Abstract
The microbial community in the human colon contains bacteria that reduce cholesterol to coprostanol, but the species responsible for this conversion are still unknown. We describe here the first isolation and characterization of a cholesterol-reducing bacterium of human intestinal origin. Strain D8 was isolated from a 10−8 dilution of a fresh stool sample provided by a senior male volunteer with a high capacity to reduce luminal cholesterol to coprostanol. Cholesterol-to-coprostanol conversion by strain D8 started on the third day, while cells were in stationary phase, and was almost complete after 7 days. Intermediate products (4-cholesten-3-one and coprostanone) were occasionally observed, suggesting an indirect pathway for cholesterol-to-coprostanol conversion. Resting-cell assays showed that strain D8 could reduce 1.5 μmol of cholesterol/mg bacterial protein/h. Strain D8 was a gram-negative, non-spore-forming, rod-shaped organism identified as a member of the genus Bacteroides closely related to Bacteroides vulgatus, based on its morphological and biochemical characteristics. The 16S rRNA gene sequence of strain D8 was most similar (>99.5%) to those of two isolates of the recently described species Bacteroides dorei. Phylogenetic tree construction confirmed that Bacteroides sp. strain D8 clustered within an independent clade together with these B. dorei strains. Nevertheless, no cholesterol-reducing activity could be detected in cultures of the B. dorei type strain. Based on Bacteroides group-specific PCR-temporal temperature gradient gel electrophoresis, there was no correlation between the presence of a band comigrating with the band of Bacteroides sp. strain D8 and cholesterol conversion in 11 human fecal samples, indicating that this strain is unlikely to be mainly responsible for cholesterol conversion in the human population.
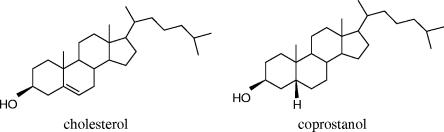
The conversion of cholesterol to the saturated product coprostanol (Fig. 1) by intestinal microorganisms was established during the 1930s (8). It was subsequently reported that the efficiency of microbial cholesterol-to-coprostanol conversion in human populations was bimodal, with a majority of high converters and a minority of low or inefficient converters (38, 40). This bacterial metabolism proceeds according to two pathways (4, 31), one involving the intermediate formation of 4-cholesten-3-one and coprostanone and the other involving the direct conversion of cholesterol into coprostanol through reduction of the 5-6 double bond. Surprisingly, the organisms responsible for this bioconversion in humans are still unknown, and only a few cholesterol-reducing strains have been isolated from the rat cecum (12), baboon feces (7), and a hog sewage lagoon (13). Isolation and characterization of cholesterol-reducing bacteria of human origin are nevertheless a prerequisite for understanding microbial cholesterol metabolism and its true impact on human health. Indeed, coprostanol, unlike cholesterol, is poorly absorbed by the human intestine (23, 35), and an inverse relationship has been observed between serum cholesterol levels and the coprostanol/cholesterol ratio in feces (35), so that the conversion of cholesterol to coprostanol has been considered a natural means of lowering serum cholesterol in humans and thus reducing the risk of cardiovascular disease. However, two studies have suggested that high fecal concentrations of coprostanol might be associated with colon carcinogenesis in humans (29) and in dimethyl hydrazine-fed rats (28).
FIG. 1.
Chemical structures of cholesterol and coprostanol.
We report here the first isolation and characterization of a cholesterol-reducing bacterium of human origin. Unlike all cholesterol-reducing strains from nonhuman sources isolated so far, the new isolate does not belong to the genus Eubacterium. Based on phenotypic and phylogenetic studies, we show that cholesterol-reducing strain D8 is closely related to the newly described species Bacteroides dorei (3).
MATERIALS AND METHODS
Bacterial strains, media, and culture conditions.
