loops and ClpP N-terminal loops
The role of the ClpX pore-2 loops has not been examined previously. There are no high-resolution ClpXP or ClpX-hexamer structures, and the pore-2 loop is disordered in the ClpX-subunit structure (Kim and Kim, 2003). However, modeling places these pore-2 loops near the N-terminal sequences that form the axial pore of ClpP (Fig. 1; Kim and Kim, 2003; Bewley et al., 2006). Our double-mutant and disulfide-crosslinking experiments support interactions between these regions of ClpX and ClpP. Severe pore-2 mutations decrease ClpP affinity only modestly in comparison to IGF mutations but would still lead to dissociation of the majority of ClpXP complexes at normal concentrations in an E. coli cell (Farrell et al., 2005). Thus, the interactions of the ClpX pore-2 loop with ClpP may be relatively weak but are physiologically important.
Many N-terminal ClpP mutations prevent detectable ClpX binding and are more deleterious than the most severe ClpX pore-2 mutations (Kang et al., 2004; Gribun et al., 2005; Bewley et al., 2006). It is possible, however, that these ClpP mutations result in unfavorable contacts with ClpX, leading to an overestimate of the importance of the wild-type contacts. We favor this model, as certain N-terminal residues in the ClpP V6A mutant, which does not bind ClpX, adopt conformations not observed in any subunits of the wild-type structure (Bewley et al., 2006). Consistent with our pore-2 results, human mitochondrial ClpP retains modest ClpX affinity when only the N-terminal residues that form a protruding loop and extend toward ClpX are deleted (Kang et al., 2004).
All of the ClpX pore-2 loop mutations that we studied cause substantial increases in the basal rate of ATP hydrolysis. Moreover, the ATPase activity of severe pore-2 mutants is not repressed by ClpP binding. Thus, the wild-type pore-2 loops appear to decrease the rate of ATP-hydrolysis in free ClpX and to a greater extent in ClpXP complexes. The Walker-B portion of the ATP-binding site is adjacent to the pore-2 loop of ClpX (Fig. 3A), and linked conformational changes could easily account for the changes in the rate of ClpX ATP hydrolysis upon ClpP binding or mutation of the pore-2 loops. ClpX-ClpP crosslinking directly supports changes in pore-2 loop conformation in response to nucleotide binding. Furthermore, the effects of ClpX pore-2 mutations differ depending on whether these mutations are placed in hydrolysis-competent subunits or those constrained to ATP-bound or ATP-free states. In wild-type ClpX, the nucleotide states of individual subunits change throughout the ATPase cycle. Hence, pore-2 loop contacts between any single ClpX subunit and ClpP are expected to vary dynamically with the nucleotide state of that subunit and its neighbors. In this regard, the relatively weak contribution of the pore-2 loops to ClpP binding may be an advantage, as stable interactions would be difficult to break at the rate of ATP hydrolysis and accompanying conformational changes (>100 min-1).
ClpP rings have seven subunits. Intriguingly, Bewley et al. (2006) found that the N-terminal loops in one ring assumed an “up” conformation in six subunits, allowing potential contacts with ClpX, and a “down” conformation in the remaining subunit. It will be important to determine whether nucleotide-dependent changes in the pore-2 loops of ClpX are dynamically coupled with structural changes in the N-terminal loops of ClpP. In other words, do movements in ClpX simply change which pore-2 loops in a hexamer contact the N-terminal loops of ClpP or do the ClpP loops move synchronously with ClpX loops, making ClpP part of the dynamic machine?
Pore-2 loop roles in ssrA-tag binding and protein unfolding
In addition to altering ClpXP binding and communication, mutations at the dynamic axial interface between ClpX and ClpP affect substrate recognition and unfolding. For example, severe pore-2 mutations in ClpX damage recognition of substrates with ssrA tags but not λO tags. Similar recognition phenotypes have been reported for sequence changes in the “RKH” and “GYVG” loops of ClpX, which line the central channel near the top and middle of the pore (Siddiqui et al., 2004; Farrell et al., 2007). It is surprising that both the RKH and pore-2 loops, which are more than 30 Å apart, play important roles in binding of the ssrA-degradation tag, because ClpX recognizes only the three C-terminal residues of this tag sequence (Flynn et al., 2001). One possibility is the ssrA tag of a substrate forms distinct complexes with ClpX in a sequential manner. For example, the C-terminus of the ssrA tag might first contact the RKH loops at the top of ClpX and then move deeper into the pore to interact with the GYVG and pore-2 loops in the channel. In the latter complex, ATP-dependent changes in loop conformation could drive protein unfolding.
