Abstract
This study identified major surface proteins of the plague bacterium Yersinia pestis. We applied a novel surface biotinylation method, followed by NeutrAvidin (NA) bead capture, on-bead digestion, and identification by liquid chromatography-tandem mass spectrometry (LC-MS-MS). The use of stachyose during biotinylation focused the reaction to the surface. Coupled with NA pulldown and immunoblot analysis, this method determined whether a protein was accessible to the surface. We applied the method to test the hypothesis that the catalase KatY is a surface protein of the plague bacterium Y. pestis. A rabbit serum recognized the catalase KatY as a major putative outer membrane-associated antigen expressed by Y. pestis cells grown at 37°C. Similar findings by other groups had led to speculations that this protein might be exposed to the surface and might be a candidate for evaluation as a protective antigen for an improved plague vaccine. KatY was obtained only in the total membrane fraction, and stachyose greatly reduced its biotinylation as well as that of the periplasmic maltose binding protein, indicating that KatY is not on the bacterial surface. LC-MS-MS analysis of on-bead digests representing ca. 109 cells identified highly abundant species, including KatY, Pal, and OmpA, as well as the lipoprotein Pcp, all of which bound in a biotin-specific manner. Pla, Lpp, and OmpX (Ail) bound to the NA beads in a non-biotin-specific manner. There was no contamination from abundant cytoplasmic proteins. We hypothesize that OmpX and Pcp are highly abundant and likely to be important for the Y. pestis pathogenic process. We speculate that a portion of KatY associates with the outer membrane in intact cells but that it is located on the periplasmic side. Consistent with this idea, it did not protect C57BL/6 mice against bubonic plague.
Yersinia pestis is the causative agent of plague and is thought to have acquired virulence factors and transmission factors to adapt for survival within its mammalian host and transmission by fleabite (7). Investigators in our laboratory are identifying surface proteins and their pathogenic functions in Y. pestis to gain an improved understanding of the physiology of virulence as well as to identify potential prophylactic and therapeutic targets for use against plague. In this study, we evaluated two approaches for characterizing a surface proteome of Y. pestis, with the goal of developing a method that can be applied to small numbers of yersiniae recovered from the lungs of experimental animals with pneumonic plague.
Previously, the extraction method of Filip et al. (16), based on the differential solubilities of inner and outer membrane proteins in sodium lauryl sarcosinate (Sarkosyl), has been used in several studies to obtain preparations of outer membrane proteins from gram-negative bacteria, including Y. pestis (e.g., see references 1, 2, 37, and 39). The method is rapid and simple but results in protein profiles comparable to those obtained by the classic isopycnic centrifugation method (35) and is amenable to downscaling to handle small samples (e.g., see references 9 and 39). The outer membrane proteins can then be resolved by two-dimensional (2-D) electrophoresis and analyzed further for surface localization in immunoblots by using antibody probes that bind to the surfaces of intact bacteria (e.g., see reference 14) or by comparing outer membrane protein profiles from intact bacteria subjected to proteolysis to those from mock-treated bacteria (e.g., see reference 40).
However, the identification of the entire surface proteome has presented a major challenge. Procedures to obtain outer membrane proteins after bacterial lysis (as in the Sarkosyl method) introduce the serious problem of contamination by abundant cytoplasmic proteins, such as ribosomal proteins and EF-Tu, that confound “blind” identification by mass spectrometry (MS). Further, any method that includes initial resolution of proteins in acrylamide gels has the drawback of missing those proteins that fail to enter the first-dimension gel due to reduced solubility in the sample buffer or to the drift of the alkaline pH extreme of the gel, and few proteins larger than ca. 100 kDa are detected, even though they are predicted potentially to be present (e.g., see reference 33). The outer membranes of gram-negative bacteria present the further challenge that the protein profile is dominated by relatively few proteins, limiting the detection of low-abundance proteins, regardless of the approach taken (see Discussion).
We believe that more than one method will need to be applied to obtain a comprehensive picture of the surface proteome of Y. pestis. In this study, we applied two approaches: the identification of immunogenic species in 2-D separations of a Sarkosyl-based preparation and a novel biotinylation method that focuses on the identification of proteins that are exposed to the surface. We validated the methods for proteins with known localization patterns and applied them to evaluate the surface localization of the 79-kDa catalase KatY (encoded by Sanger Centre coding sequence YPO3319 of Y. pestis CO92).
Originally discovered by Crumpton and Davies as antigen 5 (13), KatY is a major antigen that is present in Y. pestis cells grown at 37°C and was among the proteins identified by sera from rabbits immunized with live Y. pestis EV76 (27). Garcia et al. and Mehigh and Brubaker (18, 30) characterized it biochemically, reported that it is a weak catalase, and gave it its name. Interestingly, KatY is a major species among a set of virulence-associated proteins that are expressed at 37°C under conditions yersiniae may experience within tissues during bubonic plague (8, 12, 18). Accordingly, KatY is of great interest for vaccine development, because it may be surface located and antibody against it may protect against plague.
MATERIALS AND METHODS
Bacterial strains and growth.
The Y. pestis CO99-3015.S2 caf1 strain lacking the fibrillar capsule F1 and the Lcr virulence plasmid pCD2 (S. Forman, C. R. Wulff, S. A. Leigh, R. D. Perry, and S. C. Straley, unpublished data) was used for most of the work in this study. To test the localization of the pCD2-encoded secretin YscC, the Y. pestis CO99-3015.S5 Lcr+ Δpgm strain (Scott Bearden, Centers for Disease Control and Prevention [CDC], Ft. Collins, CO) was used. Experiments to examine F1 used the Y. pestis CO99-3015.S5 F1+ Lcr− strain (Scott Bearden, CDC, Ft. Collins, CO). For the identification of proteins from 2-D gels, Y. pestis CO92.S6 was used. This strain, derived from Y. pestis CO92 Lcr− (lacking pCD2), kindly provided by Luther Lindler (Walter Reed Army Institute of Research), contained the H208V active-site mutation in Pla (25). Pla is expressed normally by Y. pestis CO92.S6 but is proteolytically inactive, judging by its inability to lyse a fibrin film (S. C. Straley, C. Wulff, M. E. Gray, C. Cowan, R. D. Perry, and T. K. Korhonen, unpublished data). Vaccinated mice were challenged with fully virulent Y. pestis CO99-3015.S1 (CDC, Ft. Collins, CO).
The yersiniae were grown in heart infusion broth (Difco Laboratories, Detroit, MI) supplemented with 2.5 mM CaCl2 and 0.2% (wt/vol) xylose (sHIB) for at least six generations in exponential phase at 37°C with shaking at 200 rpm. On the last transfer, they were allowed to reach an optical density at 620 nm (OD620) of 1 to 2 (5 × 108 to 1 × 109 cells/ml) for use in experiments. Escherichia coli DH5α (Life Technologies, Gaithersburg, MD) was used for the cloning of the KatY-His6 tag (KatY-HT) gene and the production of KatY-HT and was grown in Luria-Bertani broth (31) at 37°C with shaking at 200 rpm or on Luria-Bertani agar. For the cloning of the KatY-HT gene and the production of KatY-HT, 100 μg of carbenicillin/ml was present in the medium.
Construction of pKatY and purification of KatY-HT.
YPO3319 (katY) was copied by PCR using ProofStart polymerase (QIAGEN, Inc., Valencia, CA) and primers KatYNcoIUp (AGTCATCCATGGAATTAAAAAAAATCTTACCCGTACTAATAACTCTCGCCATTGT) and KatYHTSalIDown (TATATGTCGACTTAATGATGATGATGATGATGGTTATTTTTTATATCAAAGCGATCAAGGTTCAT), which added NcoI and SalI restriction sites (underlined), respectively, to the product and a sequence corresponding to a C-terminal His6 tag for KatY (double underlined). The product was digested with NcoI and SalI and cloned into NcoI-SalI-digested pTrc99A (Amersham Pharmacia Biotech, Piscataway, NJ). The final clone, pKatY-HT, encoded the preprotein that was predicted to be secreted into the periplasm in E. coli and was confirmed to be correct by restriction analysis (using NcoI and SalI) and the production of the correct-sized 79-kDa protein product that was detected with anti-KatY monoclonal antibody (gift of Robert Perry, University of Kentucky, who received it from Robert Brubaker, Michigan State University) (18).
