Abstract
The microbial flora of the vagina plays a major role in preventing genital infections, including bacterial vaginosis (BV) and candidiasis (CA). An integrated approach based on PCR-denaturing gradient gel electrophoresis (PCR-DGGE) and real-time PCR was used to study the structure and dynamics of bacterial communities in vaginal fluids of healthy women and patients developing BV and CA. Universal eubacterial primers and Lactobacillus genus-specific primers, both targeted at 16S rRNA genes, were used in DGGE and real-time PCR analysis, respectively. The DGGE profiles revealed that the vaginal flora was dominated by Lactobacillus species under healthy conditions, whereas several potentially pathogenic bacteria were present in the flora of women with BV. Lactobacilli were the predominant bacterial population in the vagina for patients affected by CA, but changes in the composition of Lactobacillus species were observed. Real-time PCR analysis allowed the quantitative estimation of variations in lactobacilli associated with BV and CA diseases. A statistically significant decrease in the relative abundance of lactobacilli was found in vaginal fluids of patients with BV compared to the relative abundance of lactobacilli in the vaginal fluids of healthy women and patients with CA.
The microbial inhabitants of the human vagina constitute a finely balanced ecosystem, with the vaginal environment controlling the colonizing bacteria and the microflora in turn controlling the vaginal environment. This dynamic microbial community plays a pivotal role in preventing colonization by undesirable organisms, including those responsible for bacterial vaginosis (BV), candidiasis (CA), urinary tract infections, aerobic vaginitis, and sexually transmitted diseases (15, 20, 29, 32, 39). In women of childbearing age, the vaginal ecosystem is dominated by Lactobacillus spp., but a diverse array of other bacteria can be present in much lower numbers (24, 25). Lactobacilli are involved in maintaining the normal vaginal microflora by preventing overgrowth by pathogenic and opportunistic organisms (30). They may act by stimulating the immune system, by competing with other microorganisms for the adherence to the vaginal epithelium, and by producing lactic acid, bacteriocins, and hydrogen peroxide. This microbial metabolite represents one of the most effective protectors against bacteria, specifically against catalase-negative bacteria, which, in most cases, account for BV (5). It has been observed that 70 to 95% of lactobacilli present in the vaginal flora of healthy women produce hydrogen peroxide. This percentage for women affected by BV drops to 5% (8, 16).
The characterization of the composition and ecology of the vaginal microbial ecosystems in healthy women and in patients affected by infectious diseases, such as BV and CA, is important for an understanding of the etiology of these diseases and the development of efficient therapies. BV, widely believed to be the most common vaginal disorder affecting women of reproductive age, is defined as an alteration of the vaginal bacterial morphotypes characterized by an overgrowth of several anaerobic and microaerophilic bacteria, gram-positive cocci, and a Mycoplasma sp. and by a decreased prevalence of Lactobacillus species (17, 23). Incidence rates of BV range from 5 to 50% among both nonpregnant and pregnant women (22). CA is an infection by Candida spp., mostly Candida albicans. Yeast overgrowth can modify the normal vaginal flora. Up to 75% of women experience genital CA during their lifetime, and 5 to 8% have chronic, recurring CA, defined as four or more episodes in a 12-month period (19, 33).
In recent years, molecular techniques based on the analysis of rRNA gene sequences have been developed, providing powerful tools to reveal the phylogenetic diversity of the microorganisms found within complex ecosystems and understand community dynamics (12, 24, 41). PCR-denaturing gradient gel electrophoresis (PCR-DGGE) represents a rapid and reliable technique that has been used successfully to identify the bacterial compositions of different ecological niches, including the vaginal ecosystem (9, 10, 13, 14, 35). Sequencing of different 16S rRNA gene fragments allows the determination of which bacterial species are most common among the human specimens. Real-time PCR with genus- and species-specific primers can provide a quantification of selected bacteria present in complex microbial communities by measuring the amount of PCR products in each cycle as fluorescence (6, 11, 26, 27, 31).
In the present study, we utilized PCR-DGGE with universal primers for the bacterial 16S rrn operon to explore the composition of normal vaginal communities and assess changes related to BV and CA. In addition to using this qualitative approach, we quantified changes in Lactobacillus populations among women with different clinical profiles by using real-time PCR with 16S rRNA gene-targeted genus-specific primers.
MATERIALS AND METHODS
Study population.
Before patient recruitment, the study was considered and approved by the ethical committee of the Heilig Hart Hospital of Tienen, Belgium. After written consent had been obtained, a total of 26 randomly selected premenopausal women (23 to 48 years of age; mean, 38) who had no symptoms or signs of vaginal and urinary tract infection were recruited by telephone from the visitor list of the routine gynecology outpatient departments of the Heilig Hart Hospital, Tienen, and the Gasthuisberg University Hospital, Leuven, Belgium. None of the subjects had received oral or local antimicrobial therapy within the previous 2 weeks, required any other prescribed therapy, or was using spermicidal products. The recruits underwent examination once a month during three consecutive months. At each visit, a sterile speculum was inserted and a sample of vaginal fluid was taken from the upper lateral vaginal vault with a wooden Ayer's spatula for immediate saline microscopy, measurement of pH, and assessment for color and smell. All slides were taken in duplicate and later reviewed by a research assistant who was blinded to the patients' data or previous microscopy data. Any discrepancies were discussed, and a decision was made after a third examination of the slides. Digital photographs of all the important findings in each slide were taken. Standardized vaginal rinsings with 2 ml of saline were collected for molecular studies by flushing and reaspirating the fluid through a needle in the left, central, and right upper vaginal vaults. The vaginal rinsings were subsequently stored at −80°C until use. BV was diagnosed according to Amsel's criteria (3), with the modification that the criterion of clue-cell-positive microscopy results along with a typical granular vaginal bacterial flora in any case had to be present. The diagnosis of CA was performed by microscopy and culture on Sabouraud dextrose agar (Difco, Detroit, MI). According to this clinical evaluation, women were split into four subject groups: healthy subjects retaining a normal infection index (the NI group) (n = 11) and patients affected on this or subsequent visits by BV (n = 5), by colonization with Candida spp. (the CA group) (n = 7), or by both BV and CA (the BV-CA group) (n = 3) (Table 1). Women were classified as being in the NI group if all visits resulted in an NI, classified as being in the BV or CA group if at least one visit resulted in a diagnosis of BV or CA, respectively, and classified as being in the BV-CA group if at least one visit resulted in a diagnosis of BV and at least one visit resulted in a diagnosis of CA. The clinical history of each woman was also taken into account and reported in Table 1.
TABLE 1.
