Abstract
Soil fungal communities involved in the biodegradation of polyester polyurethane (PU) were investigated. PU coupons were buried in two sandy loam soils with different levels of organic carbon: one was acidic (pH 5.5), and the other was more neutral (pH 6.7). After 5 months of burial, the fungal communities on the surface of the PU were compared with the native soil communities using culture-based and molecular techniques. Putative PU-degrading fungi were common in both soils, as <45% of the fungal colonies cleared the colloidal PU dispersion Impranil on solid medium. Denaturing gradient gel electrophoresis showed that fungal communities on the PU were less diverse than in the soil, and only a few species in the PU communities were detectable in the soil, indicating that only a small subset of the soil fungal communities colonized the PU. Soil type influenced the composition of the PU fungal communities. Geomyces pannorum and a Phoma sp. were the dominant species recovered by culturing from the PU buried in the acidic and neutral soils, respectively. Both fungi degraded Impranil and represented >80% of cultivable colonies from each plastic. However, PU was highly susceptible to degradation in both soils, losing up to 95% of its tensile strength. Therefore, different fungi are associated with PU degradation in different soils but the physical process is independent of soil type.
The worldwide production of synthetic polymers continues to rise, resulting in an increased environmental burden through the generation of plastic waste. More than 140 million tonnes of plastic was produced worldwide in 2001 (34), and the proportion of household plastic waste in the average American home increased from 3 to 5% of total waste in 1969 (15) to more than 30% in 1995 (21) and continues to rise. Many plastics are both physically and chemically robust and cause waste management problems (10). However, several families of plastics undergo biodegradation in the environment, and an understanding of how this degradation occurs may aid in the development of strategies to exploit these processes for waste management purposes.
Microorganisms are responsible for the majority of plastic degradation (6), and abiotic factors such as photodegradation or hydrolysis play a very minor role (18, 42). Plastics vulnerable to biodegradation include the polyhydroxyalkanoates, polycaprolactone, polylactic acid, polyvinyl chloride (31, 32), and polyester polyurethane (PU). PU is used in a variety of industrial applications, including insulating foams, fibers, and synthetic leather and rubber goods. The presence of ester and urethane linkages in the backbone of PUs makes them susceptible to hydrolysis by enzymes secreted by microorganisms, releasing breakdown products which may act as a carbon source and lead to a weakening of the tensile strength (1, 13, 22, 26, 27).
Both PU-degrading fungi (5, 6, 12, 32) and bacteria (1, 20, 23) have been isolated from PU, indicating that there are potential reservoirs of PU-degrading organisms widespread in the environment. It is known that fungi and not bacteria are predominantly responsible for PU degradation in laboratory soil microcosms (5), although studies are lacking on the ecology of PU colonization and degradation in situ by fungi in the soil.
This is the first study investigating the fungal communities that develop on the surface of plastics such as PU during burial in situ in soil. PU was buried in two different soils for 5 months, and the fungal communities colonizing the surface were analyzed by culturing and denaturing gradient gel electrophoresis (DGGE). The rates of colonization and degradation of PU in both soils were compared, and the dominant organisms on the PU surface were identified by ribosomal sequencing.
MATERIALS AND METHODS
Fabrication of PU coupons.
PU pellets (Elastogran, United Kingdom) were pressed at 180°C using an electric press (Bradley & Turton Ltd., Kidderminster, United Kingdom) into sheets with a thickness of 1.5 mm. Tensile strength determinations of randomly selected pieces of PU produced this way showed that the PU had a consistent tensile strength. Rectangular coupons of PU measuring 6.25 by 4.2 by 0.15 cm were cut with a scalpel, giving a total surface area available for colonization of 55.5 cm−2.
In situ burial of PU coupons in soil.
PU coupons were surface sterilized via immersion in 70% (vol/vol) ethanol. Coupons were then buried in two contrasting garden soils near Manchester, United Kingdom (longitude, 2°15′W; latitude, 53°19′N). The soil at this site belongs to the Blackwood series (30), and it is derived from a coarse, glaciofluvial drift producing a loamy sand. One soil, which was relatively undisturbed, was beneath the canopy of a mature conifer (Thuja plicata); this soil is subsequently described as the “acidic soil.” The other soil was from a more disturbed garden location which had been previously enriched with garden compost and is referred to as the “neutral soil” (see Results). During the experimental period, there was no management intervention of any kind. Six coupons were buried in a vertical position approximately 5 cm apart in each soil type, so that the tops of the coupons were approximately 6 cm below the surface. Coupons remained buried for the 5-month period from January to May 2003.
Recovery of biomass from the surface of buried PU.
Biomass was recovered from three of the PU coupons in each soil after 5 months of burial in order to analyze the fungal communities growing on the surface. The remaining three coupons were used for tensile strength measurements. For biomass recovery, loosely adhered soil particles were first removed by agitating the PU coupons in sterile phosphate-buffered saline (PBS) (33) for 5 min. Coupons were then submerged in 20 ml sterile PBS, and the biomass was scraped from both sides of the PU into the PBS using a sterile scalpel blade (32). An aliquot (1 ml) of this biomass suspension was used for viable counting. The remainder was centrifuged at 3,000 × g for 30 min at 4°C, the supernatant was discarded, and the biomass was used for DNA extraction and DGGE analysis.
Tensile strength determination of buried PU.
The tensile strength of PU coupons after burial in soil for 5 m was determined to assess the extent of degradation. PU coupons were cut into strips measuring 4.5 by 0.5 by 0.15 cm. Replicate strips (n = 15) were stretched at a rate of 200 mm min−1, and the tensile strength was determined using an Instron 4301 (Instron Ltd., Swindon, United Kingdom). Unburied PU strips were used as a control.
Fungal viable counts.