To produce semipurified KatY-HT, E. coli DH5α(pKatY-HT) was grown to an OD620 of 1.0, IPTG (isopropyl-β-d-thiogalactopyranoside) was added to a concentration of 0.5 mM, and incubation was continued for 3 h. The cells were recovered by centrifugation, washed once with cold phosphate-buffered saline (PBS; 2.7 mM KCl, 1.2 mM KH2PO4, 138 mM NaCl, 8.1 mM Na2HPO4 [pH 7.4]), and resuspended in high-salt buffer (500 mM NaCl, 50 mM Tris HCl [pH 8.0], 10% [vol/vol] glycerol, and protease inhibitor cocktail for use with bacterial extracts [Sigma Chemical Co., St. Louis, MO]) at a 25-fold-greater concentration than in the original culture. The cells were broken by two passages through a chilled French press (pressure, 20,000 lb/in2), unbroken cells and large debris were removed by centrifugation at 13,800 × g and 4°C, and membranes were removed by ultracentrifugation for 20 min at 80,000 × g and 4°C. Nickel-nitrilotriacetic acid (Ni-NTA) resin (QIAGEN) that had been rinsed in high-salt buffer was added, mixed gently by rotation for 1 h at 4°C, and allowed to settle in a column. The column was washed with 20 column volumes of high-salt buffer containing 10 mM imidazole, and proteins were eluted with 5 column volumes containing 100 mM imidazole. Fractions were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and staining with Coomassie brilliant blue. The most concentrated elution fractions were dialyzed against PBS at 4°C, concentrated using a Centriprep 30 (Millipore Corp., Bedford, MA), and stored at −20°C in a PBS solution containing 5% glycerol and 1.5 mM dithiothreitol. The protein concentration was measured spectrophotometrically (at OD230 and OD260) (22). The final preparations also contained various amounts of a ca. 26-kDa contaminating protein and a faint non-KatY band at 32.5 kDa. The 26-kDa band was identified as the E. coli periplasmic chaperone FkpA by liquid chromatography followed by tandem MS (LC-MS-MS; performed by Carol Beach, Center for Structural Biology, University of Kentucky). Attempts to reduce the production of this band by growing the bacteria at a lower temperature or to remove the contaminant by treating the protein eluted from Ni-NTA with 0.3% SDS or 1% octylglucopyranoside followed by a second Ni-NTA chromatography step or by having 500 mM NaCl present during the entire purification procedure were not successful.
Preparation of outer membranes and outer membrane proteins.
Outer membranes were prepared by the isopycnic centrifugation method of Osborn et al. (35) as modified previously for Y. pestis (45). Protein amounts in soluble, inner membrane, and outer membrane fractions were determined by the method of Lowry et al. (29). Outer membrane proteins were isolated by the Sarkosyl procedure (16). Briefly, cells in an aliquot equivalent to 2 ml of culture at an OD600 of 1 (2 OD ml; ca. 109 cells) in 10 mM HEPES, pH 7.5, were disrupted by sonication at 4°C, large debris was removed by centrifugation at 1,000 × g for 2 min at 4°C, and total membranes were pelleted by ultracentrifugation at 337,000 × g for 15 min at 4°C. The pellet was resuspended in room temperature (RT) 0.3% (wt/vol) sodium laurylsarcosinate (Sarkosyl) in 10 mM HEPES, pH 7.5, and extracted by rotation at RT for 30 min. Insoluble material was again recovered by ultracentrifugation but at 20°C, and the pellet was dissolved in distilled water. The detergent was removed from the supernatant containing putative cytoplasmic membrane proteins and from the pellet containing putative outer membrane proteins by methanol-chloroform extraction. The aqueous phase was then extracted with methanol, and the pellet was dried and resuspended in twofold-concentrated SDS-PAGE sample buffer for SDS-PAGE or in rehydration buffer for isoelectric focusing with the ZOOM IPGRunner system (Invitrogen Corp., Carlsbad, CA). Protein amounts were determined by the bicinchoninic acid assay (Pierce Chemical Co., Rockford, IL).
For experiments to identify immunogenic membrane proteins from 2-D gels corresponding to the Sarkosyl-insoluble fraction, steps were taken to limit proteolysis and thereby reduce the complexity of the protein profile. The Y. pestis strain lacked active Pla, and immediately after the harvesting of bacteria from their culture medium, the cells were washed three times in 100 mM potassium phosphate buffer, pH 6.7, containing 1 mM MgCl2 and 1 mM phenylmethylsulfonyl fluoride to inhibit serine proteases. This wash step also removed loosely associated material from the bacterial surface.
One- and two-dimensional PAGE and visualization of proteins.
Proteins were resolved in one dimension by SDS-12% PAGE or on 4 to 15% acrylamide gradient gels (Ready gels; Bio-Rad Laboratories, Hercules, CA). Broad-range markers (New England Biolabs, Ipswich, MA) were used as molecular mass standards. Two-dimensional electrophoresis was performed using the ZOOM IPGRunner system with the ZOOM strip containing a nonlinear pH gradient from 3 to 10 according to the instructions of the manufacturer (Invitrogen) and 4 to 15% gradient Ready gels (Bio-Rad) for the second dimension. Molecular mass markers for 2-D gels were unstained Precision Plus protein standard plugs (Bio-Rad) or unstained precast SDS-PAGE molecular mass markers for 2-D gels from Genomic Solutions (Ann Arbor, MI). Proteins were visualized in gels by silver staining (41) or in immunoblots by probing with specific antibodies and using secondary antibodies coupled to horseradish peroxidase (HRP; for development by enhanced chemiluminescence [ECL] with the ECL Western blotting reagent set [GE Healthcare, Amersham Biosciences, Piscataway, NJ]) or to alkaline phosphatase (for development with nitroblue tetrazolium-5-bromo-4-chloro-3-indolylphosphate [Sigma]) for detection. When blots were to be probed for the detection of SecY, the samples were heated at 65°C for 15 min prior to electrophoresis; otherwise, samples were boiled for 5 min. Biotinylated proteins were detected in blots by NeutrAvidin (NA)-HRP (Pierce). Primary antibodies were rabbit anti-Pla (gift of Jon D. Goguen, University of Massachusetts Medical School), mouse anti-KatY (as noted above), rabbit anti-E. coli SecY (gift of Koreaki Ito, Institute for Virus Research, Kyoto University), rabbit anti-E. coli maltose binding protein (MBP; New England Biolabs), rabbit anti-E. coli GroEL (Sigma), mouse anti-F1 monoclonal antibody YPF1 (Research Diagnostics, Inc., Flanders, NJ), rabbit antibody raised against a potassium thiocyanate extract of Y. pestis KIM6 (Lcr−) grown at 37°C and pH 6 (28), and three human plague convalescent-phase sera (gift of Thomas Quan, CDC, Ft. Collins, in the 1980s). All three sera were positive for reactivity to F1 by the passive-hemagglutination test that was used diagnostically in the past, and we verified by immunoblotting that they retained activity against F1 (B. S. Murphy and S. C. Straley, unpublished data).
Identification of nonbiotinylated outer membrane proteins after 2-D electrophoresis.
Gel spots were manually excised, placed in 96-well plates, and in-gel digested using a Genomics Solutions ProGest digester according to our standard method for the identification of gel-separated proteins (5, 36). The gel extracts were lyophilized and reconstituted in 5 μl of 50% methanol-0.1% formic acid. A 0.3-μl aliquot of the extract was placed on the matrix-assisted laser desorption ionization target, followed by 0.3 μl of a saturated solution of α-cyano-4-hydroxycinnamic acid containing 50% acetonitrile, 0.1% trifluoroacetic acid, and 40 mM ammonium citrate. The spot was allowed to dry on the target at RT. Matrix-assisted laser desorption ionization-tandem time of flight spectra were acquired on an ABI 4700 proteomics analyzer mass spectrometer using the trypsin autolysis products for internal calibration. The mass spectrometer was set to acquire 15 MS-MS spectra from each spot. The MS and MS-MS data were then automatically compared to entries in the Mass Spectrometry Database by using ABI's GPS Explorer database search engine, based on the Mascot database search software (www.matrixscience.com). Search parameters were a peptide mass tolerance of 80 ppm and an MS-MS fragment ion tolerance of 0.1 Da. Peptides in the +1 charge states were included in the searches. Proteins with scores above the Mascot probability-based scoring threshold were reported as hits.