Our results show that the pore-2 loops are not required for substrate translocation but are needed for efficient unfolding of native proteins. For wild-type ClpX, ATP hydrolysis slows substantially during protein unfolding (Kenniston et al., 2003), suggesting that a lower ATPase rate allows mechanical force to be applied more efficiently. Hence, the inability of severe pore-2 ClpX mutants to slow ATP hydrolysis and allow repression by ClpP might explain their defects in protein unfolding. The pore-2 loops may also directly contact or grip the degradation tag of a substrate and contribute to the ability of the ClpX machine to exert an unfolding force by “pulling” on the tag. Interactions between the ClpP N-terminal loops and ClpX pore-2 loops would extend the substrate-binding channel, potentially providing a better grip and more efficient substrate unfolding. Strikingly, the dynamic axial ClpX-ClpP interface plays roles in substrate recognition, protein unfolding, and control of ClpX ATP hydrolysis and ClpP activity.
Bipartite determinants of binding and communication
Other AAA+ proteases may also use static and dynamic interactions. The HslU and ClpX unfoldases are homologous, but their partner peptidases, HslV and ClpP, have completely different folds. In some structures, C-terminal HslU sequences dock with the outer edge of the HslV ring and the pore-2 loops interact with residues surrounding the HslV pore (Sousa et al., 2000; Song et al., 2000; Kwon et al., 2003). Consistent with our results, the pore-2-loops of HslU adopt discrete conformations for subunits in different nucleotide states, although it remains to be determined if these loops play roles in HslV communication (Bochtler et al., 2000; Song et al., 2000; Wang et al., 2001). ClpA uses an IGL loop, similar to the IGF loop of ClpX, to make peripheral contacts with ClpP (Kim et al., 2001), and transplanting this loop allows the ClpB ATPase to interact functionally with ClpP (Weibezahn et al., 2004). Axial interactions may also stabilize ClpAP, as N-terminal ClpP mutations prevent ClpA binding (Gribun et al., 2005; Bewley et al., 2006).
Multiple types of communication between ClpX and ClpP have been reported (Jones et al., 1998; Grimaud et al., 1998; Kim et al., 2001; Joshi et al., 2004). Dynamic interactions between the pore-2 loops of ClpX and the N-terminal residues of ClpP appear to be responsible for communication that involves changes in ATP-hydrolysis rates. By contrast, static interactions between the IGF loops of ClpX and their cognate binding pockets in ClpP probably play the major role in communication that is independent of ATP hydrolysis. For example, ClpX increases the rate of peptide hydrolysis by ClpP even in the presence of poorly hydrolyzed ATP analogues (Grimaud et al., 1998), and we find that ClpX mutants with severe pore-2 mutations still enhance peptide cleavage to roughly 80% of the wild-type rate. Isolated C-terminal peptides of HslU can activate peptide cleavage by HslV (Seong et al., 2002; Ramachandran et al., 2002), and thus peripheral interactions appear to be sufficient for peptidase activation in both HslUV and ClpXP.
In principle, increased peptide cleavage could result from ClpX-dependent opening of the ClpP pore or from changes in ClpP active-site conformation, as precedents for both mechanisms exist in other AAA+ proteases (Whitby et al., 2000; Koehler et al., 2001; Sousa et al., 2002; Kwon et al., 2003; Foerster et al., 2003; Foerster et al., 2005). We favor a pore-opening mechanism, because ClpP and ClpXP cleave succinyl-Leu-Tyr-AMC at the same rate (S. Joshi, personal communication), showing that the catalytic sites in free ClpP are functional. Wang et al. (1997) originally proposed that binding of ClpA or ClpX opens the narrow axial pore of ClpP to allow denatured protein substrates to enter the degradation chamber. Our results can be understood if a “closed” pore structure dominates in free ClpP, and ClpX binding stabilizes an “open” pore structure. For example, IGF-loop binding to the hydrophobic pockets of ClpP could shift the allosteric equilibrium between these states to favor the open conformation by a small amount, resulting in a dynamic mixture of closed and open states in the population. Interactions between the N-terminal ClpP loops and the pore-2 loops of ClpX could then stabilize the open structure further, resulting in an open pore in most or all ClpXP complexes. This model may explain why deletion of even a single ClpX IGF-loop causes a reduction in ClpP activity that is larger than the effects of mutations that remove all six of the pore-2 loops of ClpX or that alter all seven of the N-terminal loops of ClpP.