Bacteroides vulgatus ATCC 8482T was obtained from the American Type Culture Collection (United States), and B. dorei 175T was obtained from the Deutsche Sammlung von Mikroorganismen und Zellkulturen (Germany). Eubacterium coprostanoligenes was provided by D. Beitz (Iowa State University). Fecal dilution was carried out in liquid casein yeast extract (LCY) medium (pH 7.0) consisting of Casitone (2 g/liter; Difco Laboratories, Detroit, MI), yeast extract (2 g/liter; Difco), NaCl (5 g/liter), and KH2PO4 (1 g/liter). Cholesterol brain agar (CBA) (pH 7.2 to 7.4) used for isolation of cholesterol-reducing strains was modified from the medium described by Brinkley et al. (6) and contained the following components (per liter): 20 g lyophilized calf brain (i.e., ∼2.4 g cholesterol), 10 g Casitone, 10 g yeast extract, 10 g lecithin (type IV-S; Sigma-Aldrich Chimie), 25 g cholesterol (Sigma-Aldrich Chimie), 5 g KH2PO4, 0.5 g sodium thioglycolate, 0.1 g trypan blue, and 14 g agar (Difco). Standard tests of cholesterol-to-coprostanol reducing activity were carried out in standard brain medium (SBM) (pH 7.2 to 7.4), which was derived from the medium described by Brinkley et al. (7) and contained the following components (per liter): 20 g lyophilized calf brain, 10 g Casitone, 10 g yeast extract, 5 g KH2PO4, and 0.5 g sodium thioglycolate. Where indicated below, activity was also assayed in a basal cholesterol (BC) medium (pH 7.5) derived from the medium described by Ren et al. (31) and containing (per liter) 10 g Casitone, 10 g yeast extract, 5 g sodium pyruvate, 0.5 g sodium thioglycolate, 1 g CaCl2 · 2H2O, 0.2 g cholesterol, and 1.0 g lecithin, as well as in brain heart infusion-yeast extract-hemin (BHI-YH) medium (pH 7.4) enriched with cholesterol (38) and containing (per liter) 10 g brain heart infusion (Difco), 10 g yeast extract, 10 ml of a 0.1% hemin solution (Sigma-Aldrich Chimie), 0.5 g l-cysteine (Sigma-Aldrich Chimie), 0.2 g cholesterol, and 1 g lecithin. Agar (14 g/liter) was added to BHI-YH medium when required. Lastly, cholesterol- and lecithin-free BHI-YH agar was used for counting total cultivable bacteria, and cholesterol- and lecithin-free BHI-YH broth was used for production of resting cells of strain D8 and for growth of B. vulgatus ATCC 8482T and preparation of its DNA.
All media were equilibrated for at least 48 h in an anaerobic Freter chamber (La Calhène, Vendôme, France) (85% N2, 10% H2, 5% CO2) before use, and all cultures were grown in a 37°C incubator placed in the anaerobic chamber.
Isolation procedure.
A fresh stool sample was provided in an anaerobic box (Anaerocult; Merck, Darmstadt, Germany) by a healthy senior (73-year-old) male volunteer consuming a normal Western diet without any evidence of gastrointestinal or hepatic disorders and without laxative or antibiotic use for the preceding 6 months. This volunteer was selected because coprostanol represented 80% of the neutral animal sterol content in his feces. Within 30 min after defecation, 1 g (wet weight) of feces was weighed in the anaerobic chamber and serially 10-fold diluted in LCY medium. One hundred microliters each of the 10−7 to 10−9 dilutions was plated on BHI-YH agar to count total cultivable bacteria. One hundred microliters of the 10−7 dilution was plated onto CBA plates, and after 3 days of culture, 104 well-isolated colonies were picked and subcultured by streaking onto CBA plates. The activity of each streak was assessed by resuspension in 2 ml SBM. After 7 days of incubation, gas chromatography (GC) analysis revealed that cholesterol was nearly totally converted to coprostanol in one culture. The corresponding culture (D8) was streaked onto CBA to check its purity.
GC analysis of sterols.
Neutral sterols were extracted from 1-ml culture aliquots using the method described by Bligh and Dyer (5) and from 2-g stool aliquots using the method described by Riottot et al. (32). Neutral sterols were analyzed by GC as their silyl derivatives (32). The fecal coprostanol content was expressed as a percentage of the total neutral animal sterols.
Physiological and biochemical characterization.
Routine tests (Gram staining, spore and catalase tests) were done as described by Smibert and Krieg (36). Resistance to atmospheric oxygen was tested by plating serial 10-fold dilutions of a strain D8 culture onto CBA or BHI-YH agar and exposing the plates to air for 1 h before incubation under anaerobic conditions. Biochemical and enzymatic tests were performed using the API 20A and API rapid ID 32A systems (BioMerieux, Marcy-l'Etoile, France) by following the manufacturer's instructions and using cells of strain D8 and B. dorei 175T grown for 7 days on cholesterol-enriched BHI-YH agar. Short-chain fatty acids were analyzed by GC using the method described by Kaneuchi et al. (18) after growth for 7 days in cholesterol-enriched BHI-YH medium.
The growth and cholesterol-reducing activity of strain D8 were studied in SBM broth at 37°C. As measurement of the optical density was impossible with SBM broth, growth in liquid cultures was monitored by plating. Cholesterol conversion was monitored in parallel by using 1-ml culture aliquots which were extracted as described above. The cholesterol-reducing activities of strain D8, B. dorei 175T, and E. coprostanoligenes ATCC 51222T were also assayed in BC medium and cholesterol-enriched BHI-YH medium. Conversion of intermediate products (4-cholesten-3-one and coprostanone) by strain D8 was assayed in BC medium in which cholesterol was replaced by the substrate tested.
Scanning electron microscopy.