Surface biotinylation and preparation of proteins for avidin capture.
Culture aliquots of 2 OD ml (ca. 109 yersiniae) were washed once in 0.05 M potassium phosphate buffer, pH 7.4, cells were resuspended in 1.125 ml of this buffer or this buffer containing 0.25 M stachyose or raffinose at a final OD620 of 1.78, and the suspensions were incubated for 1 h at RT. One sample received 125 μl of 2.5 mM EZ-Link sulfo-NHS-LC-LC-biotin (Pierce) in H2O (the final biotin concentration was 0.25 mM), and the no-biotin control sample received H2O. The samples were incubated at RT for 20 min. The biotinylation reaction was terminated by the addition of 1.25 ml of 0.05 M phosphate buffer containing 100 mM glycine, pH 7.4, to the tubes. The cells were pelleted by centrifugation at 20,000 × g for 5 min at RT and resuspended in 3 ml of cold 10 mM HEPES buffer, pH 7.5. They were disrupted by sonication at a power setting of 2.5 for 4 min (in cycles of 2 s on, 2 s off) on ice by using the microtip of a sonic dismembranator (model 550; Laboratory Equipment Division, Fisher Scientific, Pittsburgh, PA). Large debris was removed by centrifugation at 1,000 × g for 5 min at 4°C. Total membranes were pelleted at 321,000 × g for 1 h at 4°C. Soluble cellular proteins in the supernatant were precipitated with 10% (wt/vol) trichloroacetic acid. The pelleted inner and outer membranes were dissolved in PBS containing 1% (wt/vol) SDS and boiled for 5 min. Insoluble material was removed by centrifugation at 20,000 × g for 20 min at RT. The supernatant containing the dissociated total membrane proteins was diluted 10-fold with PBS to give a final concentration of 0.1% SDS.
NA pulldown assay.
Twenty microliters of immobilized NA (Pierce) slurry in PBS containing 0.1% SDS was added to 100 μl of the final preparation of dissociated total membrane proteins (representing 7.4% of the total from 2 OD-ml of culture), and the preparation was mixed by rotation for 1 h at RT. The beads were washed twice with 150 μl of 0.1% SDS in PBS by centrifugation, resuspended in 20 μl of SDS sample buffer, and boiled for 8 min. The supernatant was analyzed by SDS-PAGE as described above. To test for the biotinylation of periplasmic proteins such as MBP, the total sonicated cell mixture was subjected to the pulldown assay after the low-speed centrifugation to remove large debris. One-sixth of the preparation (0.5 ml; material from 0.33 OD-ml of culture) was diluted with 1.5 ml of 0.1% SDS in PBS and mixed with 100 μl of 1:1-diluted NA beads, and the processing continued as described above. The proteins eluted from the beads by boiling in SDS sample buffer were those pulled down by NA.
Capture and identification of biotinylated proteins.
The SDS-dissociated total membrane proteins in 1% SDS were diluted 20-fold in distilled water and placed in a compact reaction column. Twenty microliters of NA (Pierce) bead slurry was added to the solution, and the biotinylated proteins were captured onto the affinity beads. The sample was incubated with the affinity beads overnight. The beads were then washed three times with 0.1× PBS to remove any residual SDS. Loosely bound proteins were stripped off the beads with 1:1:8 ethanol-formic acid-water. The extracts were then lyophilized. Samples were reconstituted in 100 μl of 50 mM ammonium bicarbonate solution (pH 8), 20 μl of porcine trypsin (0.1 μg/μl in 50 mM ammonium bicarbonate; Promega, San Luis Obispo, CA) was added, and the samples were digested overnight at 37°C with shaking at 400 rpm. The samples were then reconstituted in 10 μl of water, and the digests were analyzed by LC-MS-MS.
To obtain tightly bound proteins from the beads, the pH of the bead supernatant was adjusted with 100 mM ammonium bicarbonate to ca. 7.5 to 8.0, and 20 μl of trypsin solution (0.1 μg/μl in 50 mM ammonium bicarbonate) was added to the NA bead slurry. The samples were digested overnight at 37°C with shaking at 400 rpm. The samples were then eluted from the compact reaction column with 500 μl of 50 mM ammonium carbonate, lyophilized, and reconstituted in 10 μl of water, and the digests were analyzed by LC-MS-MS.
LC-MS-MS.
A 6.4-μl aliquot of each digest was analyzed by LC-MS-MS on a Waters/Micromass Q-Tof API-US mass spectrometer linked through an interface to a Waters CapLC system. The high-performance liquid chromatography system was equipped with a 5-mm, 800-Å-internal-diameter C18 P3 trapping column and a 75-μm-internal-diameter C18 PepMap analytical column (Dionex Corporation, Sunnyvale, CA). A 205-min gradient from 5% A to 95% B, where A was 5:95 acetonitrile-water (both 0.1% formic acid) and B was 95:5 acetonitrile-water (both 0.1% formic acid), was used to elute the peptides at a flow rate of ca. 200 nl/min. The solvent for loading the trapping column was buffer A at 5 μl/min.
Positive ion electrospray mass spectra were acquired using data-dependent acquisition in the standard survey mode, in which an MS survey scan is acquired first, followed by MS-MS scans of four precursor ions meeting a preselected intensity threshold. For these experiments, the intensity threshold was set to 1 (the minimum allowable). MS spectra over the mass range of 400 to 1,900 Da and MS-MS spectra over the mass range of 50 to 1,900 Da were acquired at a rate of 1 s/scan. The Waters/Micromass ProteinLynx software (version 4.0) was used to create tabulated MS-MS spectra (peak lists) from the raw data. These peak lists were input into the site-licensed Mascot database-searching program, which matches the observed spectrum with those from a theoretical digest of all the proteins represented in the database (36). Searches were done using both the entire NCBI and Y. pestis databases. Proteins with scores above the Mascot scoring threshold were reported as hits.
Protection test.
Female C57BL/6 mice were immunized under the skin on the back of the neck with KatY-HT-FkpA adsorbed onto Alhydrogel (6.25% [vol/vol] final concentration; Sigma) or Alhydrogel alone on days 0, 14, and 70 with KatY-HT-FkpA doses of 80, 80, and 20 μg, respectively. One week after the final immunization, blood samples were obtained and pooled for five groups of three mice each immunized with KatY-HT-FkpA in Alhydrogel and for three groups of four mice each given Alhydrogel alone. The sera were used to determine the titers of total antigen-specific immunoglobulin G by an indirect enzyme-linked immunosorbent assay using reagents from BD Biosciences. Thirteen days after the final immunization, half of the mice, nine KatY-HT-Alhydrogel vaccinees and six Alhydrogel vaccinees, were infected subcutaneously (s.c.) with 840 CFU of Y. pestis CO99-3015.S1. Six days later (19 days after the final immunization), the other half of the mice were challenged s.c. with 357 CFU of Y. pestis CO99-3015.S1. For 14 days following challenge, the mice were monitored twice daily for survival, and signs of illness were noted. Terminally morbid mice (those that had lost their righting reflex) were humanely killed, and their deaths were counted as occurring at the next observation time point.
Statistics.
All experiments were done two or more times. For the analysis of MS-MS data, the Mascot program used a probability-based scoring algorithm to determine which proteins from the database matched the MS-MS data (38). Search parameters were a peptide mass tolerance of 200 ppm and an MS-MS fragment ion tolerance of 0.1 Da. Peptides in the +2, +3, and +4 charge states were included in the searches. The mean time to death was calculated for each group of mice, and the results for the groups were compared to determine significant differences by using Student's unpaired t test. Survival distributions were compared to determine relative risks and significant differences by using the Web-based interactive statistical calculation program on the Dartmouth College Biostat server, available through http://statpages.org/.