In conclusion, we find that stabilization of the ClpXP complex relies upon two kinds of interactions with distinct functional roles (Fig. 1). Contacts between the six IGF loops of ClpX and hydrophobic pockets near the periphery of ClpP contribute most of the binding energy, set the general positions of the ClpX and ClpP rings in the proteolytic complex, and appear to stabilize an open-pore conformation of ClpP. These strong IGF-loop interactions are static, in the sense that they are independent of the changing nucleotide states of individual subunits in working ClpX hexamers. A second set of interactions occurs between the pore-2 loops of ClpX and the N-terminal loops of ClpP. These weak axial interactions, which vary dynamically with nucleotide state, allow fine-tuning of ClpX-ClpP transactions via changes in ATP-hydrolysis rates during substrate unfolding and degradation.
Experimental Procedures
Protein expression and purification
Mutants of unlinked E. coli ClpX, ClpX-ΔN (residues 62-424), and linked single-chain variants of ClpX-ΔN were constructed by PCR and cloned into pACYCDuet-1 (Novagen). All unlinked ClpX variants, single-chain dimers, and single-chain trimers contained an N-terminal His6 tag; single-chain hexamers contained a C-terminal His6 tag (Martin et al., 2005). ClpX variants were expressed for 3 h at 22 °C in the recA- E. coli strain BLR (DE3) and were purified by Ni++-NTA affinity (Qiagen) in 25 mM HEPES (pH 7.6), 100 mM KCl, 400 mM NaCl, 10% glycerol, 10 mM β-mercaptoethanol, with 20 mM imidazole in the loading buffer and 250 mM imidazole in the elution buffer, and by size-exclusion chromatography on Sephacryl S-300 (Amersham Biosciences) in 50 mM Tris-HCl (pH 7.8), 300 mM KCl, 10 mM MgCl2, 10% glycerol, 0.1 mM EDTA, and 1 mM DTT. ClpP-His6 and untagged ClpP and mutants were expressed and purified as described (Kim et al., 2000; Joshi et al., 2004).
GFP-ssrA with a His6 tag between GFP and the ssrA tag was purified by Ni++-NTA affinity and ion-exchange chromatography (Kim et al., 2000; Bolon et al., 2004). λO-Arc, containing the N-terminal λO sequence TNTAKILNFGR, Arc repressor residues 1-53, and a C-terminal H6KNQHE sequence, was expressed from pET21b in E. coli strain BL21 and purified by Ni++-NTA affinity. Unlabeled or 35S-labeled titin-I27-ssrA was expressed and purified as described (Kenniston et al., 2003; 2005). Titin-I27-ssrA was alkylated for 3 h at 22 °C with a 100-fold excess of iodoacetic acid at pH 8.8 in 6 M GuHCl to obtain the unfolded, carboxymethylated (CM) protein.
Biochemical assays
ATPase assays were typically performed using ClpX (0.3 μM pseudo-hexamer equivalents) with or without ClpP14 (0.9 μM) and protein substrate (20 μM) at 30 °C in PD buffer (25 mM HEPES (pH 7.6) 100 mM KCl, 20 mM MgCl2, 1 mM EDTA, 10% glycerol) as described (Burton et al., 2001). For ClpX mutants with low ClpP affinity, the ClpP14 concentration was increased to 15 μM. To determine apparent ClpX-ClpP affinity by changes in ATPase rates, increasing ClpP14 was titrated against constant ClpX (50 or 300 nM pseudo-hexamer equivalents depending on the ClpP affinity).