Structure of vaginal microflora as analyzed by PCR-DGGE and real-time PCR
| Patient | Age (yr) | Clinical history | Visit | Clinical evaluation | Microbial population | 16S rrn operons (mean ± SD)
|
Lactobacillus relative abundance | |
|---|---|---|---|---|---|---|---|---|
| Lactobacillus | Total eubacteria | |||||||
| 1 | 48 | BV | I | NI | Lactobacillus sp. | 4.8E+09 ± 1.1E+09 | 1.2E+10 ± 4.9E+09 | 0.4 |
| II | CA | Lactobacillus sp. | 4.7E+09 ± 2.3E+09 | 8.7E+09 ± 4.1E+09 | 0.5 | |||
| III | NI | Lactobacillus sp. | 8.3E+09 ± 1.4E+09 | 6.4E+10 ± 1.6E+10 | 0.1 | |||
| 2 | 48 | BV, CA | I | BV | Leptotrichia amnionii, Atopobium vaginae, Gardnerella vaginalis, uncultured Prevotella sp., uncultured Chloroflexi bacterium, uncultured Megasphaera sp., uncultured Clostridium sp., Staphylococcus sp. | 6.4E+09 ± 8.0E+08 | 3.2E+10 ± 3.7E+08 | 0.2 |
| II | BV | Leptotrichia amnionii, Atopobium vaginae, Gardnerella vaginalis, uncultured Prevotella sp., uncultured Chloroflexi bacterium, uncultured Megasphaera sp., uncultured Clostridium sp., Staphylococcus sp. | 9.1E+08 ± 1.1E+08 | 2.1E+10 ± 5.9E+08 | 0.0 | |||
| III | BV | Atopobium vaginae, Gardnerella vaginalis, uncultured Prevotella sp., uncultured Chloroflexi bacterium, uncultured Megasphaera sp., uncultured Clostridium sp., Staphylococcus sp. | 1.2E+09 ± 3.3E+08 | 7.3E+10 ± 1.0E+09 | 0.0 | |||
| 3 | 48 | NI | I | NI | Lactobacillus iners | 4.8E+07 ± 1.7E+06 | 2.1E+09 ± 1.9E+07 | 0.0 |
| II | NI | Lactobacillus iners | 1.4E+10 ± 1.3E+09 | 2.2E+10 ± 3.7E+08 | 0.6 | |||
| III | NI | Lactobacillus iners | 1.3E+10 ± 1.5E+08 | 2.2E+10 ± 7.7E+08 | 0.6 | |||
| 4 | 46 | CA | I | CA | Lactobacillus sp. | 3.9E+10 ± 1.0E+10 | 4.9E+10 ± 4.1E+09 | 0.8 |
| II | CA | Lactobacillus iners, Lactobacillus sp., Lactobacillus vaginalis, Clostridium sp., Ureaplasma sp. | 9.9E+09 ± 7.0E+09 | 1.1E+10 ± 2.8E+09 | 0.9 | |||
| III | NI | Lactobacillus iners, Lactobacillus vaginalis, Clostridium sp., Ureaplasma sp. | 3.9E+09 ± 2.5E+09 | 4.1E+09 ± 1.5E+08 | 1.0 | |||
| 5 | 45 | NI | I | NI | Lactobacillus sp., Gardnerella vaginalis | 7.7E+10 ± 1.5E+10 | 6.3E+10 ± 5.6E+09 | 1.2 |
| II | NI | Lactobacillus sp., Gardnerella vaginalis | 4.4E+10 ± 7.7E+09 | 7.5E+10 ± 3.0E+10 | 0.6 | |||
| III | NI | Lactobacillus sp., Gardnerella vaginalis | 9.6E+10 ± 1.6E+10 | 9.1E+10 ± 6.6E+09 | 1.1 | |||
| 6 | 43 | BV | I | NI | Lactobacillus acidophilus, Lactobacillus iners | 1.9E+10 ± 6.9E+08 | 5.2E+10 ± 4.8E+10 | 0.4 |
| II | NI | Lactobacillus acidophilus, Lactobacillus iners | 3.0E+10 ± 7.9E+08 | 6.4E+10 ± 2.4E+09 | 0.5 | |||
| III | NI | Lactobacillus acidophilus, Lactobacillus iners | 2.3E+10 ± 2.1E+09 | 1.6E+11 ± 4.5E+10 | 0.1 | |||
| 7 | 3 | BV | I | NI | Lactobacillus iners | 5.3E+10 ± 2.4E+09 | 6.2E+10 ± 3.9E+09 | 0.9 |
| II | BV | Leptotrichia amnionii, Leptotrichia sanguinegens, Gardnerella vaginalis, uncultured Prevotella sp., uncultured Clostridium sp. | 4.4E+07 ± 1.3E+07 | 2.0E+11 ± 1.1E+10 | 0.0 | |||
| III | BV+CA | Lactobacillus iners | 5.1E+10 ± 8.3E+09 | 6.5E+10 ± 2.5E+08 | 0.8 | |||
| 8 | 41 | CA | I | NI | Lactobacillus iners | 1.8E+10 ± 1.3E+08 | 3.5E+10 ± 1.1E+10 | 0.5 |
| II | NI | Lactobacillus iners | 5.2E+09 ± 3.5E+08 | 1.9E+10 ± 1.7E+10 | 0.3 | |||
| III | NI | Lactobacillus iners | 1.4E+10 ± 2.1E+09 | 1.5E+11 ± 1.1E+11 | 0.1 | |||
| 9 | 41 | BV, CA | I | BV+CA | Lactobacillus iners, Gardnerella vaginalis | 1.8E+11 ± 1.2E+11 | 1.8E+11 ± 1.3E+10 | 1.0 |
| II | NI | Lactobacillus iners | 2.9E+10 ± 8.9E+09 | 3.2E+10 ± 3.4E+09 | 0.9 | |||
| III | CA | Lactobacillus iners | 5.2E+10 ± 3.2E+10 | 1.3E+11 ± 2.5E+09 | 0.4 | |||
| 10 | 41 | CA | I | NI | Lactobacillus gasseri, Lactobacillus acidophilus, Lactobacillus iners, Lactobacillus vaginalis | 7.4E+09 ± 2.2E+08 | 6.5E+10 ± 3.3E+10 | 0.1 |
| II | NI | Lactobacillus gasseri, Lactobacillus acidophilus, Lactobacillus iners, Lactobacillus vaginalis | 3.1E+09 ± 6.5E+08 | 1.6E+10 ± 3.5E+09 | 0.2 | |||
| III | NI | Lactobacillus gasseri, Lactobacillus acidophilus, Lactobacillus iners, Lactobacillus vaginalis | 8.6E+09 ± 5.6E+08 | 2.5E+10 ± 2.0E+09 | 0.3 | |||
| 11 | 40 | BV, CA | I | NI | Lactobacillus acidophilus | 1.4E+10 ± 7.4E+07 | 4.2E+10 ± 1.1E+10 | 0.3 |
| II | NI | Lactobacillus acidophilus | 1.6E+10 ± 9.4E+08 | 4.3E+10 ± 8.4E+09 | 0.4 | |||
| III | NI | Lactobacillus acidophilus | 1.6E+10 ± 2.7E+08 | 4.5E+11 ± 1.7E+11 | 0.0 | |||
| 12 | 40 | CA | I | NI | Lactobacillus gasseri, Atopobium vaginae | 3.4E+10 ± 1.5E+10 | 7.4E+10 ± 2.4E+09 | 0.5 |
| II | NI | Lactobacillus gasseri | 2.3E+09 ± 8.3E+07 | 4.4E+09 ± 2.4E+08 | 0.5 | |||
| III | BV | Lactobacillus gasseri, Atopobium vaginae, Veilonella montpellierensis, Prevotella sp., Streptococcus sp. | 1.6E+10 ± 8.2E+09 | 1.2E+11 ± 6.0E+09 | 0.1 | |||
| 13 | 38 | CA | I | NI | Lactobacillus sp. | 1.1E+10 ± 7.4E+07 | 2.1E+10 ± 7.2E+08 | 0.5 |
| II | CA | Lactobacillus sp. | 9.9E+09 ± 4.0E+08 | 1.4E+10 ± 1.9E+09 | 0.7 | |||
| III | NI | Lactobacillus sp. | 7.1E+09 ± 3.9E+09 | 2.0E+10 ± 1.4E+09 | 0.4 | |||
| 14 | 38 | BV, CA | I | NI | Lactobacillus gasseri, uncultured Gardnerella sp., Gardnerella vaginalis | 1.6E+10 ± 7.6E+09 | 3.4E+10 ± 1.4E+09 | 0.5 |
| II | NI | Lactobacillus gasseri, uncultured Gardnerella sp., Gardnerella vaginalis | 1.2E+10 ± 4.0E+09 | 1.7E+10 ± 1.6E+08 | 0.7 | |||
| III | NI | Lactobacillus gasseri, uncultured Gardnerella sp., Gardnerella vaginalis | 5.1E+09 ± 1.1E+07 | 2.9E+10 ± 1.1E+08 | 0.2 | |||