RESULTS
Sarkosyl-insoluble outer membrane proteins.
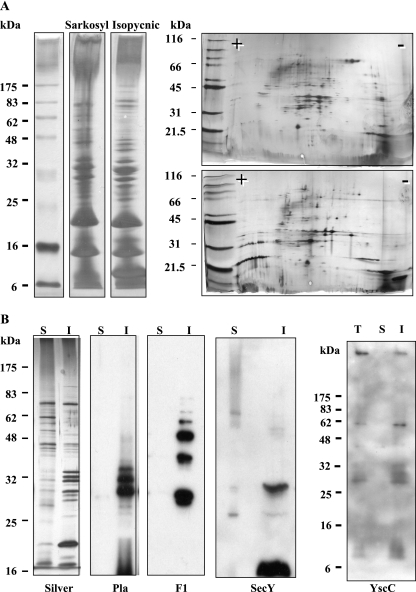
We evaluated the Sarkosyl extraction method (16) as a way to recover outer membrane proteins from small numbers of Y. pestis cells such as those that would be obtained from the lungs of mice with pneumonic plague. We found that we needed to adjust the variables of cell breakage method, detergent concentration, buffer, temperature, and duration of extraction, consistent with the existence of many variations on this procedure as described in the literature. Our main criterion was that known outer membrane proteins should localize among the Sarkosyl-insoluble proteins. The final protocol produced a profile of outer membrane proteins that was similar but not identical to that from the classic isopycnic centrifugation method (Fig. 1A), as others have also found previously (1). Figure 1B shows that the profiles of putative inner and outer membrane proteins were clearly distinct, as demonstrated by silver staining. The β-barrel outer membrane protease Pla (24) was fractionated cleanly with the Sarkosyl-insoluble outer membrane proteins and was detected as three major species as previously described (26). Similarly, the surface-associated capsular fibril F1 was present in this fraction and was resolved by SDS-PAGE as a ladder of oligomers (47) (Fig. 1B). With Lcr+ Y. pestis, the oligomeric outer membrane secretin YscC was also present among the Sarkosyl-insoluble proteins, as expected (monomer size, 64 kDa) (Fig. 1B). However, the inner membrane protein SecY mislocalized (Fig. 1B); SecY migrated as a 35-kDa species as previously reported (46). This protein was also found in both inner and outer membrane fractions after the isopycnic density centrifugation method (data not shown). If we used a higher concentration of Sarkosyl (0.5%), SecY was extracted into the Sarkosyl-soluble fraction of putative inner membrane proteins but so was Pla (data not shown). These results underscore the truism that no single membrane fractionation method works for 100% of the proteins. However, the Sarkosyl method does offer a validated approach that could be applied with the above-mentioned caveat in our characterization of Y. pestis outer membrane proteins.
FIG. 1.
Partitioning of Y. pestis membrane proteins into Sarkosyl-soluble and -insoluble fractions. Y. pestis CO99-3015.S2 cells were grown at 37°C in SHIB and broken by sonication. Total membranes recovered by centrifugation were treated with 0.3% Sarkosyl, and soluble fractions (putative inner membrane proteins) and insoluble fractions (putative outer membrane proteins) were obtained as described in Materials and Methods. For comparison, outer membranes were also prepared by the isopycnic centrifugation method (35). (A) Silver staining profiles of outer membrane proteins obtained by the two methods and resolved by SDS-PAGE (left panels) and 2-D electrophoresis (right panels). Right panels: top gel, Sarkosyl-insoluble proteins; bottom gel, outer membrane proteins from the isopycnic centrifugation method. +, acidic, and −, basic in the isoelectric focusing dimension. Locations of molecular mass standards are indicated on the left. (B) Samples from Sarkosyl fractionation of membranes representing 2 × 108 cells were resolved by SDS-PAGE and visualized by silver staining; samples representing 6 × 107 cells were analyzed in immunoblots probed with antibodies against Pla, F1, SecY, or YscC and visualized by ECL. S, Sarkosyl-soluble fraction; I, Sarkosyl-insoluble fraction; T, total membranes.
We wanted to identify highly immunogenic, abundant species in the outer membrane protein preparation that may be useful additions to the plague vaccine. Accordingly, we screened blots of 2-D gels of the Sarkosyl-insoluble proteins for recognition by several antisera. For these experiments, we used an Lcr− Y. pestis CO92 derivative that expressed a proteolytically inactive Pla. This strategy significantly reduced the number of spots present in the 2-D profile, presumably by eliminating proteolytic degradation products (S. C. Straley, C. Wulff, M. E. Gray, C. Cowan, and T. K. Korhonen, unpublished data). Prior to sonication, the yersiniae were washed three times in phosphate buffer containing MgCl2 and phenylmethylsulfonyl fluoride to inhibit any surface serine proteases and remove very loosely associated material prior to sonication and Sarkosyl extraction of membranes. A hyperimmune rabbit serum raised against cell surface material extracted with potassium thiocyanate from whole cells of Y. pestis KIM6 grown at 37°C and pH 6 strongly detected a highly abundant multilobed spot corresponding to a mass of ca. 79 kDa (Fig. 2A, circle), the known size of KatY. KatY was predicted to have a signal sequence (18), but its cellular localization was not known. It is a highly conserved protein, with 75% identity to the bifunctional catalase-peroxidase KatG in E. coli, and rabbit preimmune sera have previously been found to recognize it (27). However, none of three human plague-convalescent-phase sera recognized this protein (as illustrated for one of the sera in Fig. 2C).
FIG. 2.
Identification of strongly immunogenic proteins among the Sarkosyl-insoluble Y. pestis membrane proteins. Samples of Y. pestis putative outer membrane proteins (insoluble in 0.3% Sarkosyl) were obtained as indicated in the legend to Fig. 1, and amounts corresponding to 6 × 109 cells were resolved by 2-D electrophoresis. (A to D) The gels were analyzed by immunoblotting with alkaline phosphatase detection (A to C) or silver staining followed by MS identification of selected spots (D). The gels are oriented with the acidic (+) side to the left and the basic (−) side to the right. Primary antibodies used to probe the immunoblots were a rabbit antiserum raised against surface proteins extracted from whole Y. pestis KIM6 cells grown at 37°C and pH 6 (A), anti-GroEL of E. coli (B), and a human plague convalescent-phase serum (C). Spots circled in panels A and C were identified from the gel in panel D (corresponding circles). Numbers indicate the following proteins: 1, KatY; 2, GroEL; 3, OmpA; 4, Pla; and 5, OmpC. Panel E shows the reactivity of the human serum used to probe the blot in panel C with outer membrane proteins isolated by the isopycnic centrifugation method from ca. 109 cells of E. coli DH5α (E c) and Y. pestis CO99-3015.S2 (Y p) and detected by ECL. The locations of molecular mass markers are indicated on the left for panels A, B, and C (Precision Plus protein standard plugs from Bio-Rad) and D (precast SDS-PAGE molecular mass markers for 2-D electrophoresis from Genomic Solutions) and on the right for panel E (broad-range markers from New England Biolabs).
We selected the 79-kDa cluster and major spots corresponding to masses greater than 25 kDa that were recognized by at least two of the human sera (Fig. 2C, circles) for identification by MS. The 79-kDa cluster was divided into 12 blocks, each of which was analyzed. Four of these (Fig. 2D, region 1) were indeed identified as KatY. The others did not return identifications. All three human sera had recognized a protein that turned out to be the chaperone GroEL (Fig. 2D, region 2) and the porin OmpC (present as two charge variants in Fig. 2D, region 3). Two of the human sera recognized Pla (Fig. 2D, region 4) and OmpA (as two charge variants) (Fig. 2D, region 5). Of these proteins, the only one that is unique to Y. pestis is Pla. GroEL, OmpC, and OmpA are highly conserved among enteric bacteria, and the Y. pestis proteins have 91, 70, and 76% identity to their respective counterparts in E. coli. A recent antigenic screen of Y. pestis proteins in rabbits revealed cross-reactivity of preimmune sera to Y. pestis OmpA (27). Accordingly, it seemed possible that people would have antibodies against the proteins due simply to prior exposure to normal flora. Consistent with this idea, we verified that the Y. pestis GroEL is recognized by rabbit antibody raised against E. coli GroEL (Fig. 2B) and found that the human sera reacted with multiple species among E. coli DH5α outer membrane proteins obtained by isopycnic centrifugation, including a major band in the region of the gel that contained porins (Fig. 2E and data not shown).