Cell cultures were harvested by centrifugation (2,000 × g, 10 min, 20°C), immersed in a fixative solution (3% glutaraldehyde in 0.2 M phosphate-buffered saline) overnight at room temperature, and then deposited on isopore membrane filters (HTTP; 0.4 μm; Millipore, France). The fixative was removed, and samples were rinsed with 0.2 M phosphate-buffered saline (pH 7.4). The rinse solution was removed, and samples were postfixed by addition of 1% osmium tetroxide in 0.2 M phosphate-buffered saline (pH 7.4) for 45 min at 4°C. Samples were then dehydrated with an ethanol series (50 to 100%) for 10 min in each concentration, with three 100% ethanol changes. Samples were mounted on aluminum stubs with graphite paint and sputter coated with gold-palladium (Polaron SC7640; Elexience, Verrières-le-buisson, France) for 140 s at 10 mA and 80 mtorr. Samples were visualized by field emission gun scanning electron microscopy. They were viewed as secondary electron images (8 kV) with a Hitachi S4500 instrument (Elexience, Verrières-le-buisson, France) at the Microscopy and Imaging Platform (MIMA2, INRA, Massy, France).
Resting-cell experiments.
Resting-cell assays were conducted under anaerobic conditions in an incubator placed in the Freter chamber. Because it was impossible to harvest cells from the semiliquid brain-containing SBM, cells for resting-cell assays were produced from overnight cultures in BHI-YH medium. For each assay, a culture of strain D8 exhibiting cholesterol-reducing activity in SBM was subcultured once in 8 ml of BHI-YH medium (devoid of cholesterol-lecithin vesicles). An overnight culture was harvested by centrifugation (8,000 × g, 20 min, 20°C). The activities of the supernatant and of nondisrupted and disrupted resuspended cells were assayed. Disruption was achieved by two 1-min vortexing cycles with glass microbeads. Each assay mixture contained 1 ml supernatant or 1 ml bacterial cell suspension in reduced 25 mM sodium phosphate buffer (pH 7.5) with 45 mM sodium pyruvate and 5 mM sodium thioglycolate (31), and the assay was started by addition of 200 μl filter-sterilized cholesterol-lecithin vesicles (6 mM cholesterol and 14 mM lecithin) corresponding to a final cholesterol concentration of 1.00 mM (i.e., 0.46 mg of cholesterol per assay mixture). After incubation at 37°C for 30 min and 24 h with shaking, neutral sterols were extracted from the reaction mixtures and analyzed by GC. Protein concentrations were determined using a 2D Quant kit (GE Healthcare Europe, Orsay, France) and bovine serum albumin as a standard after bacterial cells had been lysed with detergent buffer containing 8.75 M urea, 2.50 M thiourea, 5% 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate (CHAPS), 75 mM dithiothreitol, and 31.25 mM spermine base dehydrate.
16S rRNA gene sequencing and phylogenetic analysis.
Genomic DNA was extracted from a 3-day culture in SBM using a QIAamp DNA stool mini kit (QIAGEN, Courtaboeuf, France) as specified by the manufacturer. Forward primer S-D-Bact-0008-a-S-20 (5′ AGA GTT TGA TCC TGG CTC AG 3′) and reverse primer S-*-Univ-1492-b-A-21 (5′ ACG GCT ACC TTG TTA CGA CTT 3′) were used to amplify bacterial 16S rRNA genes by PCR (39). Reaction tubes contained 5 ng (1 μl) of strain D8 genomic DNA, 1.25 U of Taq DNA polymerase (AmpliTaq Gold; Perkin-Elmer Corporation, Foster City, CA), 1× AmpliTaq Gold reaction buffer, 2.5 mM MgCl2, 200 μM of each deoxyribonucleotide triphosphate, and 0.40 μM of each primer in a 50-μl (final volume) mixture. The initial DNA denaturation and enzyme activation steps were performed at 94°C for 10 min in a PTC 150 thermocycler (MJ Research, Inc., Watertown, MA) and were followed by 35 cycles of 92°C for 1 min, 52°C for 1 min, and 72°C for 1.5 min and then a final elongation at 72°C for 15 min. The PCR product was purified and concentrated with a QIAquick spin PCR purification kit (QIAGEN). The nucleotide sequence of the PCR product was determined by MWG Biotech Company (Ebersberg, Germany). The internal primers used for sequencing were S-*-Univ-0536-a-A-18 (5′ GWA TTA CCG CGG CKG CTG 3′), S-D-Bact-1115-a-A-16 (5′ AGG GTT GCG CTC GTT R 3′), and S-D-Bact-0930-a-S-16 (5′ AGG AAT TGR CGG GGG C 3′). Each sequence was manually edited in conjunction with its chromatogram. The closest neighbors of our sequence in the Ribosomal Database Project (RDP-II) and NCBI GenBank databases were determined from local phylogenies using the maximum likelihood algorithm (online analysis at the RDP-II website http://rdp.cme.msu.edu/cgis/chimera.cgi?su=SSU and the NCBI website http://www.ncbi.nlm.nih.gov/BLAST/). Sequences were tested for possible chimeras using Chimera Check v2.7 (online analysis at the RDP-II website). Phylogenetic analysis was then performed using the Linux-based ARB software package (24). The database was downloaded from the ARB website (http://www.arb-home.de/) and consisted of 39,201 aligned full-length bacterial 16S rRNA gene prokaryotic sequences (source, RDP-II as of January 2004). The sequences were aligned using the ARB tool FastAligner v1.03 and ClustalX program. Alignments were manually checked. The tree was generated by neighbor-joining analysis and Jukes-Cantor mathematical correction and was corrected with a Bacteroidetes filter constructed in December 2004 to include only 50% of the conserved regions.