| 15 | 38 | CA | I | BV | Lactobacillus sp., Lactobacillus iners | 2.7E+10 ± 1.6E+10 | 3.1E+10 ± 1.6E+09 | 0.9 |
| II | NI | Lactobacillus sp. | 1.9E+10 ± 1.3E+10 | 3.4E+10 ± 5.7E+08 | 0.6 | |||
| III | CA | Lactobacillus sp., Lactobacillus iners | 3.7E+10 ± 7.6E+09 | 3.2E+10 ± 4.2E+08 | 1.2 | |||
| 16 | 37 | CA | I | CA | Lactobacillus acidophilus, Lactobacillus iners | 2.0E+10 ± 2.6E+08 | 2.0E+10 ± 5.1E+09 | 1.0 |
| II | NI | Lactobacillus gasseri, Lactobacillus acidophilus, Lactobacillus iners | 8.6E+09 ± 1.6E+08 | 5.9E+09 ± 3.7E+09 | 1.4 | |||
| III | NI | Lactobacillus acidophilus | 1.8E+10 ± 6.1E+08 | 2.5E+10 ± 1.2E+10 | 0.7 | |||
| 17 | 37 | BV, CA | I | NI | Lactobacillus acidophilus, Lactobacillus iners | 8.1E+10 ± 2.2E+10 | 1.4E+11 ± 9.8E+10 | 0.6 |
| II | NI | Lactobacillus acidophilus, Lactobacillus iners, Lactobacillus vaginalis | 3.2E+10 ± 3.8E+09 | 3.1E+10 ± 7.8E+09 | 1.0 | |||
| III | BV | Lactobacillus acidophilus, Lactobacillus iners, Gardnerella vaginalis | 3.2E+10 ± 2.6E+10 | 2.0E+11 ± 3.1E+10 | 0.2 | |||
| 18 | 37 | CA | I | CA | Lactobacillus iners | 4.8E+10 ± 1.8E+09 | 5.7E+10 ± 1.1E+10 | 0.8 |
| II | NI | Lactobacillus iners | 2.6E+10 ± 1.7E+10 | 4.5E+10 ± 6.0E+09 | 0.6 | |||
| III | NI | Lactobacillus iners, Shigella boydii | 4.6E+10 ± 1.0E+10 | 4.5E+10 ± 3.9E+09 | 1.0 | |||
| 19 | 36 | CA | I | NI | Lactobacillus sp. | 1.2E+10 ± 2.8E+08 | 1.3E+10 ± 3.5E+09 | 0.9 |
| II | CA | Lactobacillus sp. | 1.0E+10 ± 2.3E+09 | 1.9E+10 ± 1.1E+09 | 0.5 | |||
| III | NI | Lactobacillus sp. | 1.1E+10 ± 8.0E+08 | 1.3E+10 ± 1.3E+09 | 0.9 | |||
| 20 | 35 | BV, CA | I | NI | Lactobacillus iners | 3.8E+10 ± 1.6E+10 | 9.3E+10 ± 3.4E+10 | 0.4 |
| II | NI | Lactobacillus iners, Lactobacillus acidophilus, Megasphaera sp. | 1.2E+11 ± 2.6E+10 | 2.3E+11 ± 1.3E+11 | 0.5 | |||
| III | BV | Lactobacillus iners, Lactobacillus acidophilus, Lactobacillus plantarum, Megasphaera sp., uncultured Clostridium bacterium clone, Staphylococcus sp. | 3.2E+06 ± 2.2E+06 | 2.5E+09 ± 1.4E+09 | 0.0 | |||
| 21 | 33 | NI | I | CA | Lactobacillus iners | 5.8E+09 ± 1.8E+09 | 7.8E+09 ± 7.5E+08 | 0.7 |
| II | CA | Lactobacillus iners | 2.2E+10 ± 4.0E+09 | 2.9E+10 ± 5.7E+08 | 0.7 | |||
| III | NI | Lactobacillus iners, Lactobacillus gasseri, Gardnerella vaginalis | 6.5E+09 ± 6.8E+08 | 9.3E+09 ± 2.9E+09 | 0.7 | |||
| 22 | 33 | CA | I | NI | Lactobacillus acidophilus, Lactobacillus vaginalis | 2.1E+10 ± 3.5E+09 | 8.3E+10 ± 3.8E+09 | 0.3 |
| II | NI | Lactobacillus acidophilus, Lactobacillus vaginalis | 5.8E+09 ± 1.6E+09 | 5.6E+10 ± 7.8E+09 | 0.1 | |||
| III | NI | Lactobacillus acidophilus, Lactobacillus vaginalis | 7.7E+09 ± 1.0E+09 | 6.4E+10 ± 2.7E+09 | 0.1 | |||
| 23 | 31 | CA | I | NI | Lactobacillus acidophilus | 8.6E+09 ± 3.1E+08 | 1.9E+10 ± 9.9E+08 | 0.4 |
| II | NI | Lactobacillus acidophilus | 8.4E+09 ± 1.2E+08 | 1.7E+10 ± 1.9E+09 | 0.5 | |||
| III | NI | Lactobacillus acidophilus | 1.2E+10 ± 2.5E+07 | 2.1E+10 ± 9.4E+08 | 0.6 | |||
| 24 | 27 | NI | I | BV | Gardnerella vaginalis | 8.2E+07 ± 4.8E+07 | 1.3E+11 ± 3.2E+10 | 0.0 |
| II | NI | Lactobacillus iners | 2.5E+10 ± 3.4E+09 | 7.7E+10 ± 2.1E+09 | 0.3 | |||
| III | NI | Lactobacillus iners | 9.0E+10 ± 2.1E+10 | 2.7E+11 ± 4.2E+09 | 0.3 | |||
| 25 | 24 | CA | I | NI | Lactobacillus acidophilus, Lactobacillus gasseri, Gardnerella vaginalis | 9.4E+09 ± 1.3E+09 | 1.6E+10 ± 3.6E+09 | 0.6 |
| II | NI | Lactobacillus acidophilus, Lactobacillus gasseri, Gardnerella vaginalis | 1.6E+10 ± 1.1E+09 | 4.1E+10 ± 8.2E+09 | 0.4 | |||
| III | NI | Lactobacillus acidophilus, Lactobacillus gasseri, Gardnerella vaginalis | 2.3E+10 ± 2.6E+09 | 6.0E+10 ± 7.7E+09 | 0.5 | |||
| 26 | 23 | NI | I | NI | Lactobacillus sp. | 3.6E+09 ± 1.3E+09 | 4.1E+10 ± 1.9E+10 | 0.1 |
| II | NI | Lactobacillus sp. | 1.7E+10 ± 2.0E+08 | 8.0E+10 ± 3.5E+10 | 0.2 | |||
| III | NI | Lactobacillus sp. | 2.1E+10 ± 5.9E+09 | 1.0E+11 ± 1.0E+10 | 0.2 | |||
Extraction of bacterial DNA from vaginal rinsings.
One milliliter of vaginal rinsings was centrifuged at 10,000 rpm for 15 min, and the pellets were washed three times in saline at 40°C. Each pellet was resuspended in 180 μl of enzymatic lysis buffer (20 mM Tris-HCl, pH 8, 2 mM EDTA, 1.2% Triton X-100, 20 mg/ml lysozyme), and total bacterial DNA was extracted by using the DNeasy tissue kit (QIAGEN, Hilden, Germany) according to the manufacturer's instructions.
PCR-DGGE analysis.
Amplification of the V2-V3 region of the bacterial 16S rRNA gene was carried out using the universal eubacterial primers HDA1-GC (containing a GC clamp) and HDA2 (38), supplied by M-Medical, Milan, Italy. The amplification reactions were performed in a T Gradient thermal cycler (Biometra, Göttingen, Germany). Dynazyme II (Celbio, Milan, Italy) was used as a thermostable polymerase under conditions suggested by the supplier. The reaction mixture contained 0.5 μM of each primer, 200 μM of each deoxynucleoside triphosphate, 0.5 U of Dynazyme II, and 4 μl of the bacterial DNA template in a final volume of 50 μl. The thermal cycling program consisted of the following time and temperature profile: 95°C for 5 min; 30 cycles of 95°C for 30 s, 56°C for 30 s, and 72°C for 60 s; and 72°C for 8 min. Samples (5 μl) of the amplified products (200 bp) were subjected to gel electrophoresis in 2% agarose gels and visualized by ethidium bromide staining.