Surface biotinylation of Y. pestis.
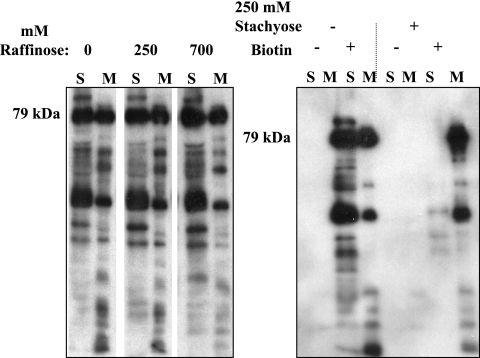
As a complementary approach, we wanted a method of identifying surface proteins that would work with small samples and not be dependent on the particular antisera we had available. The method would need to discriminate truly surface-accessible from periplasmically oriented proteins. A surface biotinylation method was developed in which biotinylation would be focused onto surface proteins by clogging the porins with oligosaccharides to prevent the permeation of the biotinylation reagent to the underside of the outer membrane, the periplasm, and the periplasmic surface of the cytoplasmic membrane. The biotinylation compound that was chosen was aqueous soluble, with a long, flexible arm to minimize steric hindrance (molecular weight, 669.75; spacer arm, 30.5 Å). Drawing upon previous studies of the permeability of E. coli porins, we tested the trisaccharide raffinose (504 Da), which had previously been found to permeate the porins poorly (34). The left panel of Fig. 3 shows the profiles of biotinylated proteins in the total soluble fractions (cytoplasm plus periplasm) and the membrane fractions (inner plus outer membranes) of cells that were biotinylated in the presence of 250 or 700 mM raffinose compared to the profiles of samples with no raffinose present. In all cases, soluble proteins were strongly biotinylated, and there was no desired effect of raffinose. In fact, higher concentrations of raffinose appeared to have the opposite effect, causing increased biotinylation of soluble proteins and decreased biotinylation of membrane proteins. We speculated that this effect might have been due to the distortion and leakage of the outer membrane due to plasmolysis.
FIG. 3.
Effects of raffinose and stachyose on biotinylation of soluble proteins in intact cells of Y. pestis. (Left panel) Y. pestis CO99-3015.S2 whole cells were washed with PBS and incubated in PBS containing the indicated concentrations of raffinose prior to biotinylation. The cells were sonicated, and the soluble fraction (S; cytoplasm and periplasm) and membranes (M; inner and outer membranes) were recovered and analyzed in blots with avidin-HRP and ECL detection. (Right panel) A similar experiment was done in the presence (+) or absence (−) of 250 mM stachyose, and soluble and membrane proteins from cells that subsequently were (+) or were not (−) biotinylated were analyzed. The position where the 79-kDa KatY would be expected to run is indicated to the left of each gel.
These tests suggested that raffinose could readily permeate the Y. pestis outer membrane. Accordingly, we tested the tetrasaccharide stachyose (666 Da), characterized by the inability to permeate E. coli porins in permeation assays and by the ability of the oxidized form to modify only surface-accessible lysines on the larger-channeled main porin, OmpF (34, 43). We used it at 250 mM, which is the isosmotic final concentration of nonpermeant sugar used in the spheroplasting step of the isopycnic centrifugation method for separating inner and outer membranes (35). The right panel of Fig. 3 shows that incubation in stachyose greatly reduced the biotinylation of soluble proteins while allowing the biotinylation of membrane proteins.
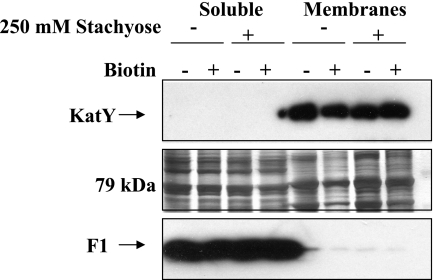
In this protocol, almost all of the capsular protein F1 was released into the soluble fraction (Fig. 4). There were prominent ca.-79-kDa species in both soluble and membrane fractions that were biotinylated in both fractions in the absence of stachyose but only in the membrane fraction in the presence of stachyose. To determine if these bands corresponded to KatY, we probed immunoblots with antibody against KatY and found that KatY reactivity was present only in the total membrane fraction (Fig. 4). Apparently, there is a major protein in the soluble fraction (a combination of the cytoplasm and the periplasm) that is similar in size to KatY and can be biotinylated in the absence of stachyose (Fig. 3 and 4), but it is not recognized by the monoclonal anti-KatY antibody. These data therefore showed that KatY is membrane associated and were consistent with the hypothesis that KatY is an outer membrane protein that is accessible to the surface.
FIG. 4.
KatY is membrane associated. (Top panel) Samples from a repetition of the experiment described in the legend to Fig. 3, right panel, were analyzed in an immunoblot probed with anti-KatY antibody and visualized by ECL. The presence (+) versus the absence (−) of stachyose and biotin is indicated, and total soluble and total membrane proteins are labeled. (Middle panel) The corresponding region of a silver-stained duplicate gel is shown. The position where the 79-kDa KatY would be expected to run is indicated on the left. (Bottom panel) A similar experiment was performed with F1+ Y. pestis CO99-3015.S5, and the locations of the 17.7-kDa F1 monomer in soluble and total membrane fractions were compared in an immunoblot probed with anti-F1 antibody and visualized by ECL.
Discrimination between surface and nonsurface biotinylation by NA pulldown assay.
To test this idea, we confirmed that stachyose in fact reduces the biotinylation of known nonsurface proteins and then determined its effect on the biotinylation of KatY. Cells that had been biotinylated in the presence or absence of stachyose were disrupted, and then NA-conjugated beads were added in the presence of 0.1% SDS and allowed to bind biotinylated proteins. Soluble proteins that were not bound by the beads and those that eluted after washing of the beads and boiling in SDS-PAGE sample buffer were analyzed in immunoblots probed with specific antibodies (Fig. 5). For known membrane-associated proteins, total membranes were obtained from the cell extracts and disrupted by boiling in the presence of 1% SDS. After dilution to give 0.1% SDS, the NA pulldown assay was carried out as described for soluble proteins. Figure 5 shows that when the cells were biotinylated in the absence of stachyose, more of the periplasmic protein MBP bound to the beads than did not, indicating that MBP could be biotinylated and that, without stachyose present, the biotinylation reagent had had access to the periplasm. In contrast, when stachyose was present, almost none of the MBP bound to the beads, indicating that the biotinylation reagent had had limited access to the periplasm. In contrast, the capsular fibril subunit F1 was biotinylated equally well in the presence or absence of stachyose, in keeping with its true surface location in intact bacteria (Fig. 5). These tests confirmed that the stachyose-focused biotinylation did discriminate between surface and non-surface-located proteins.
FIG. 5.
Stachyose reduces the biotinylation of non-surface-located protein MBP, and the biotinylation of KatY is similarly reduced. F1− Y. pestis CO99-3015.S2 and F1+ Y. pestis CO99-3015.S1 proteins in the presence or absence of stachyose were biotinylated (or not biotin treated) as in the experiments described in the legends to Fig. 3 and 4. NA-conjugated beads were used to capture biotinylated proteins from the total sonicated cell lysate (for MBP and F1) or from SDS-dissociated total membrane proteins (for KatY and Pla). The bead-bound proteins were boiled in SDS-PAGE sample buffer and analyzed in immunoblots probed with protein-specific antibodies detected by ECL. Flowthrough, proteins that did not bind to the beads; elution, proteins that bound to the beads. The presence (+) versus the absence (−) of stachyose and biotin are indicated.