Viable counts of fungi in the soil and on the surface of buried PU were determined on solid media. Samples of soil in which the PU was buried and samples of biomass recovered from the surface of buried PU were serially diluted in PBS and spread onto soil extract agar (SEA) plates (2) and Impranil agar plates (12). Colonies were counted after 5 to 7 days of incubation at 25°C. Total fungal viable counts were enumerated on SEA, while putative PU-degrading fungi were enumerated as colonies producing zones of clearance on Impranil agar. Both media included 50 μg ml−1 of chloramphenicol to inhibit bacterial growth. The number of Impranil-degrading fungi was then calculated as a percentage of the total number of colonies recovered.
DNA extraction.
The FastDNA SpinKit for soil (Q-Biogene, CA) was used to extract total DNA from 0.4-g soil samples or 0.5-g samples of biomass (wet weight) recovered from the surface of buried PU. To remove all traces of PCR inhibitory compounds, 20 μl of extracted DNA was run for ca. 15 min on a 1.0% (wt/vol) agarose-TAE (40 mM Tris base, 20 mM glacial acetic acid, 1 mM EDTA) gel. Bands of genomic DNA were then excised, and DNA was recovered using the Nucleospin extract II gel extraction kit (Machery-Nagel, Düren, Germany).
PCR amplification of fungal community DNA.
PCR was used to generate DNA fragments for fungal community DGGE analysis. The PCR DNA template consisted of approximately 50 ng per reaction of extracted DNA. Biomix red PCR master mix (Bioline, London, United Kingdom) was employed in all reactions. Primers were present in each reaction mixture at a concentration of 1 μM. Fungal DGGE fragments were generated using the GM2/JB206c primer set (GM2, 5′-CTGCGTTCTTCATCGAT-3′; JB206c, 5′-CGCCCGCCGCGCGCGGCGGGCGGGGCGGGGGCACGGGGGGAAGTAAAAGTCGTAACAA GG-3′), which amplify the internal transcribed spacer 1 (ITS1) region found in the fungal ribosomal DNA (rDNA) gene complex. The PCR regime employed was as follows: 94°C for initial denaturation for 5 min; 20 “touchdown” cycles of 94°C for 30 s, annealing for 30s at 59 to 49°C with the annealing temperature being reduced by 0.5°C per cycle and extension at 72°C for 45 s; 30 cycles at 95°C for 30 s, annealing at 49°C for 30 s, and extension at 72°C for 45 s; and 1 final extension at 72°C for 5 min.
DGGE analysis of fungal communities in the soil and on the surface of buried PU.
The compositions of the fungal communities in the soil and on the surface of buried PU were compared using DGGE (25). The D-Code universal mutation detection system (Bio-Rad, Herts, United Kingdom) was used. Gels measured 16 cm by 16 cm by 1 mm and contained 10% (vol/vol) bisacrylamide in 1× TAE. A perpendicular gel with a denaturant gradient of 25 to 55% was used. For all gels, 500 μg of PCR product was used per lane; gels were run in 1× TAE buffer at a constant temperature of 60°C for 16.5 h at 42 V. After electrophoresis was complete, gels were stained with SybrGold (Molecular Probes, The Netherlands) for 45 min and photographed under UV light.
Identification of fungal isolates with putative PU-degrading activity.
Putative PU-degrading fungal isolates were recovered from the surface of soil-buried PU. They were detected by their ability to produce zones of clearance on Impranil plates and then grown in malt extract broth (Oxoid, United Kingdom). Genomic DNA was extracted (4), and the ITS1-5.8S-ITS2 region of the fungal rRNA gene complex was PCR amplified using the ITS1/ITS4 primer set (ITS1, 5′-TCCGTAGGTGAACCTGCGG-3′; ITS4, 5′-TCCTCCGCTTATTGATATGC-3′) using the Expand high-fidelity PCR system (Roche, Mannheim, Germany). This region of the fungal genome has been used previously both for identifying members of fungal communities and also for determining phylogenetic relationships between fungi (37). The PCR regime was as follows: 94°C for 3 min; 35 cycles of 94°C for 1 min, 56°C for 1 min, and 72°C for 1 min; and a final extension at 72°C for 5 min. PCR products were then sequenced using in-house facilities. Sequences were used to interrogate the EMBL fungal database using the blastn algorithm (www.ncbi.nlm.nih.gov).
Identification of fungi on the surface of PU via cloning and sequencing of ITS1 DGGE products.
To identify fungi on the surface of buried PU in a culture-independent manner, ITS1 DGGE fragments generated from DNA extracted from fungal communities on buried PU were cloned into the pGEM-T Easy plasmid (Promega, United Kingdom) and transformed into Escherichia coli strain JM109 as per the manufacturer's instructions. Individual clones were screened using colony PCR to reamplify the ITS1 fragments contained within them using the DGGE PCR regimen described above. These fragments were then run on DGGE alongside whole PU community DGGE products. Clones producing bands that migrated to the same position as bands within the PU community profiles were then selected for sequencing. These sequences were used to interrogate the EMBL fungal database as described previously.
Phylogenetic analysis of fungal isolates.
In order to determine the reliability of the initial identifications obtained from the EMBL database, the identities of the fungi were verified by phylogenetic analysis. For each fungus identified, the most closely related species were determined using the Taxonomy Browser provided by the National Centre for Biotechnology Information (www.ncbi.nlm.nih.gov). Sequences from these closely related species were obtained and aligned to the sequences recovered in this work using the ClustalW implementation in the MEGA 3.1 software package (24). Maximum parsimony trees (bootstrap corrected using 1,000 samples) were constructed using the aligned sequences, also using MEGA 3.1. Trees were rooted using Candida albicans, Kluyveromyces lactis, Aspergillus fumigatus, Leucostoma persoonii, Boletus satanas, and Russula compacta as outliers. The identities obtained from the EMBL database were considered reliable if the strains clustered with those of closely related fungi in the phylogenetic tree.