DGGE analysis was performed using the D-Code universal mutation detection system apparatus (Bio-Rad, Hercules, CA) with 20-cm by 20-cm by 0.75-mm gels. The sequence-specific separation of the PCR fragments was obtained in 8% (wt/vol) polyacrylamide (acrylamide-N,N′bisacrylamide; 40:3 [wt/vol]) gels in 0.5 × TAE buffer (20 mM Tris, 10 mM glacial acetic acid, 0.5 mM EDTA, pH 8). The denaturing gels contained a 30% to 50% gradient of urea and formamide increasing in the direction of electrophoresis. A 100% denaturing solution contained 40% (vol/vol) formamide and 7 M urea. A stacking gel containing 8% (wt/vol) polyacrylamide was applied onto the denaturing gel. A volume of 8 to 16 μl of PCR samples was loaded onto the stacking gel. Electrophoresis was conducted at a constant voltage of 130 V and a temperature of 60°C for approximately 6 to 7 h. Following electrophoresis, the gel was silver stained by using the protocol of Bassam et al. (7).
Sequencing of the V2-V3 region of the 16S rRNA gene.
DNA fragments of interest were excised from the denaturing gels with a sterile scalpel, washed once in 1 × PCR buffer, and incubated in 20 μl of the same buffer overnight at 4°C. Four microliters of the buffer solution was used as the template for PCR. Reamplification of the V2-V3 region was conducted as described above by employing the primers HDA1 (without the 5′ GC clamp) and HDA2. The reamplified fragments were purified by using the QIAquick PCR purification kit (QIAGEN) and then subjected to automated sequence analysis of both DNA strands with HDA1 and HDA2 primers. BigDye terminators (ABI-PerkinElmer, Foster City, CA) were used with a PRISM 377 sequencer (ABI). The sequence identities were determined by comparison with rRNA gene sequences deposited in the GenBank database by using the BLAST algorithm (2).
Real-time quantitative PCR.
Quantitative PCR was performed with a LightCycler instrument (Roche, Mannheim, Germany), and SYBR green I fluorophore was used to correlate the amount of PCR product with the fluorescence signal. Total bacterial DNA extracted from vaginal fluids was amplified with the universal primer set HDA1/HDA2 (38) and the Lactobacillus genus-specific primer set Bact-0011f/Lab-0677r (21), which amplifies a 700-bp region inside the 16S rRNA genes. Three subsamples of each DNA extract were amplified in a final volume of 20 μl containing 4 mM of MgCl2, 0.5 μM of each primer, 2 μl of LightCycler-FastStart DNA Master SYBR green I (Roche), and 2 μl of either template or water (no-template control). The thermal cycling conditions used were as follows: an initial denaturation step at 95°C for 10 min followed by 40 cycles of denaturation at 95°C for 15 s, primer annealing at 56°C (HDA1/HDA2) or 63°C (Bact-0011f/Lab-0677r) for 25 s, extension at 72°C for 40 s, and an additional incubation step at 85°C for 5 s for fluorescence acquisition. For each step, the temperature transition rate was 20°C s−1. Melting curve analysis for PCR product identification was obtained by heating at 20°C s−1 to 95°C, cooling at 20°C s−1 to 60°C with a 15-s hold, and then slowly heating at 0.2°C s−1 to 99°C. Fluorescence readings were collected continuously during this heating to monitor the denaturation of PCR products.
Quantification of rrn operons of total eubacteria and lactobacilli was done by using standard curves made with known concentrations of Lactobacillus acidophilus NCFM genomic DNA, which has a size of 1.994 Mb and four rrn operons (1). Chromosomal DNA of L. acidophilus NCFM was extracted by using the DNeasy tissue kit (QIAGEN) and serially diluted from 105 to 102 molecules μl−1. Results obtained by PCR were converted to the average estimate of total eubacterial and Lactobacillus sp. rrn operons present in 1 ml of vaginal rinsing, and standard deviations (SD) were calculated. The ratio of Lactobacillus sp. rrn operons to total eubacterial 16S rRNA gene copies (relative abundance) was evaluated for each vaginal sample.
Statistical analysis.
Differences in the numbers of total eubacterial and lactobacillus rrn operons and relative abundances of lactobacilli among NI, BV, and CA groups were analyzed. Data were assessed for normal distribution by using the Kolmogorov-Smirnov test, and the Kruskal-Wallis test was performed to simultaneously compare the analysis groups. When significant differences (P < 0.05) were obtained, multiple-comparison Dunn's procedure was used to isolate the groups that differed from the others. Statistical analysis was carried out with SigmaStat software (Systat Sofware Inc., San Jose, CA).
Nucleotide sequence accession numbers.
The nucleotide sequences of the V2-V3 DGGE fragments have been deposited in the DDBJ nucleotide sequence database under accession numbers AB279893 to AB279983.
RESULTS
Qualitative analysis of vaginal bacterial communities by PCR-DGGE.
The DGGE profiles of the four subject groups (NI, BV, CA, and BV-CA) are shown in Fig. 1 to 4, and a summary of the identifications obtained by BLAST search of the GenBank database with the 16S V2-V3 region sequences is presented in Table 1. It should be noted that universal eubacterial primers were used for PCR-DGGE analysis; hence, only abundant populations were represented in this study.
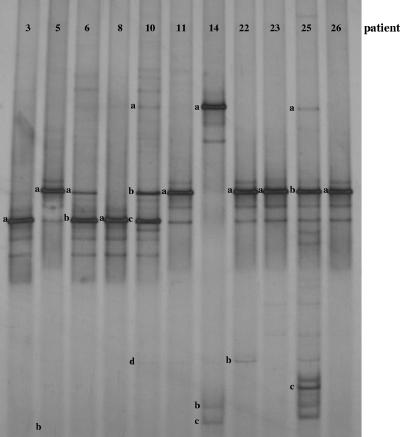
FIG. 1.
DGGE profiles of healthy women (NI group). A single DGGE profile was reported for each woman in the NI group. The bands correspond to bacterial taxa and GenBank accession numbers as follows: Lactobacillus iners, 3a (AB279893), 6b (AB279894), 8a (AB279895), and 10c (AB279896); Lactobacillus sp., 5a (AB279907) and 26a (AB279908); Lactobacillus acidophilus, 6a (AB279917), 10b (AB279918), 11a (AB279919), 22a (AB279920), 23a (AB279921), and 25b (AB279922); Lactobacillus gasseri, 10a (AB279926), 14a (AB279927), and 25a (AB279928); Lactobacillus vaginalis, 10d (AB279933) and 22b (AB279934); Gardnerella vaginalis, 5b (AB279938), 14c (AB279939), and 25c (AB279940); uncultured Gardnerella sp., 14b (AB279952).
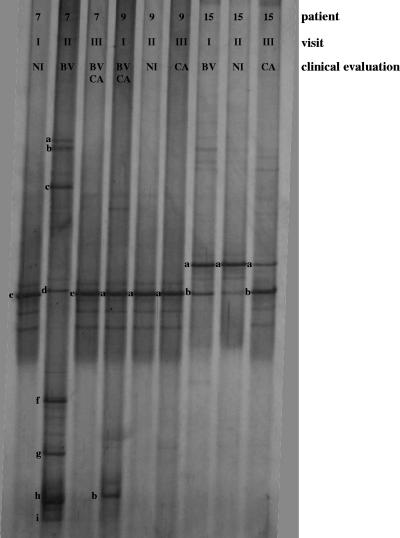
FIG. 4.
DGGE profiles of patients developing both BV and CV (BV-CA). Three DGGE courses corresponding to the three monthly gynecologic visits were reported. The bands correspond to bacterial taxa and GenBank accession numbers as follows: Lactobacillus iners, 7e (AB279904), 9a (AB279905), and 15b (AB279906); Lactobacillus sp., 15a (AB279916); Gardnerella vaginalis, 7h (AB279949), 7i (AB279950), and 9b (AB279951); Leptotrichia amnionii, 7a (AB279958) and 7b (AB279959); Leptotrichia sanguinegens, 7c (AB279961); uncultured Prevotella sp., 7d (AB279966) and 7f (AB279967); uncultured Clostridium sp., 7g (AB279975).