Stachyose greatly reduced the amount of KatY that bound to the NA beads, similar to its effect on the NA pulldown of MBP (Fig. 5). This finding indicated that free amines on KatY were not available to the biotinylation reagent. The true outer membrane protein Pla bound the NA-conjugated beads poorly, indicating that its potentially biotinylatable residues were not accessible to the cross-linking reagent (Fig. 5). These tests indicated that KatY is not truly surface located but also illustrated a known limitation of our method, that not all surface proteins will be biotinylated.
Mass spectrometric analysis of surface-biotinylated proteins.
To learn what surface proteins were being biotinylated and evaluate the sensitivity of the coupled biotinylation-MS approach to obtaining a surface proteome, we carried out NA pulldown assays of proteins in total membranes from 109 biotinylated and 109 nonbiotinylated Y. pestis cells. After rigorous washing, the bead-bound proteins were digested with trypsin, and the tryptic peptides were analyzed by LC-MS-MS. Sequences were compared to entries in the available databases for all species as well as the database for Y. pestis CO92 (Table 1). The top hit was KatY, even though it may not be a true surface protein (see Discussion). The next six hits were all known to be outer membrane proteins or outer membrane-associated proteins that were also obtained when all available databases were queried and were likely to be real hits. Three of these proteins were also obtained from nonbiotinylated cells and bound the beads tightly in a non-biotin-specific manner. Of these non-biotin-specific proteins, Lpp and Ail were included as hits for biotinylated and nonbiotinylated proteins from both all-species and Y. pestis databases. The final three hits listed in Table 1 had marginal scores and were obtained only when the Y. pestis database was queried. Caf1M is a periplasmic chaperone involved in the assembly of the F1 capsular fibril (17), and YPO0524 is predicted to encode the periplasmic component of an ABC transport system. YPO1397 encodes a conserved hypothetical protein of unknown function.
TABLE 1.
Identification of Y. pestis proteins by LC-MS-MS analysis of on-bead digests after NA bead capture of dissociated total membrane proteins
| Proteina | GenBank accession no. | Scoreb | No. of peptides matchedc | Biotin specificd |
|---|---|---|---|---|
| KatY (YPO3319) | CAC92551 | 403 | 13 (2) | Yes |
| OmpX/Ail (YPO2905) | CAC92156 | 264 | 6 (0) | No |
| Pla (YPPCP1.07) | CAB53170 | 222 | 8 (1) | Noe |
| Lpp (YPO2394) | CAC91199 | 99 | 2 (0) | No |
| Pcp/SlyB (YPO2373) | CAC91178 | 83 | 1 (0) | Yes |
| Pal (YPO1125) | CAC89968 | 56 | 1 (2) | Yes |
| OmpA (YPO1435) | CAC90263 | 56 | 1 (0) | Yes |
| Caf1M (YPMT1.82) | CAB55264 | 26 | 1 (0) | Yes |
| Hypothetical (YPO0524) | CAC89382 | 23 | 0 (1) | Yes |
| Hypothetical (YPO1397) | CAC90226 | 23 | 0 (1) | Yes |
Sanger Centre coding sequence designations for the corresponding genes are given in parentheses.
The protein score is a ranking based on the ion (peptide) scores obtained.
Shown is the number of matches with a P value of <0.01 that the hit is a random event; in parentheses is the number of additional matches with P values of >0.01 and <0.05.
“Yes” indicates that the hit was obtained only from sequences from biotinylated yersiniae; “no” indicates that the hit was obtained from sequences from both biotinylated and nonbiotinylated yersiniae.
Pla was obtained as a hit for nonbiotinylated proteins only when the Y. pestis database was searched (i.e., it was not obtained as a hit from the all-species search).
Loosely bound proteins that had been stripped off the beads with 1:1:8 ethanol-formic acid-water prior to the on-bead digestion of tightly bound proteins did not return any identifications as Y. pestis proteins when all databases were queried (data not shown).
Test of KatY for protection against bubonic plague in mice.
Because KatY is a highly abundant and strongly antigenic outer membrane-associated protein, we carried out a study in parallel with the surface biotinylation study described above to determine whether KatY is a protective antigen. A His6-tagged version, KatY-HT, was expressed in E. coli and recovered by metal chelation chromatography. The preparation also contained a contaminating 26-kDa species that we were not able to remove. This species was identified as the E. coli periplasmic chaperone FkpA (see Materials and Methods). We felt that its presence was unlikely to cause a false-negative result and used the protein mixture to immunize mice. Eighteen mice were immunized s.c. with KatY-HT-FkpA adsorbed onto Alhydrogel, and 12 received Alhydrogel alone. The five pools of sera from mice immunized with KatY-HT-FkpA all showed a statistically significant anti-KatY total immunoglobulin G response (mean OD450 ± standard deviation of 0.719 ± 0.140 versus a mean OD450 ± standard deviation of 0.183 ± 0.031 for mice given Alhydrogel alone). Thirteen and 19 days after the final immunization, half of each immunization group was challenged s.c. with fully virulent Y. pestis CO99-3015. The first group of mice received 840 CFU; two of the nine mice that received KatY-HT-FkpA survived, while none of the six that received only the Alhydrogel adjuvant survived. In the second test, the challenge dose was 357 CFU; four of nine mice vaccinated with KatY-HT-FkpA survived, while one of five that received Alhydrogel alone survived. There was no difference in the mean time to death for KatY-FkpA and Alhydrogel-only groups, and the levels of protection were not statistically significant, indicating that KatY is not a protective antigen for bubonic plague.
DISCUSSION
In this study, we developed and tested a surface-focused biotinylation method. The general idea of surface biotinylation followed by biotin capture and MS identification has been applied in other systems, as well as in a Y. pestis study contemporaneous with the present study (44). However, other studies have not limited biotinylation to the surface, and the proteins identified from 2-D gels of whole biotinylated yersiniae were KatY, OmpA, DnaK, GroEL, enolase, EF-Tu, and OmpC (44). We had previously identified DnaK, GroEL, enolase, and EF-Tu among the Sarkosyl-insoluble Y. pestis proteins by LC-MS-MS (our unpublished data). As noted by Smither et al. (44), these proteins have been found previously in preparations of outer membrane proteins (32) and can truly be present on the surfaces of some bacteria (e.g., see references 20 and 21). However, EF-Tu is one of the most abundant protein species in the cell and, along with the abundant chaperone DnaK, can be transferred to the periplasm in E. coli upon osmotic shock (4). It can be present in the outer membranes of certain ribosomal mutants of E. coli (15) but normally is a cytoplasmic protein (42). The procedure for obtaining Sarkosyl-insoluble membrane proteins involves bacterial lysis, which creates an opportunity for the contamination of membranes by cytosolic proteins. Because we did not obtain these highly abundant proteins in our MS identification of surface-biotinylated proteins, we believe that they are not truly on the Y. pestis surface.
In our study, the seven top hits from the MS analysis of avidin-captured surface-biotinylated proteins included two outer membrane-associated proteins that are known to be highly abundant in E. coli, OmpA (23) and Pal (11), as well as the hugely abundant KatY. The 18-kDa lipoprotein Pal functions to stabilize the outer membrane through noncovalent interactions with peptidoglycan and other outer membrane proteins, including OmpA and the Braun lipoprotein Lpp (11). We would not expect Pal to be accessible to the surface, and the possibility that it reassociated with OmpA when the SDS was diluted to carry out the avidin capture should be evaluated in future work. We also obtained the 15-kDa outer membrane lipoprotein Pcp (SlyB in E. coli) in a biotin-specific manner. The function of this protein is not known, and its role in the pathogenesis of Y. pestis has not been studied. Its recovery in our system raises the possibility that it may be especially abundant, although again, unless it has an outer membrane translocation mechanism, it, like Pal, is not expected to be surface located.