DNA-DNA reassociation.
The degree of DNA-DNA binding was determined quantitatively by spectrophotometry from renaturation rates using a modification of the method described by De Ley et al. (9). The renaturation temperature was 25°C below the midpoint (i.e., 61.1°C). DNA-DNA relatedness values were calculated after incubation for 21 and 24 min, following removal from the calculation of the first 3 min of renaturation. The G+C compositions of strain D8 and B. vulgatus ATCC 8482T were determined as described by Marmur and Doty (26).
DNA isolation, PCR, and TTGE analysis.
Total DNA was extracted for PCR-temporal temperature gradient gel electrophoresis (TTGE) as previously described (15) from 0.2-g frozen and thawed stool aliquots which had been obtained from 11 healthy human subjects and for which we had previously assessed the population level of cultivable coprostanoligenic bacteria and the cholesterol-to-coprostanol ratio (38). The concentration and integrity of the nucleic acids were determined visually by electrophoresis on a 1% agarose gel containing ethidium bromide. The V6 region of the Bacteroides-Porphyromonas-Prevotella group 16S rRNA gene was selectively amplified using primers S-D-Bact-0933-a-S-22 (5′ GCA CAA GCG GTG GAG CAT GTG G 3′) and S-*-Bacto-1080-a-A-18 (5′ GCA CTT AAG CCG ACA CCT 3′) (10). For TTGE analysis of the amplicons, a 40-bp GC clamp (5′ CGCCCGGGGCGCGCCCCGGGCGGGGCGGGGGCACGGGGGG 3′) was attached to the 5′ end of the forward primer. TTGE was performed as previously described (14), with minor modifications. Briefly, electrophoresis was performed using the DCode universal mutation detection system (Bio-Rad, Paris, France) and a 1-mm-thick 10% polyacrylamide gel (16 by 16 cm) at a fixed voltage of 65 V for 16 h with an initial temperature of 65.5°C and a ramp rate of 0.3°C/h. The gel was stained in the dark by immersion for 30 min in a solution of SYBR gold nucleic acid gel stain (Invitrogen, Eugene, OR) and was read with a Storm system (Molecular Dynamics, Bondoufle, France).
Nucleotide sequence accession number.
The 16S rRNA gene sequence of strain D8 has been deposited in the GenBank database under accession number DQ217839.
RESULTS
Isolation.
Strain D8 was obtained from a serial dilution in LCY medium of a fresh feces sample provided by a healthy senior male volunteer known to be a high cholesterol converter. The total count of cultivable bacteria on BHI-YH agar was 2.6 × 1010 CFU/g (wet weight) of stool. A 100-μl inoculum originating from the 10−7 dilution yielded well-separated colonies on CBA. After growth of each colony in SBM broth, cholesterol-reducing activity was detected in one culture. No cholesterol-reducing activity was detected in any other assay, including SBM controls. Microscopic examination revealed the presence of a unique bacterial morphotype corresponding to strain D8. Purity was checked using the 10-fold serial dilution technique, which in several independent experiments invariably demonstrated that the highest dilution showing bacterial growth in SBM was associated with coprostanol production. Purity was further confirmed by the observation that only one 16S rRNA gene sequence was obtained from the highest dilution.
Physiological and biochemical properties.
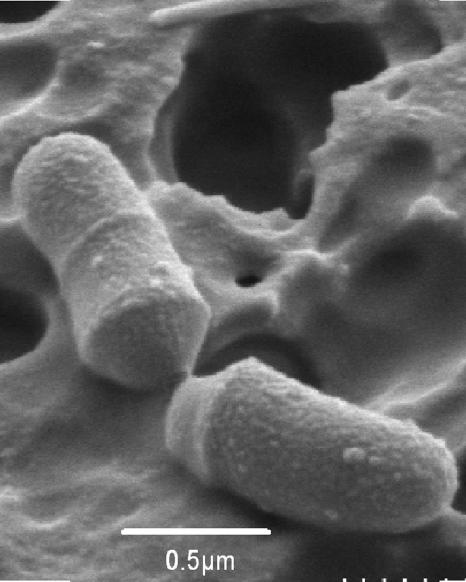
Cells of strain D8 were gram-negative and non-spore-forming cells and occurred singly or in pairs. The short rods or rod-shaped cells were 0.5 μm wide, and the length was variable, ranging from 1 to 4 μm (Fig. 2). Under anaerobic conditions, strain D8 grew in 1- to 2-mm, semiopaque, slightly convex, S-type colonies on CBA or BHI-YH agar. When serial 10-fold dilutions of strain D8 were plated onto CBA or BHI-YH agar, the colony number and morphology remained unchanged after 1 h of exposure to air. Strain D8 was found to be catalase negative. The biochemical properties of strain D8, B. dorei 175T, and their close neighbors are listed in Table 1. Using the API 20A and API rapid ID 32A systems, no difference was observed between strain D8 and B. dorei 175T phenotypic properties. Among related Bacteroides species, strain D8 and B. dorei 175T were found to be phenotypically closer to B. vulgatus, which differs only in arginine dihydrolase and β-glucosidase activities and indole production (Table 1). In cholesterol-enriched BHI-YH broth, major amounts of propionic (7.5 mM) and acetic (5.0 mM) acids and minor amounts of isovaleric (1.2 mM) and isobutyric (0.3 mM) acids were produced by strain D8.