For each healthy woman, identical DGGE patterns were obtained at the three monthly gynecologic visits, indicating that the dominant bacterial populations remained remarkably stable over time in absence of pathological events. Thus, a single profile for each woman belonging to the NI subject group is shown in Fig. 1. According to the data reported in previous studies (9, 12, 24, 41), a low level of bacterial diversity appeared to characterize the communities of healthy vaginas. Lactobacilli represented the dominant inhabitants of the normal vaginal tract, as all the women in the NI group had at least one sequence homologous to a sequence of a Lactobacillus species. In patients 3, 8, 11, 23, and 26, only one dominant DGGE fragment was detected, and it was related to Lactobacillus iners, L. acidophilus, and Lactobacillus sp. These lactobacilli were also found in the other healthy women, in which more than one dominant DGGE fragments were visualized. Other Lactobacillus species detected in these women were Lactobacillus gasseri and Lactobacillus vaginalis. Interestingly, the microorganism Gardnerella vaginalis, which is commonly associated with BV, was found in vaginal samples of three healthy women, confirming that its presence should not be strictly linked to a pathological condition.
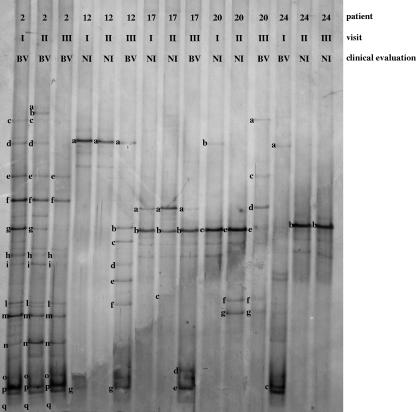
L. iners, L. acidophilus, Lactobacillus plantarum, L. gasseri, and L. vaginalis remained the predominant species in the vaginas of the women in the BV subject group when the clinical evaluation registered an NI (Fig. 2). The PCR-DGGE patterns of women during visits during which BV was diagnosed (BV visits) showed a number of dominant fragments that was higher than that for women during NI visits, indicating that high bacterial diversity is associated with BV status, in accordance to what has been previously reported (9, 14, 36, 40). Generally, we observed a correlation between the BV condition and a relative lack of lactobacilli, accompanied by a profound increase in the quantity of other vaginal anaerobic bacteria related to Gardnerella vaginalis, Atopobium vaginae, Leptotrichia amnioni, Leptotrichia sanguinegens, Prevotella sp., Megasphaera sp., an uncultured Chloroflexi bacterium, an uncultured Clostridium sp., Streptococcus sp., Staphylococcus sp., and Veillonella montpellierensis. Subject 2, who had the clinical symptoms for BV at every visit, presented the most complex and diversified vaginal microbiota. Minor variations in DGGE profiles were observed between samples from BV and NI visits for subjects 12, 17, 20, and 24, for whom BV was diagnosed in one visit only. Notably, Gardnerella vaginalis was found in all vaginal samples corresponding to BV visits except for the third visit for subjects 20 and 12.
FIG. 2.
DGGE profiles of patients developing BV. Three DGGE courses corresponding to the three monthly gynecologic visits were reported. The bands correspond to bacterial taxa and GenBank accession numbers as follows: Lactobacillus iners, 17b (AB279897), 20e (AB279898), and 24b (AB279899); Lactobacillus acidophilus, 17a (AB279923) and 20d (AB279924); Lactobacillus plantarum, 20a (AB279937); Lactobacillus gasseri, 12a (AB279929) and 20b (AB279930); Lactobacillus vaginalis, 17c (AB279935); Atopobium vaginae, 2q (AB279953) and 12g (AB279954); Gardnerella vaginalis, 2m (AB279941), 2o (AB279942), 2p (AB279943), 17d (AB279944), 17e (AB279945), and 24c (AB279946); Leptotrichia amnionii, 2a (AB279955), 2b (AB279956), and 2d (AB279957); Leptotrichia sanguinegens, 24a (AB279960); uncultured Prevotella sp., 2c (AB279962), 2f (AB279963), 2g (AB279964), and 2h (AB279965); Prevotella sp., 12b (AB279968) and 12e (AB279969); uncultured Megasphaera sp., 2l (AB279970), 20f (AB279971), and 20g (AB279972); uncultured Chloroflexi bacterium, 2i (AB279973); uncultured Clostridium sp., 2n (AB279974); Streptococcus sp., 12c (AB279977); Staphylococcus sp., 2e (AB279978) and 20c (AB279979); Veillonella montpellierensis, 12d (AB279980) and 12f (AB279981).
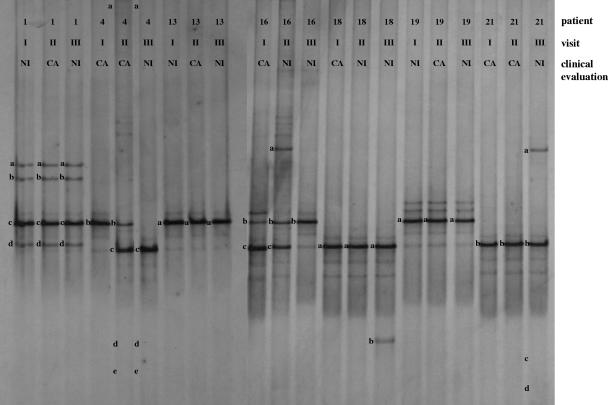
PCR-DGGE analysis of vaginal bacterial flora for women affected by CA led to the interesting result of the predominant presence of Lactobacillus species (Fig. 3). We did not find changes in DGGE profiles between vaginal fluids from women during CA and NI visits, and Lactobacillus sp., L. iners, L. gasseri, L. acidophilus, and L. vaginalis were the most frequent species detected. Other microorganisms related to Gardnerella vaginalis, Ureaplasma sp., Clostridium sp., and Shigella boydii were identified. However, these species cannot be associated with CA status, because they were found in vaginal fluids recovered from women during both NI and CA visits.
FIG. 3.
DGGE profiles of patients developing CA. Three DGGE courses corresponding to the three monthly gynecologic visits were reported. The bands correspond to bacterial taxa and GenBank accession numbers as follows: Lactobacillus sp., 1a (AB279909), 1b (AB279910), 1c (AB279911), 1d (AB279912), 4b (AB279913), 13a (AB279914), and 19a (AB279915); Lactobacillus iners, 4c (AB279900), 16c (AB279901), 18a (AB279902), and 21b (AB279903); Lactobacillus gassseri, 16a (AB279931) and 21a (AB279932); Lactobacillus acidophilus, 16b (AB279925); Lactobacillus vaginalis, 4d (AB279936); Gardnerella vaginalis, 21c (AB279947) and 21d (AB279948); uncultured Ureaplasma sp., 4a (AB279982); uncultured Clostridium sp., 4e (AB279976); Shigella boydii, 18b (AB279983).
Patients affected by both vaginal infections (BV-CA) showed PCR-DGGE patterns similar to those of CA subject group (Fig. 4). L. iners and Lactobacillus sp. were the dominant species of the vaginal ecosystem at the NI and CA visits. BV-related bacterial species were detected only in sample 7II (i.e., from subject 7 during the second visit) and not in samples 7III, 9I, and 15I, although BV was diagnosed at these visits.
Multiple fragments of different sizes were sometimes visible in the DGGE gels (for example, bands 1a, 1b, 1c, and 1d of Fig. 3). After the minor DNA fragments were sequenced, they were found to be homologous to the species corresponding to the closest major fragment. Similar to findings reported by other authors (10, 38), the major fragment indicated the identity of the isolate, whereas the minor ones were probably PCR artifacts resulting from the highly folded (loops and stems) structure of V2-V3 region.
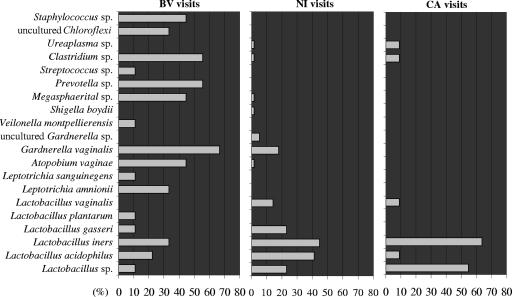
Figure 5 shows the frequency of the occurrence (in percentages) of the vaginal bacteria, detected by PCR-DGGE, in relation to the clinical status. Samples 7III and 9I were not included in this analysis and in the subsequent statistical evaluations because of the simultaneous presence of BV and CA conditions. The histograms indicate the increased complexity of vaginal ecology under BV conditions, characterized by the appearance of several anaerobic species and the decrease of L. vaginalis, L. gasseri, L. acidophilus, and Lactobacillus sp. The figure also shows that the composition of the vaginal bacterial flora did not undergo significant changes as a consequence of Candida infection, even if a diverse distribution of Lactobacillus species appeared. In particular, we observed increased frequencies of L. iners and Lactobacillus sp. and decreased frequencies of L. acidophilus, L. gasseri, and L. vaginalis. Notably, H2O2 is commonly produced by strains of L. acidophilus, L. gasseri, and L. vaginalis, while it is an uncommon metabolite among strains of L. iners (5, 34, 41).