We obtained three outer membrane proteins in a non-biotin-specific manner: Lpp, which in E. coli is probably numerically the most abundant protein species in the cell; one of four OmpX family members in Y. pestis; and the very abundant plasmid-encoded surface protease Pla. OmpX family members such as Ail of Y. enterocolitica are exposed on the surface and have virulence roles in a number of pathogens (23). Their role in plague has not been examined. Pla is a clear example of a protein that is missed by our biotin capture approach. It was not recovered when the NA pulldown assay was done in the presence of 0.1% SDS (Fig. 5) but bound nonspecifically to avidin beads when the SDS concentration was lowered to 0.05% for the avidin capture step prior to LC-MS-MS analysis, despite a subsequent sodium carbonate wash step.
Our study found that KatY was secreted and detected as a major protein in both our Sarkosyl-insoluble and surface-biotinylated fractions as well as in the recent study by Smither et al. (44). KatY has long been considered a potential virulence property of Y. pestis (e.g., see reference 6), because it is expressed strongly at 37°C (13, 18) and may play a role at the surface to detoxify reactive oxygen intermediates produced by polymorphonuclear neutrophils and monocytes/macrophages in plague (18). However, its final localization was not known, and the latest version of PSORT for bacterial proteins (PSORTb version 2.0 [19]) was not able to predict its location. Our study revealed that KatY is associated with a membrane but did not identify which membrane. KatY has 60 lysines scattered through the predicted mature protein, and it was recovered as the top hit among our surface-biotinylated proteins. However, the level of biotinylation was greatly reduced by the presence of stachyose, indicating that it likely was not truly on the surface. Consistent with that idea, it was purified from E. coli in association with the periplasmic chaperone FkpA (3). Immunization with KatY did not protect mice against bubonic plague, again consistent with a periplasmic orientation of the antigen. The surface-focusing effect of stachyose was not 100% (Fig. 5) and did vary from experiment to experiment, although stachyose always reduced the biotinylation of soluble proteins. We speculate that a small fraction of the periplasmically oriented KatY is biotinylated, and owing to the great abundance of this protein and its membrane association, it was captured by the avidin beads in our experiments. Accordingly, we conclude that KatY is neither surface located nor protective against bubonic plague.
In sum, our findings have ruled out KatY as a surface-localized protein and identified Ail (OmpX) and Pcp as two proteins likely to be very abundant in the cell envelope of Y. pestis and worthy of investigation for possible pathogenic roles. Because Ail (OmpX) is likely to be a truly surface-accessible protein, it warrants evaluation as a protective antigen. The surface-focused biotinylation method we applied in this study is adaptable to other bacteria once the optimal oligosaccharide to block the dominant porins is identified. Future work is needed to move past the dominant outer membrane proteins to identify the rest of the ca. 50 that are present (10). We are presently evaluating depletion methods for the detection of lower-abundance membrane proteins.
Acknowledgments
This work was supported by PHS (NIAID) grants AI061432 and UF4 AI057175 for project 4.1 of region IV of the Center of Excellence for Biodefense and Emerging Infectious Diseases. The University of North Carolina-Duke Proteomics Center was partially funded by a gift in honor of Michael Hooker from an anonymous donor to support research in proteomics and cystic fibrosis.
We thank Annette Uittenbogaard for talented technical help with the protection experiment and acknowledge the excellent service by Carol M. Beach at the University of Kentucky Center for Structural Biology Proteomics Core Facility for MS identification of FkpA expressed in E. coli.
Footnotes
Published ahead of print on 20 July 2007.
REFERENCES
- 1.Abath, F. G., and L. C. Ferreira. 1990. Comparative studies of Yersinia pestis outer membrane isolation techniques and their potential use in plague epidemiology. Rev. Inst. Med. Trop. Sao Paulo 3:78-83. [DOI] [PubMed] [Google Scholar]
- 2.Baik, S.-C., K.-M. Kim, S.-M. Song, D.-S. Kim, J.-S. Jun, S.-G. Lee, J.-Y. Song, J. U. Park, H.-L. Kang, W.-K. Lee, M.-J. Cho, H.-S. Youn, G.-H. Ko, and K.-H. Rhee. 2004. Proteomic analysis of the sarcosine-insoluble outer membrane fraction of Helicobacter pylori strain 26695. J. Bacteriol. 186:949-955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baneyx, F., and M. Mujacic. 2004. Recombinant protein folding and misfolding in Escherichia coli. Nat. Biotechnol. 22:1399-1408. [DOI] [PubMed] [Google Scholar]
- 4.Berrier, C., A. Garrigues, G. Richarme, and A. Ghazi. 2000. Elongation factor Tu and DnaK are transferred from the cytoplasm to the periplasm of Escherichia coli during osmotic downshock presumably by the mechanosensitive channel MscL. J. Bacteriol. 182:248-251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Borchers, C., J. F. Peter, M. C. Hall, T. A. Kunkel, and K. B. Tomer. 2000. Identification of in-gel digested proteins by complementary peptide-mass fingerprinting and tandem mass spectrometry data obtained on an electrospray ionization quadrupole time-of-flight mass spectrometer. Anal. Chem. 72:1163-1168. [DOI] [PubMed] [Google Scholar]
- 6.Brubaker, R. R. 1972. The genus Yersinia: biochemistry and genetics of virulence. Curr. Top. Microbiol. 57:111-158. [DOI] [PubMed] [Google Scholar]
- 7.Brubaker, R. R. 2004. The recent emergence of plague: a process of felonious evolution. Microb. Ecol. 47:293-299. [DOI] [PubMed] [Google Scholar]
- 8.Brubaker, R. R. 2005. Influence of Na+, dicarboxylic amino acids, and pH in modulating the low-calcium response of Yersinia pestis. Infect. Immun. 73:4743-4752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carlone, G. M., M. L. Thomas, H. S. Rumschlag, and F. O. Sottnek. 1986. Rapid microprocedure for isolating detergent-insoluble outer membrane proteins from Haemophilus species. J. Clin. Microbiol. 24:330-332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Casadio, R., P. Fariselli, G. Finocchiaro, and P. L. Martelli. 2003. Fishing new proteins in the twilight zone of genomes: the test case of outer membrane proteins in Escherichia coli K12, Escherichia coli O157:H7, and other Gram-negative bacteria. Protein Sci. 12:1158-1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cascales, E., A. Bernadac, M. Gavioli, J.-C. Lazzaroni, and R. Lloubes. 2002. Pal lipoprotein of Escherichia coli plays a major role in outer membrane integrity. J. Bacteriol. 184:754-759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chromy, B. A., M. W. Choi, G. A. Murphy, A. D. Gonzales, C. H. Corzett, B. C. Chang, J. P. Fitch, and S. L. McCutchen-Maloney. 2005. Proteomic characterization of Yersinia pestis virulence. J. Bacteriol. 187:8172-8180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Crumpton, M. J., and D. A. L. Davies. 1956. An antigenic analysis of Pasteurella pestis by diffusion of antigens and antibodies in agar. Proc. R. Soc. B 145:109-134. [DOI] [PubMed] [Google Scholar]
- 14.Doig, P., and T. J. Trust. 1994. Identification of surface-exposed outer membrane antigens of Helicobacter pylori. Infect. Immun. 62:4526-4533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dombou, M., S. V. Bhide, and S. Mizushima. 1981. Appearance of elongation factor Tu in the outer membrane of sucrose-dependent spectinomycin-resistant mutants of Escherichia coli. Eur. J. Biochem. 113:397-403. [DOI] [PubMed] [Google Scholar]