FIG. 2.
Scanning electron micrograph of strain D8 grown on BHI-YH medium enriched with cholesterol.
TABLE 1.
Biochemical properties of Bacteroides sp. strain D8, B. dorei 175T, and phenotypically related members of the genus Bacteroidesa
| Test | Strain D8 | B. dorei | B. vulgatus | B. fragilis | B. ovatus | B. distasonis | B. thetaiotaomicron |
|---|---|---|---|---|---|---|---|
| Arabinose | + | + | + | − | + | −/+ | + |
| Cellobiose | − | − | − | −/+ | + | +/− | + |
| Esculin hydrolysis | − | − | −/+ | + | + | + | + |
| Gelatin hydrolysis | − | − | −/+ | − | −/+ | − | − |
| Melezitose | − | − | − | − | − | −/+ | −/+ |
| Rhamnose | + | + | + | − | +/− | + | + |
| Salicilin | − | − | − | − | + | + | −/+ |
| Trehalose | − | − | − | − | + | + | + |
| Arginine dihydrolase | + | + | − | −/+ | − | − | − |
| 6-Phospho-β-galactosidase | + | + | −/+ | − | − | −/+ | − |
| β-Glucosidase | + | + | − | +/− | + | + | + |
| α-Arabinosidase | + | + | + | − | + | + | + |
| β-Glucuronidase | + | + | −/+ | −/+ | − | − | −/+ |
| Indole production | + | + | − | − | + | − | + |
| Arginine arylamidase | − | − | −/+ | +/− | − | + | +/− |
| Phenylalanine arylamidase | − | − | − | −/+ | − | +/− | −/+ |
| Leucine arylamidase | − | − | − | +/− | −/+ | + | + |
| Pyroglutamic acid arylamidase | − | − | −/+ | − | − | +/− | − |
| Glycine arylamidase | + | + | + | − | − | +/− | +/− |
| Histidine arylamidase | − | − | − | −/+ | − | + | −/+ |
| α-Fucosidase | + | + | + | + | + | − | + |
The data for B. vulgatus, B. fragilis, B. ovatus, B. distasonis, and B. thetaiotaomicron were adapted from the identification tables of the API 20A and API 32A test kits (bioMérieux, France). All the strains can utilize glucose, lactose, maltose, mannose, raffinose, sucrose, and xylose. All strains do not ferment glycerol, mannitol, and sorbitol. All strains are positive for α-galactosidase, β-galactosidase, α-glucosidase, N-acetyl-β-glucosaminidase, glutamic acid decarboxylase, alkaline phosphatase, leucyl glycine arylamidase, alanine arylamidase, and glutamyl glutamic acid arylamidase. All strains are negative for urease, reduction of nitrates, proline arylamidase, tyrosine arylamidase, and serine arylamidase. +, positive; −, negative; +/−, most strains positive; −/+, most strains negative.
Phylogenetic analyses.
An almost complete 16S rRNA gene sequence comprising 1,487 nucleotides was determined. Sequence comparisons of the GenBank and RDP libraries for validly described species revealed that the unknown organism was a member of the phylum Bacteroidetes and the genus Bacteroides. Pairwise comparisons based on more than 1,400 nucleotides revealed approximately 5% sequence divergence between strain D8 and B. vulgatus and indicated that the 16S rRNA gene of our isolate was most similar to those of the recently described (3) strains B. dorei 175T (99.7%) and B. dorei 219 (99.6%). Interestingly, the closest uncultured sequences matching these sequences were all from humans. When we used the RDP-II database with only cultivated strains, we also detected a strain originating from the bovine rumen (strain Ar20) (data not shown). A tree was generated by the neighbor-joining method and showed the phylogenetic position of Bacteroides sp. strain D8 among recognized members of the genus Bacteroides (Fig. 3). The tree also confirmed the phylogenetic affinity with B. vulgatus and indicated that Bacteroides sp. strain D8 was part of a clade with the two B. dorei isolates.