FIG. 5.
Frequency of the occurrence of bacterial species in relation to clinical status of the subject. Frequencies were calculated as the ratios (percentage) of the number of visits characterized by the presence of each species to the total number of visits belonging to each visit group (NI, BV, and CA).
Quantification of vaginal lactobacilli by real-time PCR.
Quantitative variations in vaginal lactobacillus populations related to BV or CA were evaluated by real-time PCR analysis. For each vaginal fluid sample for each woman enrolled in the trial, the numbers of 16S rrn operons of total eubacteria and lactobacilli were determined by using universal and genus-specific primers, respectively (Table 1). The reliability of the real-time PCR data was supported by calibration curves with an r of >0.99 (data not shown) and acceptable SD. Relative abundances of lactobacilli were calculated in order to assess whether variations in lactobacillus populations were linked to changes in the total concentration of bacteria (Table 1). In accordance with the PCR-DGGE results, no evident differences in rRNA gene copy numbers of lactobacilli, as well as in their relative abundances, were observed between vaginal fluids of women during CA and NI visits, while decreases in both parameters occurred in cases in which BV was diagnosed. However, in samples from subjects 12 and 17 during BV visits, the reduction in the relative abundance of lactobacilli did not reflect a decline in the corresponding rRNA genes. Notably, vaginal fluids collected at the third visit for subject 7, the first visit for subject 9, and the first visit for subject 15 (patients belonging to the BV-CA subject group) presented a high number of Lactobacillus 16S rRNA gene copies and a high relative abundance of lactobacilli in spite of the BV diagnosis. These results are in agreement with the PCR-DGGE profiles, which showed the predominance of lactobacilli in the vaginal tracts of these patients. Therefore, the colonization patterns of lactobacilli in these women developing either BV or CA, or BV and CA at the same time, differ from the normal BV pattern, but they rather resemble the typical pattern observed for women affected by CA.
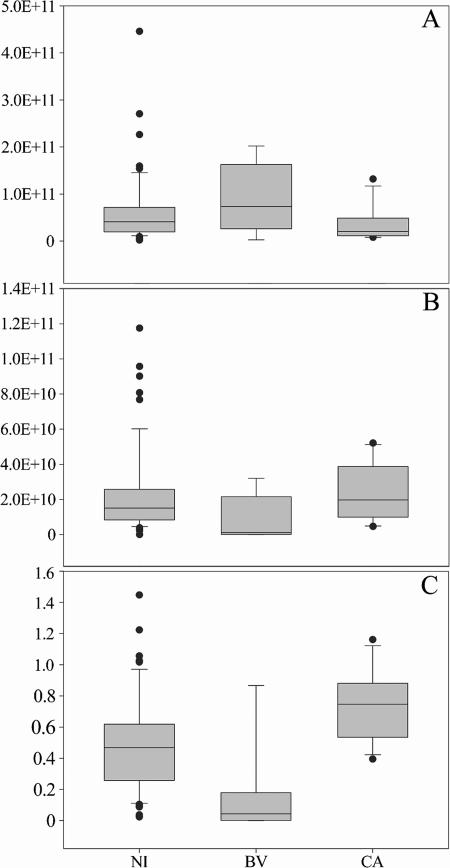
To test the significance of quantitative changes in Lactobacillus populations related to BV and CA infections, the numbers of 16S rRNA gene copies of total eubacteria and lactobacilli and the relative abundance of lactobacilli in samples from women during NI, BV, or CA visits (plotted in Fig. 6) were analyzed by a Kruskal-Wallis test due to nonnormal data distribution. The only significant difference among the groups was found for relative abundances of lactobacilli (P < 0.001). Dunn's procedure confirmed the existence of a significant difference in relative abundances of lactobacilli (P < 0.05) for each possible comparison (BV versus NI, CA versus NI, and CA versus BV). Figure 6C highlights the reduction of the relative abundance of lactobacilli in the vaginal flora of women during BV visits compared to those in the vaginal flora of women during NI and CA visits and the increase in the same parameter for women during CA visits compared to women during NI visits.
FIG. 6.
Real-time PCR evaluation of 16S rrn operons of total eubacteria (A) and lactobacilli (B) and relative abundance of lactobacilli (C) related to each group of visits characterized by a specific clinical status (NI, BV, and CA). The number of operons in 1 ml of vaginal rinsing is expressed as an absolute value. Lactobacillus relative abundance is expressed as the ratio of lactobacillus rrn operons to total eubacterial 16S rrn operons. The box for each group represents the interquartile range (25th to 75th percentile), and the line within this box is the median value. The bottom and top bars indicate the 10th and 90th percentiles, respectively. Outlier values are indicated (black circles).
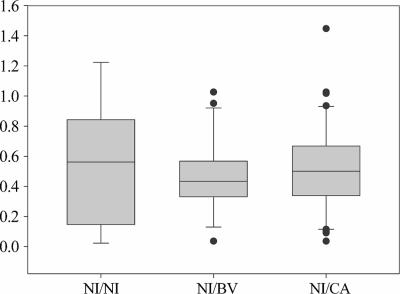
The relative abundance of lactobacilli in the vaginal flora of women during NI visits was evaluated, taking into consideration the general clinical picture of the women. Three NI visit subgroups were identified: NI visits by women with only NI visits and NI history (NI/NI), NI visits by women with at least one BV visit or BV history (NI/BV), and NI visits by women with at least one CA visit or CA history (NI/CA) (Fig. 7). Kruskal-Wallis test did not show any significant difference among NI/NI, NI/BV, and NI/CA subgroups, suggesting that the vaginal lactobacillus concentration is not influenced by the past clinical situation.
FIG. 7.
Relative abundances of lactobacilli in the vaginal flora of women during NI visits, relative to the clinical history of the women. Lactobacillus relative abundance is expressed as the ratio of lactobacillus 16S rrn operons to total eubacterial 16S rrn operons. The box for each group represents the interquartile range (25th to 75th percentile), and the line within this box is the median value. The bottom and top bars indicate the 10th and 90th percentiles, respectively. Outlier values are indicated (black circles).
DISCUSSION
The health status of the human female urogenital tract is largely a function of the vaginal microflora (25). Lactobacilli have been demonstrated to dominate the normal vaginal environment. They play a key role in determining the overall structure of the community and in providing protection against invasion by overt pathogens or against overgrowth by potentially pathogenic species among the normal flora through the production of hydrogen peroxide, bacteriocins, and lactic acid (6, 16, 28, 30).
The present study provides basic information concerning the structure of bacterial communities in healthy vaginas and qualitative/quantitative changes in numerically dominant bacterial populations related to BV and CA diseases. Particular attention was paid to dynamics of lactobacilli. Two molecular techniques, PCR-DGGE and real-time PCR, were combined to achieve a detailed picture of vaginal ecology under normal and abnormal conditions. This integrated approach has been recently proposed for monitoring the effect of prebiotics, probiotics, and symbiotics on the fecal microbiota of healthy humans (37).