Statistical analysis.
Where appropriate, data were subjected to analysis of variance to determine statistical significance, with the significance threshold set at P < 0.05.
RESULTS
Soil chemical analysis.
The two soils employed in this work were analyzed for pH and for phosphorus, potassium, magnesium, and organic carbon content (analysis carried out by Adas Laboratories, Wolverhampton, United Kingdom) (Table 1). The two soils could be clearly distinguished: soil 1 (neutral soil) was black and had a coarse texture and a neutral pH (6.7), while soil 2 (acidic soil) was much paler, with a fine, sandy consistency and an acidic pH (5.5), and contained 45% less organic carbon. Each soil had different levels of phosphorus, potassium, and magnesium.
TABLE 1.
Chemical analysis of the two soils used for burial of PU coupons
| Soil | pH | Content in soil:
|
|||
|---|---|---|---|---|---|
| Organic carbon (% [wt/wt]) | Phosphorus (mg/liter) | Potassium (mg/liter) | Magnesium (mg/liter) | ||
| 1 (neutral) | 6.7 | 7.85 | 37 | 171 | 244 |
| 2 (acidic) | 5.5 | 4.34 | 141 | 243 | 119 |
Total viable and putative PU-degrading fungi recovered from soil and from the surface of buried PU.
The numbers of viable fungi and putative PU-degrading fungi in the two soils and on the surface of PU coupons buried for 5 months were determined (Table 2). Zones of clearance on Impranil agar were very obvious and extended <1 cm outwards from the colony edge. There was no significant difference in the numbers of viable fungi in the two soils (P < 0.05). However, 5.5-fold more fungal CFU were recovered from the surface of PU buried in the acidic soil (5.5 × 103) compared to PU buried in the neutral soil (9.9 × 102). In the acidic soil, there was no significant difference (P > 0.05) between the percentages of Impranil-degrading fungi on PU (41.2%) compared with the soil itself (37.4%), whereas in the neutral soil there was a significant increase (P < 0.05) in the percentage of Impranil-degrading fungi on the PU (58.5%) compared to the soil (45.1%). The ability to degrade colloidal PU was a very common property among the fungi in all of the communities investigated, ranging from 37.4% of the fungi in the acidic soil communities to 58.5% in the communities growing on the surface of PU buried in neutral soil.
TABLE 2.
Total numbers of viable fungi and percentages of viable fungi able to degrade Impranil in soil and on the surface of buried PU
| Soil type | Viable count or Impranil clearance for fungia:
|
|||||
|---|---|---|---|---|---|---|
| In soil
|
On surface of buried PU
|
|||||
| Viable (CFU g−1) | Impranil degrading (CFU g−1) | Colonies clearing Impranil (%) | Viable (CFU cm−2) | Impranil degrading (CFU cm−2) | Colonies clearing Impranil (%) | |
| Neutral | 6.3 × 105 | 2.9 × 105 | 45.1 | 9.9 × 102 | 5.7 × 102 | 58.5 |
| Acidic | 5.5 × 105 | 2.0 × 105 | 37.4 | 5.5 × 103 | 2.3 × 103 | 41.2 |
In all cases, n = 3.
Community analysis using DGGE.
Sequence-dependent separation of PCR-amplified ITS1 rRNA genes using DGGE was used to analyze and compare the species compositions of fungal communities in each type of soil (the native soil communities) and communities colonizing the surface of the PU after 5 months of burial (Fig. 1). The band migration behaviors in each DGGE profile revealed clear differences in the species compositions of all of the communities investigated (Fig. 1). Approximately 35 to 40 bands were present in each native soil community profile (Fig. 1, lanes A and C), indicating a considerable diversity of fungi in these consortia. Only a few bands migrated to the same position in the DGGE profiles of both types of soil, indicating that the two communities were distinct, with the majority of the detectable fungi unique to each soil type. Replicate soil samples from both soils gave highly reproducible DGGE profiles (data not shown), indicating that there was spatial homogeneity in the fungal communities in these soils.
FIG. 1.