Apparent ClpX-ClpP affinities were also measured by changes in the rate of cleavage of the decapeptide Abz-KASPVSLGYNO2D, in which 2-aminobenzoic acid (Abz) is a fluorogenic group and 3-nitrotyrosine (YNO2) is a quencher. The rate of cleavage of 6.5 μM decapeptide by 50 or 200 nM ClpP14 in the presence of 1 mM ATPγS and different concentrations of ClpX was monitored by the increase in fluorescence (excitation 320 nm; emission 420 nm) using a QM-2000-4SE spectrofluorimeter (Photon Technology International).
Degradation of 35S-labeled titin-ssrA by ClpX variants (0.3 μM pseudo-hexamer equivalents) and ClpP14 (0.9 μM) was carried out at 30 °C in PD buffer with an ATP regeneration system and was assayed by the release of acid-soluble peptides (Kenniston et al., 2003). GFP-ssrA degradation was monitored by the loss of GFP fluorescence (Kim et al., 2000). For affinity measurements, ClpP14 was titrated against constant ClpX (50 or 300 nM pseudo-hexamer) and the rate of degradation of 15 μM GFP-ssrA was determined. ClpX unfolding of GFP-ssrA was assayed using the D87K GroEL trap mutant (Weber-Ban et al., 1999; Kim et al., 2000) or trypsin to degrade the unfolded substrate. Degradation of GFP-ssrA was not observed with the trypsin concentrations (1-2 μM) and times (< 2 min) used in these experiments. Trypsin did degrade ClpX slowly under these conditions but did not affect the initial rates of ClpX-mediated GFP-ssrA denaturation.
Pull-down assays were performed in PD buffer as described in Joshi et al. (2004) using 1 μM His6-tagged ClpX variants and 1 μM ClpP in the presence of 1 mM ATPγS or 1 mM ADP. ClpX-ClpP binding was detected using either ATPγS or ATP but was not detected using GTP, UTP or CTP.
Crosslinking
Cysteines in the pore loop of ClpX single-chain hexamers were activated by incubation with 1.5 mM 5,5′-dithiobis-(2-nitrobenzoic acid) (DTNB) in 25 mM HEPES (pH 7.8), 300 mM KCl, 20 mM MgCl2, 1 mM EDTA, 10% glycerol for 5 min at 22 °C. After separation of free DTNB by buffer exchange using Micro Bio-Spin columns (BIO-RAD), DTNB-activated ClpX (1 μM pseudo-hexamer equivalents) was incubated with ClpP14 S16C (1 μM) in the presence of 1 mM ATPγS for 3 min at 22 °C. Crosslinking reactions were stopped by alkylation of free cysteines for 20 min with 140 mM iodoacetic acid in 420 mM Tris-HCl (pH 8.5), 6.5 M urea, 3 mM EDTA. Following non-reducing SDS-PAGE, the disulfide-crosslinked ClpX-ClpP complexes were analyzed by western blotting using an anti-ClpP antibody.