- 16.Filip, C., G. Fletcher, J. L. Wulff, and C. F. Earhart. 1973. Solubilization of the cytoplasmic membrane of Escherichia coli by the ionic detergent sodium-lauryl sarcosinate. J. Bacteriol. 115:717-722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Galyov, E. E., A. V. Karlishev, T. V. Chernovskaya, D. A. Dolgikh, O. Y. Smirnov, and K. I. Volkovoy. 1991. Expression of the envelope antigen F1 of Yersinia pestis is mediated by the product of caf1M gene having homology with the chaperone protein PapD of Escherichia coli. FEBS Lett. 286:79-82. [DOI] [PubMed] [Google Scholar]
- 18.Garcia, E., Y. A. Nedialkov, J. Elliott, V. L. Motin, and R. R. Brubaker. 1999. Molecular characterization of KatY (antigen 5), a thermoregulated chromosomally encoded catalase-peroxidase of Yersinia pestis. J. Bacteriol. 181:3114-3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gardy, J. L., M. R. Laird, F. Chen, S. Rey, C. J. Walsh, M. Ester, and F. S. L. Brinkman. 2005. PSORTb v. 2.0: expanded prediction of bacterial protein subcellular localization and insights gained from comparative proteome analysis. Bioinformatics 21:617-623. [DOI] [PubMed] [Google Scholar]
- 20.Granato, D., G. E. Bergonzelli, R. D. Pridmore, L. Marvin, M. Rouvet, and I. E. Corthézey-Theulaz. 2003. Cell surface-associated elongation factor Tu mediates the attachment of Lactobacillus johnsonii NCC533 (La1) to human intestinal cells and mucins. Infect. Immun. 72:2160-2169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hoffman, P. S., and R. A. Garduno. 1999. Surface-associated heat shock proteins of Legionella pneumophila and Helicobacter pylori: roles in pathogenesis and immunity. Infect. Dis. Obstet. Gynecol. 7:58-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kalb, V. F., and R. W. Bernlohr. 1977. A new spectrophotometric assay for protein in cell extracts. Anal. Biochem. 82:362-371. [DOI] [PubMed] [Google Scholar]
- 23.Koebnik, R., K. P. Locher, and P. van Gelder. 2000. Structure and function of bacterial outer membrane proteins: barrels in a nutshell. Mol. Microbiol. 37:239-253. [DOI] [PubMed] [Google Scholar]
- 24.Kukkonen, M., and T. K. Korhonen. 2004. The omptin family of enterobacterial surface proteases/adhesins: from housekeeping in Escherichia coli to systemic spread of Yersinia pestis. Int. J. Med. Microbiol. 294:7-14. [DOI] [PubMed] [Google Scholar]
- 25.Kukkonen, M., K. Lähteenmäki, M. Suomalainen, N. Kalkkinen, L. Emödy, H. Lång, and T. K. Korhonen. 2001. Protein regions important for plasminogen activation and inactivation of α2-antiplasmin in the surface protease Pla of Yersinia pestis. Mol. Microbiol. 40:1097-1112. [DOI] [PubMed] [Google Scholar]
- 26.Kutyrev, V., V. L. Motin, M. S. Pokrovskaya, G. B. Smirnov, and R. R. Brubaker. 1999. Expression of the plague plasminogen activator in Yersinia pseudotuberculosis and Escherichia coli. Infect. Immun. 67:1359-1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li, B., L. Jiang, Q. Song, J. Yang, Z. Chen, Z. Guo, D. Zhou, Z. Du, Y. Song, J. Wang, H. Wang, S. Yu, J. Wang, and R. Yang. 2005. Protein microarray for profiling antibody responses to Yersinia pestis live vaccine. Infect. Immun. 73:3734-3739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lindler, L. E., M. S. Klempner, and S. C. Straley. 1990. Yersinia pestis pH 6 antigen: genetic, biochemical, and virulence characterization of a protein involved in the pathogenesis of bubonic plague. Infect. Immun. 58:2569-2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lowry, O. H., N. J. Rosebrough, A. L. Farr, and R. J. Randall. 1951. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 193:265-275. [PubMed] [Google Scholar]
- 30.Mehigh, R. J., and R. R. Brubaker. 1993. Major stable peptides of Yersinia pestis synthesized during the low-calcium response. Infect. Immun. 61:13-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miller, J. H. 1992. A short course in bacterial genetics: a laboratory manual and handbook for Escherichia coli and related bacteria. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 32.Molloy, M., N. Phadke, J. R. Maddock, and P. C. Andrews. 2001. Two-dimensional electrophoresis and peptide mass fingerprinting of bacterial outer membrane proteins. Electrophoresis 22:1686-1696. [DOI] [PubMed] [Google Scholar]
- 33.Molloy, M. P., B. R. Herbert, M. B. Slade, T. Rabilloud, A. S. Nouwens, K. L. Williams, and A. A. Gooley. 2000. Proteomic analysis of the Escherichia coli outer membrane. Eur. J. Biochem. 267:2871-2881. [DOI] [PubMed] [Google Scholar]
- 34.Nikaido, H., and E. Y. Rosenberg. 1983. Porin channels in Escherichia coli: studies with liposomes reconstituted from purified proteins. J. Bacteriol. 153:241-252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Osborn, M. J., J. E. Gander, E. Parsi, and J. Carson. 1972. Mechanism of the assembly of the outer membrane of Salmonella typhimurium. Isolation and characterization of cytoplasmic and outer membrane. J. Biol. Chem. 247:3962-3972. [PubMed] [Google Scholar]
- 36.Parker, C. E., M. R. Warren, D. R. Loiselle, N. N. Dicheva, C. O. Scarlett, and C. H. Borchers. 2005. Identification of components of protein complexes, p. 117-151. In C. Patterson and D. M. Cyr (ed.), Methods in molecular biology, vol. 301. Ubiquitin-proteasome protocols. Humana Press Inc., Totowa, NJ. [DOI] [PubMed] [Google Scholar]
- 37.Peng, X., X. Ye, and S. Wang. 2004. Identification of novel immunogenic proteins of Shigella flexneri 2a by proteomic methodologies. Vaccine 22:2750-2756. [DOI] [PubMed] [Google Scholar]
- 38.Perkins, D. N., D. J. Pappin, D. M. Creasy, and J. S. Cottrell. 1999. Probability-base protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 20:3551-3567. [DOI] [PubMed] [Google Scholar]
- 39.Ravaoainoro, M., C. Ciurli, E. Toma, and R. Morisset. 2005. Rapid method for isolating detergent-insoluble outer membrane proteins from Pseudomonas aeruginosa. Electrophoresis 15:594-596. [DOI] [PubMed] [Google Scholar]
- 40.Rhomberg, T. A. 2004. Proteomic analysis of the sarcosine-insoluble outer membrane fraction of the bacterial pathogen Bartonella henselae. Proteomics 4:3021-3033. [DOI] [PubMed] [Google Scholar]
- 41.Schevchenko, A., M. Wilm, O. Vorm, and M. Mann. 1996. Mass-spectrometric sequencing of proteins from silver-stained polyacrylamide gels. Anal. Chem. 72:2482-2489. [DOI] [PubMed] [Google Scholar]
- 42.Schilstra, M. J., J. W. Slot, P. H. van der Meide, G. Posthuma, A. F. Cremers, and L. Bosch. 1984. Immunochemical localization of the elongation factor Tu in E. coli cells. FEBS Lett. 165:175-179. [DOI] [PubMed] [Google Scholar]
- 43.Schlaeppi, J.-M., S. Ichihara, and H. Nikaido. 1985. Accessibility of lysyl residues of Escherichia coli B/r porin (OmpF) to covalent labeling reagents of different sizes. Approach for a three-dimensional structure of a channel-forming protein. J. Biol. Chem. 260:9775-9783. [PubMed] [Google Scholar]
- 44.Smither, S. J., J. Hill, B. L. M. van Baar, A. G. Hulst, A. L. de Jong, and R. W. Titball. 2007. Identification of outer membrane proteins of Yersinia pestis through biotinylation. J. Microbiol. Methods 68:26-31. [DOI] [PubMed] [Google Scholar]
- 45.Straley, S. C., and Robert R. Brubaker. 1981. Cytoplasmic and membrane proteins of yersiniae cultivated under conditions simulating mammalian intracellular environment. Proc. Natl. Acad. Sci. USA 78:1224-1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Watanabe, M., and G. Blobel. 1989. Site-specific antibodies against the PrlA (SecY) protein of Escherichia coli inhibit protein export by interfering with plasma membrane binding of preproteins. Proc. Natl. Acad. Sci. USA 86:1895-1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zavialov, A. V., J. Berglund, A. F. Pudney, L. J. Fooks, T. M. Ibrahim, S. MacIntyre, and S. D. Knight. 2003. Structure and biogenesis of the capsular F1 antigen of Yersinia pestis: preserved folding energy drives fiber formation. Cell 113:587-596. [DOI] [PubMed] [Google Scholar]