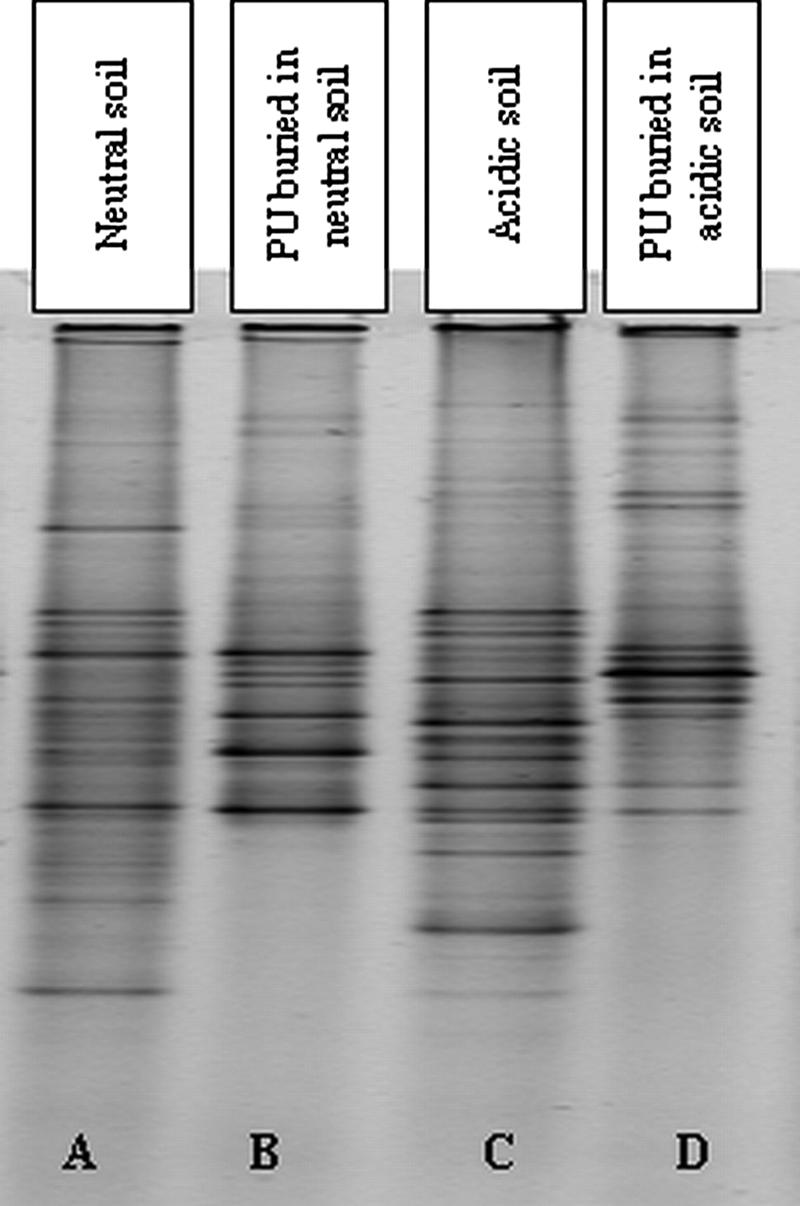
Comparison of DGGE profiles of soil fungal communities and fungal communities growing on the surface of PU buried in both soil types.
When the fungal communities on the plastic surface (Fig. 1, lanes B and D) were compared to those in the soil (Fig. 1, lanes A and C), fewer bands were visible in profiles of plastic communities (≤30) compared to soil communities (≤40), indicating a lower diversity of fungi on the surface of the buried PU. Furthermore, many of the bands from the PU community profiles were not detectable in the corresponding native soil community DGGEs, with ≤5 bands in either of the PU-associated community profiles also visible in their respective soil profiles. Thus, only a small number of specific members of the native soil fungal communities were enriched for during growth on buried plastic, with many of these fungi being minor members of the native soil communities.
Identification of isolates recovered from the surface of soil-buried PU by ITS sequencing and phylogenetic analysis.
In order to identify cultivable fungi colonizing the surface of buried PU, the predominant colony morphotypes were isolated from the SEA plates used to count the viable fungi on the surface of buried PU. Isolates were subcultured onto Impranil agar to determine putative PU-degrading ability, and their identities were determined by ITS sequencing. In total, nine distinct colony morphotypes were recovered: five from PU in acidic soil and four from PU in neutral soil (Table 3). The two most dominant fungi recovered from PU in acidic soil (ASIGP1 and ASIN2) had the highest homology to Geomyces pannorum and a Nectria sp., respectively. Fungi present in lower numbers on PU from acidic soil (ASICP1, ASIPI1, and ASIPC1) had the highest homology to Cylindrocladiella parva, Penicillium inflatum, and Plectosphaerella cucumerin, respectively. G. pannorum and P. inflatum from PU in acidic soil were able to clear Impranil.
TABLE 3.
Fungi isolated and purified from the surface of PU coupons buried in neutral and acidic soil as identified via ITS1-5.8S-ITS sequence homology
| Isolatea | Closest match in EMBL database | Soil type | Observed frequency of morphotype on SEA platesb | % Homology | Impranil clearancec |
|---|---|---|---|---|---|
| ASIGP1 (DQ779788) | Geomyces pannorum (AF015789) | Acidic | +++ | 99.2 | + |
| ASIN2 (DQ779785) | Nectria sp. strain BC11 (DQ317342) | Acidic | ++ | 100 | − |
| ASICP1 (DQ779786) | Cylindrocladiella parva (AY793455) | Acidic | + | 99.6 | − |
| ASIPI1 (DQ779783) | Penicillium inflatum (AY373920) | Acidic | + | 99.7 | + |
| ASIPC1 (DQ779781) | Plectosphaerella cucumerina (AJ246154) | Acidic | + | 99.8 | − |
| NSIA1 (DQ779787) | Alternaria sp. strain 18-2 (AY148445)d | Neutral | +++ | 98.8 | + |
| NSIP2 (DQ779784) | Penicillium venetum (AY373939) | Neutral | + | 100 | − |
| NSINR1 (DQ779780) | Neonectria ramulariae (AY677291) | Neutral | + | 99.1 | + |
| NSIPV1 (DQ779779) | Penicillium viridicatum (AY373935) | Neutral | + | 99.11 | + |
GenBank accession numbers for each sequence are given in parentheses. Strains with the prefix AS are from acid soil, and those with the prefix NS are from the neutral soil.
Subjective measure of the frequency with which each morphotype was observed on the SEA plates: +, morphology of <5% of the colonies; ++, morphologies of 10 to 20% of the colonies present; +++, the dominant morphotype, representing >80% of colonies present on the plates.
Production of clear zones on Impranil agar.
Subsequently named as a Phoma sp. strain by phylogenetic analysis.
The most dominant fungal isolate recovered from PU in neutral soil (NSIA1) was most homologous to an Alternaria sp. Isolates NSIP2, NSINR1, and NSIPV1 were recovered in much smaller numbers and were most homologous to Penicillium venetum, Neonectria ramulariae, and Penicillium viridicatum, respectively. Alternaria sp., N. ramulariae, and P. viridicatum from PU in neutral soil were able to clear Impranil.