Consistent with data from previous studies (4, 10, 12, 24, 41), our DGGE data showed that the vaginal bacterial floras in healthy women were stable over time and dominated by a single species or group of closely related species of Lactobacillus (L. iners, L. acidophilus, L. gasseri, and L. vaginalis). The coexistence of multiple species of lactobacilli is rare because of the competitive exclusion of one species by another, preemptive colonization by a particular species or host factors that strongly influence which species are able to colonize the environment (41). However, in samples from a small number of healthy women, G. vaginalis was detected in conjunction with Lactobacillus species. This result suggests that the presence of lactobacilli may not exclude potential pathogens from the vagina; likewise, the presence of G. vaginalis is not strictly related to clinically manifested disturbances. In healthy vaginas, G. vaginalis could represent a “sentinel species,” a term used to refer to an indigenous species that is particularly sensitive to changes in biological, physical, or chemical characteristics of the environment and responds through an increase in population size (12). The identification of potential sentinel species can help to predict whether there is an increased risk of an individual contracting an infection due to shifts in the composition of the microbial communities or changes in the abundance of specific populations. The sentinel species concept may be also used for prevention and early diagnosis of BV disturbance, which is characterized by the outgrowth of several potentially pathogenic microorganisms normally present but at much lower numbers (9, 14, 20, 40). Actually, the patients affected by BV enrolled in this clinical trial showed a relative scarcity of lactobacilli, together with an increased complexity of vaginal communities dominated by anaerobic bacteria related to Gardnerella vaginalis, Atopobium vaginae, Leptotrichia sp., Prevotella sp., Megasphaera sp., Chloroflexi, Clostridium sp., Streptococcus sp., Staphylococcus sp., and Veillonella sp. All these organisms have already been identified as components of the abnormal vaginal flora under conditions of BV disturbance (9, 20), except for Chloroflexi, Streptococcus sp., and Veillonella sp., which represent newly recognized populations not previously associated with BV. By contrast, we did not find an abnormal bacterial vaginal ecosystem in patients affected by CA, as the predominant presence of lactobacilli was observed with our DGGE-based qualitative analysis. However, a different frequency of particular Lactobacillus species was found in respect to the frequency detected under healthy conditions. In particular, we observed a decrease in H2O2-producing species (L. acidophilus, L. gasseri, and L. vaginalis) and an increase of non-H2O2-producing L. iners. This finding suggests some hypotheses regarding the role of lactobacilli in protecting women from Candida infection: (i) women with a vaginal flora dominated by H2O2-producing Lactobacillus species have a minor risk to contract CA; (ii) H2O2, rather than other metabolites produced by lactobacilli, such as lactic acid, could be responsible for the control of Candida overgrowth (33). In patients prone to developing CA as well as BV, the DGGE pattern of CA-associated lactobacilli was dominant, while the DGGE pattern of BV-associated bacteria was not detected in this group with mixed infections.
An important advantage of the DGGE technique is the ability to detect several bacterial populations not readily culturable. For example, L. iners does not grow on the major selective media for isolation of Lactobacillus, including MRS and Rogosa media (18). Likewise, the strict anaerobes Atopobium vaginae, Megasphaera, and Leptotrichia require specialized media and often grow slowly. The finding of these organisms as members of vaginal flora demonstrates how cultivation-based methods can be misleading.
Since DGGE can be considered a semiquantitative tool for monitoring bacterial populations, additional analysis with real-time PCR was carried out to obtain a quantitative estimation of changes in lactobacillus concentrations associated with BV and CA diseases. An analogous approach was used by Bartosch et al. (6) to monitor changes in intestinal bacterial community structure among different groups of elderly people. A statistically significant decrease in Lactobacillus relative abundance during BV visits was registered compared to the relative abundances during NI and CA visits, confirming the qualitative observations derived from DGGE analysis. Notably, the relative abundance of lactobacilli was significantly increased for women during CA visits, further supporting our hypothesis that the predisposition to Candida infection is not correlated with the predominance of lactobacilli but to the presence of particular Lactobacillus species. We also investigated the possibility of a link between the clinical history and the relative abundance of lactobacilli in vaginal fluids of women during NI visits, but no significant correlations were detected.
In conclusion, it has been demonstrated for the first time that the integrated use of DGGE and real-time PCR represents a successful strategy to monitor the qualitative and quantitative changes of vaginal bacterial communities correlated to different infectious diseases, such as BV and CA. In particular, the major findings obtained in this study are the following: (i) Lactobacillus species are the major constituents in vagina of healthy women and patients developing CA but not in patients developing BV; (ii) new components of vaginal microbiota in BV have been discovered; (iii) women prone to developing CA mainly host non-H2O2-producing Lactobacillus species; and (iv) in patients developing a mixed BV and CA infection, the colonization pattern of lactobacilli resembles the CA pattern.
Footnotes
Published ahead of print on 20 July 2007.
REFERENCES
- 1.Altermann, E., W. M. Russell, M. A. Azcarate-Peril, R. Barrangou, B. L. Buck, O. McAuliffe, N. Souther, A. Dobson, T. Duong, M. Callanan, S. Lick, A. Hamrick, R. Cano, and T. R. Klaenhammer. 2005. Complete genome sequence of the probiotic lactic acid bacterium Lactobacillus acidophilus NCFM. Proc. Natl. Acad. Sci. USA 102:3906-3912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, V. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search program. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amsel, R., P. A. Totten, C. A. Spiegel, C. K. Chen, D. Eschenbach, and K. K. Holmes. 1983. Nonspecific vaginitis: diagnostic criteria and microbial and epidemiological associations. Am. J. Med. 74:14-22. [DOI] [PubMed] [Google Scholar]
- 4.Antonio, M. A., S. E. Hawes, and S. L. Hillier. 1999. The identification of vaginal Lactobacillus species and the demographic and microbiologic characteristics of women colonized by these species. J. Infect. Dis. 180:1950-1956. [DOI] [PubMed] [Google Scholar]
- 5.Aroutcheva, A., D. Gariti, M. Simon, S. Shott, J. Faro, J. A. Simoes, A. Gurguis, and S. Faro. 2001. Defense factors of vaginal lactobacilli. Am. J. Obstet. Gynecol. 185:375-379. [DOI] [PubMed] [Google Scholar]
- 6.Bartosch, S., F. Alemu, G. T. Macfarlane, and M. E. T. McMurdo. 2004. Characterization of bacterial communities in feces from healthy elderly volunteers and hospitalized patients by using real-time PCR and effects of antibiotic treatment on the fecal microbiota. Appl. Environ. Microbiol. 70:3575-3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bassam, B. J., G. Caetano-Anolles, and P. M. Gresshoff. 1991. Fast and sensitive silver staining of DNA in polyacrylamide gels. Anal. Biochem. 196:80-83. [DOI] [PubMed] [Google Scholar]
- 8.Beigi, R. H., H. C. Wiesenfeld, S. L. Hillier, T. Straw, and M. A. Krohn. 2005. Factors associated with absence of H2O2-producing Lactobacillus among women with bacterial vaginosis. J. Infect. Dis. 191:924-929. [DOI] [PubMed] [Google Scholar]
- 9.Burton, J. P., and G. Reid. 2002. Evaluation of the bacterial vaginal flora of 20 postmenopausal women by direct (Nugent score) and molecular (polymerase chain reaction and denaturing gradient gel electrophoresis) techniques. J. Infect. Dis. 186:1770-1780. [DOI] [PubMed] [Google Scholar]