Acknowledgements
We thank M. Lee for the ClpP S16C mutant and for developing the decapeptide-cleavage assay, M. Lee and S. Joshi for communication of unpublished results, C. Farrell for λO-tagged substrates, B. Burton for the GroEL trap mutant, and S. Bell, P. Chien, J. Davis, S. Glynn, A. Keating, M. Laub, M. Lee, T. Schwartz, and K. Wang for helpful suggestions. Supported by NIH grants AI-15706 and AI-16892. A.M. was supported by a Merck/MIT CSBi postdoctoral fellowship. T.A.B. is an H.H.M.I. employee.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Beuron F, Maurizi MR, Belnap DM, Kocsis E, Booy FP, Kessel M, Steven AC. At sixes and sevens: characterization of the symmetry mismatch of the ClpAP chaperone-assisted protease. J. Struct. Biol. 1998;123:248–259. doi: 10.1006/jsbi.1998.4039. [DOI] [PubMed] [Google Scholar]
- Bewley MC, Graziano V, Griffin K, Flanagan JM. The asymmetry in the mature amino-terminus of ClpP facilitates a local symmetry match in ClpAP and ClpXP complexes. J. Struct. Biol. 2006;153:113–128. doi: 10.1016/j.jsb.2005.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bochtler M, Ditzel L, Groll M, Huber R. Crystal structure of heat shock locus V (HslV) from Escherichia coli. Proc. Natl. Acad. Sci. USA. 1997;94:6070–6074. doi: 10.1073/pnas.94.12.6070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bochtler M, Hartmann C, Song HK, Bourenkov GP, Bartunik HD, Huber R. The structures of HsIU and the ATP-dependent protease HsIU-HsIV. Nature. 2000;403:800–805. doi: 10.1038/35001629. [DOI] [PubMed] [Google Scholar]
- Bolon DN, Grant RA, Baker TA, Sauer RT. Nucleotide-dependent substrate handoff from the SspB adaptor to the AAA+ ClpXP protease. Mol. Cell. 2004;16:343–350. doi: 10.1016/j.molcel.2004.10.001. [DOI] [PubMed] [Google Scholar]
- Burton RE, Siddiqui SM, Kim YI, Baker TA, Sauer RT. Effects of protein stability and structure on substrate processing by the ClpXP unfolding and degradation machine. EMBO J. 2001;20:3092–3100. doi: 10.1093/emboj/20.12.3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrell CM, Baker TA, Sauer RT. Altered specificity of a AAA+ protease. Mol. Cell. 2007;25:161–166. doi: 10.1016/j.molcel.2006.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynn JM, Levchenko I, Seidel M, Wickner SH, Sauer RT, Baker TA. Overlapping recognition determinants within the ssrA degradation tag allow modulation of proteolysis. Proc. Natl. Acad. Sci. USA. 2001;11:10584–10589. doi: 10.1073/pnas.191375298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynn JM, Neher SB, Kim YI, Sauer RT, Baker TA. Proteomic discovery of cellular substrates of the ClpXP protease reveals five classes of ClpX-recognition signals. Mol. Cell. 2003;11:671–683. doi: 10.1016/s1097-2765(03)00060-1. [DOI] [PubMed] [Google Scholar]
- Foerster A, Whitby FG, Hill CP. The pore of activated 20S proteasomes has an ordered 7-fold symmetric conformation. EMBO J. 2003;22:4356–4364. doi: 10.1093/emboj/cdg436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foerster A, Masters EI, Whitby FG, Robinson H, Hill CP. The 1.9 Å structure of a proteasome-11S activator complex and implications for the proteasome-PAN/PA700 interactions. Mol. Cell. 2005;18:589–599. doi: 10.1016/j.molcel.2005.04.016. [DOI] [PubMed] [Google Scholar]
- Gottesman S, Maurizi MR, Wickner S. Regulatory subunits of energy-dependent proteases. Cell. 1997;91:435–438. doi: 10.1016/s0092-8674(00)80428-6. [DOI] [PubMed] [Google Scholar]
- Gribun A, Kimber MS, Ching R, Sprangers R, Fiebig KM, Houry WA. The ClpP double ring tetradecameric protease exhibits plastic ring-ring interactions, and the N termini of its subunits form flexible loops that are essential for ClpXP and ClpAP complex formation. J. Biol. Chem. 2005;280:16185–16196. doi: 10.1074/jbc.M414124200. [DOI] [PubMed] [Google Scholar]