Phylogenetic analysis (Fig. 2) confirmed the genus and species EMBL database identifications for eight of the nine strains. However, in the case of Alternaria sp. (NSIA1), phylogenetic analyses showed that this strain clustered with members of the genus Phoma. Strain NSIA1 will therefore be referred to as a Phoma sp. strain.
FIG. 2.
Phylogenetic analysis of isolates recovered from the surface of buried PU (boldface). ITS1-5.8S-ITS2 sequences from the isolates were compared to putatively closely related species using a maximum parsimony phylogenetic tree (bootstrap corrected with 1,000 samples). The tree is rooted using the outlying fungi Boletus satanas, Russula compacta, Leucostoma persoonii, Aspergillus fumigatus, Candida albicans, and Kluyveromyces lactis.
DGGE analysis of isolates recovered from the surface of buried PU.
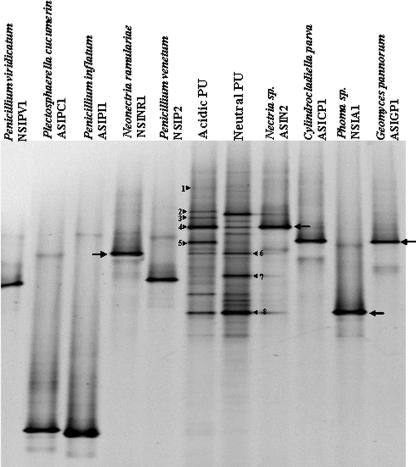
Each of the isolates from the surface of the buried PU was subjected to DGGE, and their band positions were compared to the DGGE profiles of the fungal communities growing on the surface of the buried PU (Fig. 3). All of the pure culture isolates produced a single intense band after DGGE, with only faint secondary bands indicating that no significant heterogeneities existed between ITS1 copies within a single organism.
FIG. 3.
Comparison of DGGE bands produced by fungi isolated in pure culture from buried PU with bands produced by fungal communities growing on PU buried in neutral and acidic soils. DGGE bands of the isolates indicated by lined arrows migrate to the same position as bands within the DGGE profiles of fungal communities from PU. Numbered bands were cloned into E. coli, sequenced, and identified.
Of the nine colony morphotypes recovered, N. ramulariae, the Nectria sp., the Phoma sp., and G. pannorum migrated to the same position as bands in the PU community profiles (lined arrows in Fig. 3). G. pannorum and the Nectria sp. were the dominant cultivable fungi on PU in acidic soil (Table 3), and these isolates produced bands that comigrated with the most intense bands in the DGGE profile of the fungal community on PU buried in acidic soil. Similarly, the Phoma sp., which was the dominant cultivable organism on PU buried in neutral soil, produced a DGGE band that comigrated to the same position as the most intense band in the DGGE profile of the fungal community on PU buried in neutral soil.
In addition, four fungi (G. pannorum, N. ramulariae, the Nectria sp., and the Phoma sp.) produced clear bands in the DGGE profiles of fungal communities on PU buried in both soil types and probably represent fungi well adapted to growth on the surface of PU. However, the bands representing these isolates differed in intensity between the two community profiles, indicating that soil type affected the abundance of these potentially well-adapted isolates.
The remaining five isolates (P. viridicatum, P. cucumerin, P. inflatum, P. venetum, and C. parva) did not comigrate with any of the bands in either of the PU community profiles. All of these colony morphotypes were recovered on SEA plates in low numbers from the PU. There were also numerous bands within each PU community profile that did not comigrate with any of the isolated cultivable species, and some of these bands were very intense, suggesting that they represented species that are noncultivable on SEA plates but were important members of the PU community.
Identifying community members from DGGE amplicons.
In order to identify the fungi on PU buried in the acidic and neutral soils by a cultivation-independent method, PCR using the DGGE primers was performed on DNA from fungal communities colonizing the surface of buried PU. DGGE-PCR products were cloned into E. coli, and over a hundred transformants were screened by DGGE. In total, eight different ITS1 sequences that migrated to different positions on the DGGE gel were recovered: five from fungi on the surface of PU buried in the acidic soil and three from PU buried in the neutral soil. These fragments were then sequenced in order to determine their putative identities. Of the eight ITS1 fragments cloned, four were found to produce bands (Fig. 3, bands 4, 5, 6, and 8) that migrated to the same position as bands produced by the isolates Nectria sp., G. pannorum, N. ramularia, and Phoma sp. Sequencing revealed that these clones also had 100% homology to these isolates.
Of the four remaining cloned ITS1 fragments (Table 4), three (Fig. 3, bands 1 [faint], 3 and 7) returned no significant matches upon database interrogation (≤93% homology) or were homologous to uncultured soil fungi. Also, bands produced from these clones (data not shown) did not comigrate with any of the cultivable isolates, indicating that these sequences represented noncultivable members of the PU fungal community. The final clone (Fig. 3, band 2) was putatively identified as a Sarcosomatacea sp. Colonies with a morphotype typical of this species were not isolated from the surface of PU buried in either soil type.
TABLE 4.
Identification of noncultivable members of the fungal communities colonizing PU buried in acidic and neutral soila
| Clone no.b | Soil type for bands in community DGGE profiles from buried PU | Putative identity | % Homology |
|---|---|---|---|
| 1 (DQ779777) | Acidic | No matches in database | <90 |
| 2 (DQ779774) | Acidic | Sarcosomataceae sp. strain sd2bN1c | 100 |
| 3 (DQ779777) | Acidic | Uncultured soil fungus | 99.7 |
| 7 (DQ779778) | Neutral | Uncultured ascomycete | 98.1 |
ITS1 fragments recovered from bands excised from DGGE profiles of PU communities were sequenced and used to interrogate the EMBL database.