FIG. 3.
Neighbor-joining tree showing the phylogenetic position of Bacteroides sp. strain D8 among recognized members of the genus Bacteroides based on 16S rRNA gene sequences. The 16S rRNA gene sequence of Prevotella melaninogenica was used as an outgroup to root the tree. Bootstrap values are indicated at branch points of interest. A GenBank database accession number is included for each sequence in the phylogenetic tree. Bar = 10% sequence divergence.
A DNA-DNA reassociation value of 59% for Bacteroides sp. strain D8 and B. vulgatus ATCC 8482T was determined, confirming that these strains belong to related but distinct species. The G+C contents of DNA were determined to be 40 mol% for strain D8 and 41 mol% for B. vulgatus ATCC 8482T.
Growth and cholesterol reduction.
The anaerobic growth and cholesterol-reducing activity of Bacteroides sp. strain D8 were studied in SBM at 37°C. Bacteroides sp. strain D8 grew to a concentration of 1010 CFU/ml in 2 days with a doubling time of 2.5 h in the log phase of growth. Cholesterol-to-coprostanol conversion began on day 3, while cells were in stationary phase, and was almost complete after 7 days (Fig. 4). 4-Cholesten-3-one and coprostanone, intermediate products of the indirect pathway of cholesterol-to-coprostanol conversion (31), were occasionally observed at low levels (<10% of the total neutral sterols). When Bacteroides sp. strain D8 was grown in BC medium containing 4-cholesten-3-one or coprostanone, these products were converted into coprostanol. No cholesterol conversion was obtained with B. dorei 175T grown in SBM, BC medium, or cholesterol-enriched BHI-YH medium. SBM cultures of Bacteroides sp. strain D8 could be kept for up to 1 year at 4°C without any loss of growth or cholesterol-reducing activity when they were inoculated into fresh SBM. Inversely, when an SBM inoculum was subcultured in BC or cholesterol-enriched BHI-YH medium, growth was sustained but the cholesterol-reducing activity progressively declined until extinction after two to five subcultures. Cholesterol conversion returned to its previous level after two subcultures when these cultures were switched back to SBM. In contrast, the cholesterol-reducing activity of E. coprostanoligenes ATCC 51222T was sustained over repeated subcultures in BC medium, whereas growth and activity rapidly declined when this bacterium was transferred into SBM (data not shown).
FIG. 4.
Growth and kinetics of cholesterol-to-coprostanol conversion by Bacteroides sp. strain D8 in SBM at 37°C. Symbols: ▪, cholesterol content; □, coprostanol content; ▴, log CFU per ml.
Resting-cell experiments.
The results of resting-cell assays are summarized in Fig. 5. When nondisrupted cells (approximately 200 μg bacterial protein per assay mixture) were incubated in the presence of cholesterol-lecithin vesicles (0.46 mg cholesterol per assay mixture) and under anaerobic conditions, 12.4% of the cholesterol substrate was reduced to coprostanol during the first 30 min, corresponding to 0.57 mg (1.5 μmol) cholesterol reduced/mg bacterial protein/h. When cells were disrupted, the yield was three times lower. A weak cholesterol-to-coprostanol reducing activity (2.5% of the cholesterol substrate reduced to coprostanol) was found in the cell-free supernatant. With prolonged incubation (24 h), 20% of the cholesterol substrate was reduced to coprostanol by nondisrupted cells. Again, the yield was three times lower with disrupted cells, and less than 3% of the cholesterol was reduced in the presence of the supernatant alone. No intermediate products could be detected over 30-min or 24-h incubation periods. No background reducing activity could be detected in control assay mixtures devoid of cells or supernatant.
FIG. 5.
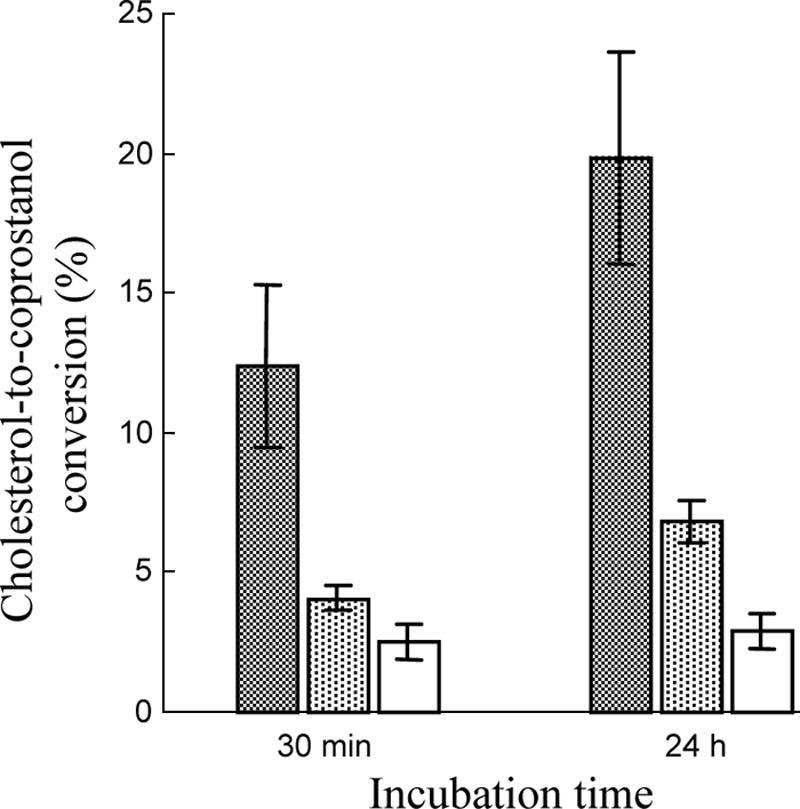
Cholesterol-to-coprostanol conversion by nondisrupted cells (cross-hatched bars), disrupted cells (dotted bars), and supernatants (open bars) produced from 24-h-old Bacteroides sp. strain D8 cultures in BHI-YH medium. The data are means ± standard errors of experiments performed in triplicate. The conditions used for the resting-cell assays are described in Materials and Methods.