- Grimaud R, Kessel M, Beuron F, Steven AC, Maurizi MR. Enzymatic and structural similarities between the Escherichia coli ATP-dependent proteases, ClpXP and ClpAP. J. Biol. Chem. 1998;273:12476–12481. doi: 10.1074/jbc.273.20.12476. [DOI] [PubMed] [Google Scholar]
- Groll M, Ditzel L, Löwe J, Stock D, Bochtler M, Bartunik HD, Huber R. Structure of 20S proteasome from yeast at 2.4 Å resolution. Nature. 1997;386:463–471. doi: 10.1038/386463a0. [DOI] [PubMed] [Google Scholar]
- Hersch GL, Burton RE, Bolon DN, Baker TA, Sauer RT. Asymmetric interactions of ATP with the AAA+ ClpX6 unfoldase: allosteric control of a protein machine. Cell. 2005;121:1017–1027. doi: 10.1016/j.cell.2005.05.024. [DOI] [PubMed] [Google Scholar]
- Jones JM, Welty DJ, Nakai H. Versatile action of Escherichia coli ClpXP as protease or molecular chaperone for bacteriophage Mu transposition. J. Biol. Chem. 1998;273:459–465. doi: 10.1074/jbc.273.1.459. [DOI] [PubMed] [Google Scholar]
- Joshi SA, Hersch GL, Baker TA, Sauer RT. Communication between ClpX and ClpP during substrate processing and degradation. Nat. Struct. Mol. Biol. 2004;11:404–411. doi: 10.1038/nsmb752. [DOI] [PubMed] [Google Scholar]
- Kang SG, Maurizi MR, Thompson M, Mueser T, Ahvazi B. Crystallography and mutagenesis point to an essential role for the N-terminus of human mitochondrial ClpP. J. Struct. Biol. 2004;148:338–352. doi: 10.1016/j.jsb.2004.07.004. [DOI] [PubMed] [Google Scholar]
- Kenniston JA, Baker TA, Fernandez JM, Sauer RT. Linkage between ATP consumption and mechanical unfolding during the protein processing reactions of a AAA+ degradation machine. Cell. 2003;114:511–520. doi: 10.1016/s0092-8674(03)00612-3. [DOI] [PubMed] [Google Scholar]
- Kim DY, Kim KK. Crystal structure of ClpX molecular chaperone from Helicobacter pylori. J. Biol. Chem. 2003;278:50664–50670. doi: 10.1074/jbc.M305882200. [DOI] [PubMed] [Google Scholar]
- Kim YI, Burton RE, Burton BM, Sauer RT, Baker TA. Dynamics of substrate denaturation and translocation by the ClpXP degradation machine. Mol. Cell. 2000;5:639–648. doi: 10.1016/s1097-2765(00)80243-9. [DOI] [PubMed] [Google Scholar]
- Kim YI, Levchenko I, Fraczkowska K, Woodruff RV, Sauer RT, Baker TA. Molecular determinants of complex formation between Clp/Hsp100 ATPases and the ClpP peptidase. Nat. Struct. Biol. 2001;8:230–233. doi: 10.1038/84967. [DOI] [PubMed] [Google Scholar]
- Koehler A, Cascio P, Leggett DS, Woo KM, Goldberg AL, Finley D. The axial channel of the proteasome core particle is gated by the Rpt2 ATPase and controls both substrate entry and product release. Mol. Cell. 2001;7:1143–1152. doi: 10.1016/s1097-2765(01)00274-x. [DOI] [PubMed] [Google Scholar]
- Kwon AR, Kessler BM, Overkleeft HS, McKay DB. Structure and reactivity of an asymmetric complex between HslV and I-domain deleted HslU, a prokaryotic homolog of the eukaryotic proteasome. J. Mol. Biol. 2003;330:185–195. doi: 10.1016/s0022-2836(03)00580-1. [DOI] [PubMed] [Google Scholar]
- Loewe J, Stock D, Jap B, Zwickl P, Baumeister W, Huber R. Crystal structure of the 20S proteasome from the archaeon T. acidophilum at 3.4 Å resolution. Science. 1995;268:533–539. doi: 10.1126/science.7725097. [DOI] [PubMed] [Google Scholar]
- Martin A, Baker TA, Sauer RT. Rebuilt AAA+ motors reveal operating principles for ATP-fueled machines. Nature. 2005;437:1115–1120. doi: 10.1038/nature04031. [DOI] [PubMed] [Google Scholar]
- Neuwald AF, Aravind L, Spouge JL, Koonin EV. AAA+: A class of chaperone-like ATPases associated with the assembly, operation, and disassembly of protein complexes. Genome Res. 1999;9:27–43. [PubMed] [Google Scholar]