For details about the clone numbers, see Fig. 3. GenBank accession numbers for each sequence are given in parentheses.
Degradation of PU buried in two soil types for 5 months.
The degree of degradation of PU after 5 months of burial was determined by measuring the tensile strength of the PU coupons. As cleavage of the PU backbone during degradation weakens the plastic, the extent of degradation is inversely proportional to tensile strength. Burial of PU in either the neutral or the acidic soil led to severe degradation after 5 months. The tensile strength of the control PU before burial was approximately 360 N, while after burial, its tensile strength had decreased approximately 15-fold in both soils. Thus, PU degradation was extensive in both soil types.
DISCUSSION
This work has studied the fungal communities associated with in situ degradation of PU in natural soils, and degradation was extensive in both soils tested, with the PU losing up to 95% of its tensile strength after 5 months. PU is known to be highly susceptible to degradation in a number of laboratory microcosm studies (5, 6, 13); however, this is the first work to quantify the degradation of PU in situ in the environment. We previously reported (5) that the maximum loss of tensile strength of PU after 1.5 months of burial in laboratory microcosms was <60%, compared with 95% in this study. Although PU in this work was buried for a much longer period of time, much of the burial period was during the winter and early spring months, when soil temperatures and fungal activity are low and are likely to retard degradation of PU compared to the 25°C laboratory microcosm (5). Nonetheless, the extensive PU degradation in this in situ study, even under suboptimal conditions, suggests biodegradation of PU in waste remediation would occur under a variety of landfill conditions.
A number of previous studies of PU degradation have focused on bacterial degraders (1, 20, 23), isolated using enrichment and screening strategies. Although bacteria were recovered from the PU surface after burial in this study, only very few could degrade Impranil, with very narrow, faint clearance zones (data not presented). Previously, we found that PU pieces buried for 44 days in a laboratory soil microcosm had bacterial counts on them of <107 CFU cm−2, but only 2 Impranil-degrading colonies were ever found (5). In addition, there are many more reports of fungal species being isolated from the surface of PU in comparison to bacterial species (12, 28, 31, 36).
A high percentage of the cultivable fungi from the acidic and neutral soil (37% and 45%, respectively) were putative PU degraders, a proportion similar to that reported previously for a laboratory soil microcosm (5). Environmental soils therefore contain a large reservoir of fungi with the potential to degrade PU. PU contains many molecular bonds that are analogous to those found in biological macromolecules, and fungal genes encode a broad range of secreted hydrolases, increasing the likelihood of fortuitous PU degradation due to such enzymes (26).
The most dominant cultivable organism isolated from the surface of PU buried in the neutral soil was identified by phylogenetic analysis as a Phoma sp., while G. pannorum and to a lesser extent a Nectria sp. were the dominant cultivable fungi on PU buried in the acidic soil. G. pannorum was the dominant fungus in plasticized polyvinyl chloride (pPVC) degradation in a laboratory microcosm (5), and it was also important in pPVC degradation in Bulgarian grassland soil (32). This fungus may therefore prove to be important for plastic waste remediation in the future.
The Phoma sp. and G. pannorum cleared Impranil, but other PU isolates found in smaller numbers lacked this ability. Thus, only some members of the PU community could degrade the polymer. Previous longitudinal studies on the colonization of plasticized pPVC buried in soil (32) and pPVC exposed to the air (41) showed that early colonizers degraded the plasticizer, but other nondegrading fungi appeared later in community development. We suggested that breakdown products from the primary colonizers might act as a carbon source for nondegraders, which could also explain the presence of nondegraders on the PU after 5 months of burial in the present study.
Since culture-based techniques have limited use in identification and quantification of fungi (11, 29, 43), we used DGGE to study the composition of the PU and soil communities. DGGE has been used to analyze fungal communities from a variety of environments (8, 9, 17, 38, 40), and here DGGE revealed that only a subset of the fungal species present in either soil were present on the buried PU (Fig. 1). Both culture-based methods and the DGGE profiles showed that the two soils possessed distinct fungal communities which resulted in different fungal species colonizing the buried PU. Also, DGGE showed that the PU community was different from the surrounding soil community, indicating an enrichment of species that colonized and/or degraded PU (Fig. 1). Soil conditions influence the composition of fungal communities on the surface of buried pPVC, and in forest soil, pPVC supported a different range of fungi compared to pPVC in grassland soil (32). Also, changing the water-holding capacity within the same soil also altered the fungal communities on buried PU (5). Soil organic carbon content and pH influence the structure of soil microbial communities (14, 16), and the soils used here differed in these parameters (Table 1). Attachment of microorganisms to buried PU is mediated by nonspecific hydrophobic interactions (7), and local environmental conditions influence the surface hydrophobicity of fungi (35) and bacteria (3). Therefore, differences in the physicochemical properties of the two soils may influence which microbes successfully colonize the surface of the PU.
Only a very few of the putative PU-degrading fungi in the soils colonized the surface of the PU. It has been suggested that some enzymes that degrade the colloidal PU dispersion Impranil are unable to degrade solid PU due to physiochemical differences between the two forms of the plastic (1). Therefore, some putative PU-degrading fungi defined by the Impranil clearing assay may not grow on and degrade PU. However, the Impranil clearance assay is the only method available to detect the potential PU-degrading abilities of both bacteria and fungi.
The three major species isolated from the surface of buried PU (G pannorum, a Phoma sp., and a Nectria sp.) produced bands that comigrated with bands from the whole PU community DGGE profile for each soil. However, some clear DGGE bands were not represented by any recovered isolate, indicating that important members of the PU community were not cultivable. Only about 17% of fungi in the environment can be grown in culture (19), and in this study, we found three unidentifiable fungi when DGGE amplicons were transformed into E. coli and the inserts screened by DGGE. DGGE did not detect PU-degrading fungi present in low numbers on SEA, and this insensitivity has been well documented, with estimates of the detection threshold varying between 0.1% (39) and 5% (22) of the total population However, such rare members of the fungal communities are unlikely to contribute significantly to the degradation of the PU.