TTGE profile of Bacteroides population.
In a previous study, we established that the population level of cultivable fecal coprostanoligenic bacteria correlated with the intensity of cholesterol-to-coprostanol conversion in the human gut. Indeed, we found that populations containing less than 106, between 106 and 108, and more than 108 coprostanoligenic bacteria/g (wet weight) of stool are associated with inefficient, intermediate, and high cholesterol conversion patterns, respectively (38). In the present study, fecal communities belonging to the Bacteroides-Porphyromonas-Prevotella group from 11 humans with different cholesterol conversion patterns were profiled by TTGE of amplified 16S rRNA gene fragments in order to estimate the incidence of Bacteroides sp. strain D8 in humans. PCR with specific primers amplified a 205-bp fragment of the V6 region of the 16S rRNA gene. TTGE of the amplicons obtained resulted in profiles exhibiting low (less than 5 bands) to moderate (around 10 bands) complexity (Fig. 6). Variation in the composition of the fingerprints could be observed, although some bands were shared by several individuals. As expected from their 16S rRNA gene sequences, Bacteroides sp. strain D8 and B. dorei 175T produced the same profile with one major band (Fig. 6). A band comigrating with this major band was obtained from three fecal samples from individuals exhibiting inefficient, intermediate, and high cholesterol conversion patterns, respectively (Fig. 6).
FIG. 6.
Bacteroides spp. group-specific TTGE profiles of fecal samples from 11 individuals with different cholesterol conversion patterns. Lanes 1 to 3, low or inefficient cholesterol converters; lanes 4 to 6, intermediate cholesterol converters; lanes 7 to 11, high cholesterol converters; lane 12, Bacteroides sp. strain D8; lane 13, B. dorei 175T. The arrow indicates the main band obtained with Bacteroides sp. strain D8 and B. dorei 175T. Circles indicate the bands comigrating with this band. Lane M contained markers corresponding to mixed PCR products from pure cultures.
DISCUSSION
Although cholesterol-to-coprostanol conversion in the human gut is a well-established phenomenon defined as a microbiota-associated characteristic, the bacteria responsible for this metabolism are still virtually unknown. Only a few cholesterol-reducing strains have been isolated from rat or baboon feces (7, 12). All these strains exhibited similar properties and have tentatively been assigned to the genus Eubacterium. More recently, a small anaerobic gram-positive coccobacillus that reduces cholesterol to coprostanol was isolated from a hog sewage lagoon (13). Because of its unique morphological and physiological properties, the authors proposed this organism as the type strain (ATCC 51222) of a new species, E. coprostanoligenes. The present paper is the first published report of the isolation and characterization of a cholesterol-reducing bacterium of human origin.
Differences in cholesterol-to-coprostanol conversion among human populations have been reported in different studies which included North American individuals consuming a normal mixed Western diet (40), omnivorous and vegetarian women (20), and healthy Norwegian subjects (27). Of these individuals, approximately 20% had a coprostanol content representing less than one-third of their fecal neutral sterols, whereas in the remaining subjects there was almost complete cholesterol conversion. It has been suggested that low or inefficient conversion could be due to a lack of mucosal receptors for coprostanol-producing bacteria or to inhibition of these bacteria by other members of the gut microbiota (27). Only recently has it been demonstrated that the rate of cholesterol-to-coprostanol conversion in the human gut results mainly from the abundance of coprostanoligenic bacteria (14, 38). The level of cholesterol-reducing bacteria must be at least 106 cells/g (wet weight) of stool to efficiently convert cholesterol in the human gut, while a population containing more than 108 cells/g (wet weight) of stool leads to nearly complete conversion (38). As inferred from the isolation procedure, Bacteroides sp. strain D8 occurred at a concentration of ≥108 CFU/g (wet weight) of stool, while the total concentration of cultivable bacteria was 2.6 × 1010 CFU/g (wet weight) of stool. The new coprostanoligenic strain was therefore part of the dominant fecal microbiota and by itself could account for the overall cholesterol-to-coprostanol conversion in the volunteer in whom 80% of the total neutral animal sterols were represented by coprostanol.