- Ortega J, Lee HS, Maurizi MR, Steven AC. Alternating translocation of protein substrates from both ends of ClpXP protease. EMBO J. 2002;21:4938–4949. doi: 10.1093/emboj/cdf483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortega J, Singh SK, Ishikawa T, Maurizi MR, Steven AC. Visualization of substrate binding and translocation by the ATP-dependent protease, ClpXP. Mol. Cell. 2000;6:1515–1521. doi: 10.1016/s1097-2765(00)00148-9. [DOI] [PubMed] [Google Scholar]
- Ramachandran R, Hartmann C, Song HK, Huber R, Bochtler M. Functional interactions of HslV (ClpQ) with the ATPase HslU (ClpY) Proc. Natl. Acad. Sci. USA. 2002;99:7396–73401. doi: 10.1073/pnas.102188799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer RT, et al. Sculpting the proteome with AAA+ proteases and disassembly machines. Cell. 2004;119:9–18. doi: 10.1016/j.cell.2004.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seong IS, Kang MS, Choi MK, Lee JW, Koh OJ, Wang J, Eom SH, Chung CH. The C-terminal tails of HslU ATPase act as a molecular switch for activation of HslV peptidase. J. Biol. Chem. 2002;277:25976–25982. doi: 10.1074/jbc.M202793200. [DOI] [PubMed] [Google Scholar]
- Siddiqui SM, Sauer RT, Baker TA. Role of the protein-processing pore of ClpX, a AAA+ ATPase, in recognition and engagement of specific protein substrates. Genes Dev. 2004;18:369–374. doi: 10.1101/gad.1170304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh SK, Rozycki J, Ortega J, Ishikawa T, Lo J, Steven AC, Maurizi MR. Functional domains of the ClpA and ClpX molecular chaperones identified by limited proteolysis and deletion analysis. J. Biol. Chem. 2001;276:29420–29429. doi: 10.1074/jbc.M103489200. [DOI] [PubMed] [Google Scholar]
- Song HK, Hartmann C, Ramachandran R, Bochtler M, Behrendt R, Moroder L, Huber R. Mutational studies on HslU and its docking mode with HslV. Proc. Natl. Acad. Sci. USA. 2000;97:14103–14108. doi: 10.1073/pnas.250491797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sousa MC, Kessler BM, Overkleeft HS, McKay DB. Crystal structure of HslUV complexed with a vinyl sulfone inhibitor: corroboration of a proposed mechanism of allosteric activation of HslV by HslU. J. Mol. Biol. 2002;318:779–785. doi: 10.1016/S0022-2836(02)00145-6. [DOI] [PubMed] [Google Scholar]
- Sousa MC, Trame CB, Tsuruta H, Wilbanks SM, Reddy VS, McKay DB. Crystal and solution structures of an HslUV protease-chaperone complex. Cell. 2000;103:633–643. doi: 10.1016/s0092-8674(00)00166-5. [DOI] [PubMed] [Google Scholar]
- Wang J, Hartling JA, Flanagan JM. The structure of ClpP at 2.3 Å resolution suggests a model for ATP-dependent proteolysis. Cell. 1997;91:447–456. doi: 10.1016/s0092-8674(00)80431-6. [DOI] [PubMed] [Google Scholar]
- Wang J, Song JJ, Seong IS, Franklin MC, Kamtekar S, Eom SH, Chung CH. Nucleotide-dependent conformational changes in a protease-associated ATPase HsIU. Structure. 2001;9:1107–1116. doi: 10.1016/s0969-2126(01)00670-0. [DOI] [PubMed] [Google Scholar]
- Weber-Ban EU, Reid BG, Miranker AD, Horwich AL. Global unfolding of a substrate protein by the Hsp100 chaperone ClpA. Nature. 1999;401:90–93. doi: 10.1038/43481. [DOI] [PubMed] [Google Scholar]
- Weibezahn J, et al. Thermotolerance requires refolding of aggregated proteins by substrate translocation through the central pore of ClpB. Cell. 2004;119:653–665. doi: 10.1016/j.cell.2004.11.027. [DOI] [PubMed] [Google Scholar]
- Whitby FG, Masters EI, Kramer L, Knowlton JR, Yao Y, Wang CC, Hill CP. Structural basis for the activation of the 20S proteasomes by 11S regulators. Nature. 2000;408:115–120. doi: 10.1038/35040607. [DOI] [PubMed] [Google Scholar]