This study has extended our knowledge of fungi with the potential to degrade PU under different environmental conditions. However, the ability of fungi to biodegrade plastics has not yet been exploited to its full potential, and the development of microbial consortia with proven biodegradation properties could improve plastic waste reduction and should be investigated further.
Acknowledgments
This study was supported by a Biotechnology and Biological Sciences Research Council (BBSRC) CASE award provided by Arch UK Biocides Ltd., United Kingdom.
The expertise of Roland Ennos in the use of the Instron for tensile strength measurements is gratefully acknowledged.
Footnotes
Published ahead of print on 27 July 2007.
REFERENCES
- 1.Akutsu, Y., T. Nakajima-Kambe, N. Nomura, and T. Nakahara. 1998. Purification and properties of a polyester polyurethane-degrading enzyme from Comamonas acidovorans TB-35. Appl. Environ. Microbiol. 64:62-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alef, K., and P. Nannipieri. 1995. Methods in applied soil microbiology and biochemistry. Academic Press, London, United Kingdom.
- 3.An, Y. H., and R. J. Friedman. 1998. Concise review of mechanisms of bacterial adhesion to biomaterial surfaces. J. Biomed. Mater. Res. 43:338-348. [DOI] [PubMed] [Google Scholar]
- 4.Anderson, M. J., K. Gull, and D. W. Denning. 1996. Molecular typing by randomly amplified polymorphic DNA and M13 Southern hybridization of related paired isolates of Aspergillus fumigatus. J. Clin. Microbiol. 34:87-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barratt, S. R., A. R. Ennos, M. Greenhalgh, G. D. Robson, and P. S. Handley. 2003. Fungi are the predominant micro-organisms responsible for the degradation of soil-buried polyester polyurethane over a range of soil water holding capacities. J. Appl. Microbiol. 94:1-8. [DOI] [PubMed] [Google Scholar]
- 6.Bentham, R. H., L. G. H. Morton, and N. G. Allen. 1987. Rapid assessment of the microbial deterioration of polyurethanes. Int. Biodeterior. Biodegrad. 23:377-386. [Google Scholar]
- 7.Bos, R., H. C. van der Mei, and H. J. Busscher. 1999. Physico-chemistry of initial microbial adhesive interactions—its mechanisms and methods for study. FEMS Microbiol. Rev. 23:179-230. [DOI] [PubMed] [Google Scholar]
- 8.Bougoure, D. S., and J. W. G. Cairney. 2005. Assemblages of ericoid mycorrhizal and other root-associated fungi from Epacris pulchella (Ericaceae) as determined by culturing and direct DNA extraction from roots. Environ. Microbiol. 7:819-827. [DOI] [PubMed] [Google Scholar]
- 9.Bougoure, D. S., and J. W. G. Cairney. 2005. Fungi associated with hair roots of Rhododendron lochiae (Ericaceae) in an Australian tropical cloud forest revealed by culturing and culture-independent molecular methods. Environ. Microbiol. 7:1743-1754. [DOI] [PubMed] [Google Scholar]
- 10.Bouwer, E. J. 1992. Bioremediation of organic contaminants in the subsurface, p. 287-318. In R. Mitchell (ed.), Environmental microbiology. Wiley-Liss, New York, NY.
- 11.Bridge, P., and B. Spooner. 2001. Soil fungi: diversity and detection. Plant Soil 232:147-154. [Google Scholar]
- 12.Crabbe, J. R., J. R. Campbell, L. Thompson, S. L. Walz, and W. W. Schultz. 1994. Biodegradation of colloidal ester-based polyurethane by soil fungi. Int. Biodeterior. Biodegrad. 33:103-113. [Google Scholar]
- 13.Dale, R., and D. J. Squirrell. 1990. A rapid method for assessing the resistance of polyurethanes to biodeterioration. Int. Biodeterior. Biodegrad. 26:355-367. [Google Scholar]
- 14.Drenovsky, R. E., D. Vo, K. J. Graham, and K. M. Scow. 2004. Soil water and organic carbon availability are major determinants of soil microbial community composition. Microb. Ecol. 48:424-430. [DOI] [PubMed] [Google Scholar]
- 15.Eggins, H. O., J. Mills, A. Holt, and G. Scott. 1971. Biodeterioration and biodegradation of synthetic polymers. Soc. Appl. Bacteriol. Symp. Ser. 1:267-279. [DOI] [PubMed] [Google Scholar]
- 16.Garbeva, P., J. A. van Veen, and J. D. van Elsas. 2004. Microbial diversity in soil: selection of microbial populations by plant and soil type and implications for disease suppressiveness. Annu. Rev. Phytopathol. 42:243-270. [DOI] [PubMed] [Google Scholar]
- 17.Gomes, N. C. M., O. Fagbola, R. Costa, N. G. Rumjanek, A. Buchner, L. Mendona-Hagler, and K. Smalla. 2003. Dynamics of fungal communities in bulk and maize rhizosphere soil in the tropics. Appl. Environ. Microbiol. 69:3758-3766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Griffin, G. J. L. 1980. Synthetic polymers and the living environment. Pure Appl. Chem. 52:399-407. [Google Scholar]
- 19.Hawksworth, D. L. 1991. The fungal dimension of biodiversity: magnitude, significance and conservation. Mycol. Res. 95:641-655. [Google Scholar]