- 10.Burton, J. P., P. A. Cadieux, and G. Reid. 2003. Improved understanding of the bacterial vaginal microbiota of women before and after probiotic instillation. Appl. Environ. Microbiol. 69:97-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Convert, M., G. Martinetti Lucchini, M. Dolina, and J. C. Piffaretti. 2005. Comparison of LightCycler PCR and culture for detection of group B streptococci from vaginal swabs. Clin. Microbiol. Infect. 11:1022-1026. [DOI] [PubMed] [Google Scholar]
- 12.Coolen, M. J. L., E. Post, C. C. Davis, and L. J. Forney. 2005. Characterization of microbial communities found in the human vagina by analysis of terminal restriction fragment length polymorphisms of 16S rRNA genes. Appl. Environ. Microbiol. 71:8729-8737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Devillard, E., J. P. Burton, J. Hammon, D. Lam, and G. Reid. 2004. Novel insight into the vaginal microflora in postmenopausal women under hormone replacement therapy as analyzed by PCR-denaturing gradient gel electrophoresis. Eur. J. Obstet. Gynecol. Reprod. Biol. 117:76-81. [DOI] [PubMed] [Google Scholar]
- 14.Devillard, E., J. P. Burton, and G. Reid. 2005. Complexity of vaginal microflora as analyzed by PCR denaturing gradient gel electrophoresis in a patient with recurrent bacterial vaginosis. Infect. Dis. Obstet. Gynecol. 13:25-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Donders, G. G. G., A. Vereecken, E. Bosmans, A. Dekeersmaecker, G. Salembier, and B. Spitz. 2002. Definition of a type of abnormal vaginal flora that is distinct from bacterial vaginosis: aerobic vaginitis. BJOG 109:34-43. [DOI] [PubMed] [Google Scholar]
- 16.Eschenbach, D. A., P. R. Davick, B. L. Williams, S. J. Klebanoff, K. Young-Smith, C. M. Critchlow, and K. K. Holmes. 1989. Prevalence of hydrogen peroxide-producing Lactobacillus species in normal women and women with bacterial vaginosis. J. Clin. Microbiol. 27:251-256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eschenbach, D. A. 1993. History and review of bacterial vaginosis. Am. J. Obstet. Gynecol. 169:441-445. [DOI] [PubMed] [Google Scholar]
- 18.Falsen, E., C. Pascual, B. Sjoden, M. Ohlen, and M. D. Collins. 1999. Phenotypic and phylogenetic characterization of a novel Lactobacillus species from human sources: description of Lactobacillus iners sp. nov. Int. J. Syst. Bacteriol. 49:217-221. [DOI] [PubMed] [Google Scholar]
- 19.Foxman, B., R. Barlow, H. D'Arcy, B. Gillespie, and J. D. Sobel. 2000. Candida vaginitis: self-reported incidence and associated costs. Sex. Transm. Dis. 27:230-235. [DOI] [PubMed] [Google Scholar]
- 20.Fredericks, D. N., T. L. Fiedler, and J. M. Marrazzo. 2005. Molecular identification of bacteria associated with bacterial vaginosis. N. Engl. J. Med. 353:1899-1911. [DOI] [PubMed] [Google Scholar]
- 21.Heilig, H. G. H. J., E. G. Zoetendal, E. E. Vaughan, P. Marteau, A. D. L. Akkermans, and W. M. de Vos. 2002. Molecular diversity of Lactobacillus spp. and other lactic acid bacteria in the human intestine as determined by specific amplification of 16S ribosomal DNA. Appl. Environ. Microbiol. 68:114-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hill, L. H., H. Ruparelia, and J. A. Embel. 1983. Non specific vaginitis and other genital infections in three clinic populations. Sex. Transm. Dis. 10:114-118. [DOI] [PubMed] [Google Scholar]
- 23.Hillier, S. H., and K. K. Holmers. 1990. Bacterial vaginosis, p. 547-559. In K. K. Holmes, P. A. Mardh, and P. F. Sparling (ed.), Sexually transmitted diseases, 2nd ed. McGraw-Hill, New York, NY.
- 24.Hyman, R. W., M. Fukushima, L. Diamond, J. Kumm, L. C. Giudice, and R. W. Davis. 2005. Microbes of human vaginal epithelium. Proc. Natl. Acad. Sci. USA 102:7952-7957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Larsen, B., and G. R. Monif. 2001. Understanding the bacterial flora of female genital tract. Clin. Infect. Dis. 32:e69-e77. [DOI] [PubMed] [Google Scholar]
- 26.Malinen, E., T. Rinttila, K. Kajander, J. Matto, A. Kassinen, L. Krogius, M. Saarela, R. Korpela, and A. Palva. 2005. Analysis of the fecal microbiota of irritable bowel syndrome patients and healthy controls with real-time PCR. Am. J. Gastroenterol. 100:373-382. [DOI] [PubMed] [Google Scholar]
- 27.Matsuki, T., K. Watanabe, J. Fujimoto, Y. Kado, T. Takada, K. Matsumoto, and R. Tanaka. 2004. Quantitative PCR with 16S rRNA-gene-targeted species-specific primers for analysis of human intestinal bifidobacteria. Appl. Environ. Microbiol. 70:167-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McLean, N. W., and I. J. Rosenstein. 2000. Characterization and selection of a Lactobacillus species to re-colonize the vagina of women with recurrent bacterial vaginosis. J. Med. Microbiol. 49:543-552. [DOI] [PubMed] [Google Scholar]
- 29.Myer, L., L. Kuhn, Z. A. Stein, T. C. Wright, and L. Denny. 2005. Intravaginal practices, bacterial vaginosis, and women's susceptibility to HIV infection: epidemiological evidence and biological mechanisms. Lancet Infect. Dis. 5:786-794. [DOI] [PubMed] [Google Scholar]
- 30.Ronnqvist, P. D., U. B. Forsgren-Brusk, and E. E. Grahn-Hakansson. 2006. Lactobacilli in the female genital tract in relation to other genital microbes and vaginal pH. Acta Obstet. Gynecol. Scand. 85:726-735. [DOI] [PubMed] [Google Scholar]
- 31.Sakamoto, M., Y. Takeuchi, M. Umeda, I. Ishikawa, and Y. Benno. 2001. Rapid detection and quantification of five periodontopathic bacteria by real-time PCR. Microbiol. Immunol. 45:39-44. [DOI] [PubMed] [Google Scholar]
- 32.Sobel, J. D. 2002. Pathogenesis of recurrent vulvovaginal candidiasis. Curr. Infect. Dis. Rep. 4:514-519. [DOI] [PubMed] [Google Scholar]
- 33.Sobel, J. D., and W. Chaim. 1996. Vaginal microbiology of women with acute recurrent vulvovaginal candidiasis. J. Clin. Microbiol. 34:2497-2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Song, Y.-L., N. Kato, Y. Matsumiya, C.-X. Liu, H. Katu, and K. Watanabe. 1999. Identification of and hydrogen peroxide production by fecal and vaginal lactobacilli isolated from Japanese women and newborn infants. J. Clin. Microbiol. 37:3062-3064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vasquez, A., T. Jakobsson, S. Ahrne, U. Forsum, and G. Molin. 2002. Vaginal Lactobacillus flora of healthy Swedish women. J. Clin. Microbiol. 40:2746-2749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Verstraelen, H., R. Verhelst, G. Claeys, M. Temmerman, and M. Vaneechoutte. 2004. Culture-independent analysis of vaginal microflora: the unrecognized association of Atopobium vaginae with bacterial vaginosis. Am. J. Obstet. Gynecol. 191:1130-1132. [DOI] [PubMed] [Google Scholar]
- 37.Vonhoutte, T., V. D. Preter, E. D. Brandt, K. Verbeke, J. Swings, and G. Huys. 2006. Molecular monitoring of the fecal microbiota of healthy human subjects during administration of lactulose and Saccharomyces boulardii. Appl. Environ. Microbiol. 72:5990-5997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Walter, J., G. W. Tannock, A. Tilsala-Timisjarvi, S. Rodtong, D. M. Loach, K. Munro, and Y. Alatossava. 2000. Detection and identification of gastrointestinal Lactobacillus species by using denaturing gradient gel electrophoresis and species-specific PCR primers. Appl. Environ. Microbiol. 66:297-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Watts, D. H., M. Fazarri, H. Minkoff, S. L. Hillier, B. Sha, M. Glesby, A. M. Levine, R. Burk, J. M. Palefsky, M. Moxley, L. Ahdieh-Grant, and H. D. Strickler. 2005. Effects of bacterial vaginosis and other genital infections on the natural history of human Papillomavirus infection in HIV-1-infected and high-risk HIV-1-uninfected women. J. Infect. Dis. 191:1129-1139. [DOI] [PubMed] [Google Scholar]
- 40.Zariffard, M. R., M. Saifuddin, B. E. Sha, and G. T. Spear. 2002. Detection of bacterial vaginosis-related organisms by real-time PCR for lactobacilli, Gardnerella vaginalis and Mycoplasma hominis. FEMS Immunol. Med. Microbiol. 34:277-281. [DOI] [PubMed] [Google Scholar]
- 41.Zhou, X., S. J. Bent, M. G. Schneider, C. C. Davis, M. R. Islam, and L. J. Forney. 2004. Characterization of vaginal microbial communities in adult healthy women using cultivation-independent methods. Microbiology 150:2565-2573. [DOI] [PubMed] [Google Scholar]