- 20.Howard, G. T., C. Ruiz, and N. P. Hilliard. 1999. Growth of Pseudomonas chlororaphis on a polyester-polyurethane and the purification and characterization of a polyurethanase-esterase enzyme. Int. Biodeterior. Biodegrad. 43:7-12. [Google Scholar]
- 21.Kawai, F. 1995. Breakdown of plastics and polymers by microorganisms. Adv. Biochem. Eng. Biotechnol. 52:151-194. [DOI] [PubMed] [Google Scholar]
- 22.Kawai, F. 1995. Bacterial degradation of glycol ethers. Appl. Microbiol. Biotechnol. 44:532-538. [DOI] [PubMed] [Google Scholar]
- 23.Kay, M. J., L. H. G. Morton, and E. L. Prince. 1991. Bacterial degradation of polyester polyurethane. Int. Biodeterior. Biodegrad. 27:205-222. [Google Scholar]
- 24.Kumar, S. T. K., and M. Nei. 2004. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief. Bioinform. 5:150-163. [DOI] [PubMed] [Google Scholar]
- 25.Muyzer, G., E. C. de Waal, and A. G. Uitterlinden. 1993. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 59:695-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nakajima-Kambe, T., Y. Shigeno-Akutsu, N. Nomura, F. Onuma, and T. Nakahara. 1999. Microbial degradation of polyurethane, polyester polyurethanes and polyether polyurethanes. Appl. Microbiol. Biotechnol. 51:134-140. [DOI] [PubMed] [Google Scholar]
- 27.Pathirana, R. A., and K. J. Seal. 1985. Studies on polyurethane deteriorating fungi. Int. Biodeterior. Biodegrad. 21:41-49. [Google Scholar]
- 28.Pommer, E. H., and G. Lorenz. 1985. The behaviour of polyester and polyether polyurethanes towards microorganisms, p. 77-86. In K. J. Seal (ed.), Biodeterioration and biodegradation of polymers. International Biodeterioration and Biodegradation Society, Manchester, United Kingdom.
- 29.Pugh, G. J. F. 1969. Some problems in the classification of soil fungi, p. 119-130. In J. G. Sheals (ed.), The soil ecosystem, a symposium. Systematics Association publication no. 8. Academic Press, London, United Kingdom.
- 30.Ragg, J. M., G. R. Beard, H. George, F. W. Heaven, J. M. Hollis, R. J. A. Jones, R. C. Palmer, M. J. Reeve, J. D. Robson, and W. A. D. Whitfield. 1984. Soils and their use in Midland and Western England: soil survey of England and Wales, bulletin no. 12. Lawes Agricultural Trust, Whitstable, Kent, United Kingdom.
- 31.Sabev, H. A., S. R. Barratt, M. Greenhalgh, P. S. Handley, and G. D. Robson. 2006. Biodegradation of manmade polymeric materials, p. 212-235. In G. M. Gadd (ed.) Fungi in geochemical cycles. Cambridge University Press, Cambridge, United Kingdom.
- 32.Sabev, H. A., P. S. Handley, and G. D. Robson. 2006b. Fungal colonization of soil-buried plasticized polyvinyl chloride (pPVC) and the impact of incorporated biocides. Microbiology 152:1731-1739. [DOI] [PubMed] [Google Scholar]
- 33.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 34.Shimao, M. 2001. Biodegradation of plastics. Curr. Opin. Biotechnol. 12:242-247. [DOI] [PubMed] [Google Scholar]
- 35.Smits, T. H. M., L. Y. Wick, H. Harms, and C. Keel. 2003. Characterization of the surface hydrophobicity of filamentous fungi. Environ. Microbiol. 5:85-91. [DOI] [PubMed] [Google Scholar]
- 36.Stranger-Johannessen, M. 1985. Microbial degradation of polyurethane products in service, p. 93-102. In K. J. Seal (ed.), Biodeterioration and biodegradation of polymers. International Biodeterioration and Biodegradation Society, Manchester, United Kingdom.
- 37.Tuckwell, D. S., M. J. Nicholson, C. S. McSweeney, M. K. Theodorou, and J. L. Brookman. 2005. The rapid assignment of ruminant fungi to presumptive genera using ITS1 and ITS2 RNA secondary structures to produce group-specific fingerprints. Microbiology 151:1557-1567. [DOI] [PubMed] [Google Scholar]
- 38.Vainio, E. J., and J. Hantula. 2000. Direct analysis of wood-inhabiting fungi using denaturing gradient gel electrophoresis of amplified ribosomal DNA. Mycol. Res. 104:927-936. [Google Scholar]
- 39.van Elsas, J. D., G. F. Duarte, A. Keijzer-Wolters, and E. Smit. 2000. Analysis of the dynamics of fungal communities in soil via fungal-specific PCR of soil DNA followed by denaturing gradient gel electrophoresis. J. Microbiol. Methods 43:133-151. [DOI] [PubMed] [Google Scholar]
- 40.Viebahn, M., C. Veenman, K. Wernars, L. C. van Loon, E. Smit, and P. A. H. M. Bakker. 2005. Assessment of differences in ascomycete communities in the rhizosphere of field-grown wheat and potato. FEMS Microbiol. Ecol. 53:245-253. [DOI] [PubMed] [Google Scholar]
- 41.Webb, J. S., M. Nixon, I. M. Eastwood, M. Greenhalgh, G. D. Robson, and P. S. Handley. 2000. Fungal colonization and biodeterioration of plasticized polyvinyl chloride. Appl. Environ. Microbiol. 66:3194-3200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Woods, G. 1990. The ICI polyurethanes book, 2nd ed. John Wiley and Sons, Chichester, United Kingdom.
- 43.Zak, J. C., and S.Visser. 1996. An appraisal of soil fungal biodiversity: the crossroads between taxonomic and functional biodiversity. Biodivers. Conserv. 5:169-183. [Google Scholar]