Recently, our understanding of complex microbial communities has been greatly enhanced by the introduction of molecular techniques. In particular, denaturing gel electrophoresis profiling of 16S rRNA gene amplicons has been used for successful analysis of the human gut microbiota (21, 38, 41). These methods separate PCR amplicons that are the same length on the basis of differences in base composition. When universal bacterial primers are used, a complex banding pattern is obtained, corresponding to the dominant species of the microbiota. The use of genus-specific primers allows a more in-depth view of subpopulations as it results in less complex banding patterns that display only the diversity of the targeted group. In this study, a PCR-TTGE method was developed to generate a profile of Bacteroides spp. in fecal samples from 11 healthy human subjects whose cholesterol conversion patterns were previously determined (38). The Bacteroides-Porphyromonas-Prevotella-specific primers used were designed as 16S rRNA-targeted oligonucleotide probes for fluorescence in situ hybridization experiments (10) and were later used as primers in PCRs (17). Using this technique, the same major band was obtained with Bacteroides sp. strain D8 and B. dorei 175T. A band comigrating with this band was observed in one individual from each group (low, intermediate, and high cholesterol converters). This result indicates that B. dorei may be a common Bacteroides species in the human gut but that Bacteroides sp. strain D8 is unlikely to be mainly responsible for cholesterol conversion in the human population and that other bacterial species also have this function.
The mechanism of cholesterol-to-coprostanol reduction has been investigated for many years. Two pathways, direct and indirect, have been proposed for this conversion. The direct pathway theory was supported by the fact that no significant change occurred in the labeling pattern of suitably marked cholesterol (33). Nevertheless, the conclusions were called into question because intermediate cofactors might remove and restore the label (4). More recently, it was shown that isomerization of the 5-6 double bond to a 4-5 double bond occurred via a mechanism involving the transfer of C-4 H to the C-6 position during the cholesterol-to-coprostanol conversion by E. coprostanoligenes, indicating that there was an indirect pathway involving the formation of 4-cholesten-3-one (31). In the present study, 4-cholesten-3-one and coprostanone were detected during cholesterol-to-coprostanol conversion by Bacteroides sp. strain D8 grown on SBM. Moreover, we showed that Bacteroides sp. strain D8 is able to convert 4-cholesten-3-one and coprostanone to coprostanol. Thus, our results are consistent with the presence of such an indirect pathway for coprostanol production by Bacteroides sp. strain D8.
The cholesterol-to-coprostanol reduction efficiency of undisrupted resting cells of Bacteroides sp. strain D8 was found to be 0.57 mg (1.5 μmol) cholesterol reduced/mg bacterial protein/h. This yield is higher than the maximum yields previously obtained with the E. coprostanoligenes ATCC 51222T strain (1.2 μmol cholesterol reduced/mg protein/h) (22) and would probably have been still higher if Bacteroides sp. strain D8 resting cells could have been obtained directly from SBM rather than via subculture in BHY-YH medium. In E. coprostanoligenes ATCC 51222T considerable loss of activity was also observed as soon as cells were disrupted by sonication, passage through a French press, or enzymatic digestion. In the latter study, resting cells (but not actively growing cultures) of E. coprostanoligenes ATCC 51222T were shown to produce intermediate reaction products (4-cholesten-3-one and coprostanone), while coprostanol was the only measurable product in our resting-cell assays.
Because the cholesterol-reducing activity of Bacteroides sp. strain D8 was sustained only in SBM, our results suggest that this activity would be inducible by brain tissue-specific constituents which were not present in BC and cholesterol-enriched BHI-YH media or inhibited by constituents or metabolic products present in the latter media. Conversely, we were able to verify that the cholesterol-reducing activity of E. coprostanoligenes ATCC 51222T was sustained over repeated subcultures in BC medium, whereas growth and activity rapidly declined when this bacterium was transferred into SBM (data not shown). It therefore appears that the growth and activity requirements of cholesterol-reducing bacteria are highly strain and medium dependent. However, it is notable that both Bacteroides sp. strain D8 and E. coprostanoligenes ATCC 51222T (13) started to reduce cholesterol to coprostanol on the third day of growth in their respective media and needed 7 days to achieve complete cholesterol conversion.
Unlike all other cholesterol-reducing strains isolated so far, phenotypic and phylogenetic analyses revealed that strain D8 belongs to the Cytophaga-Flavobacter-Bacteroides phylum and is part of the Bacteroides fragilis cluster. Bacteroides is the most commonly cultivated genus in the human fecal microbiota, accounting for approximately 30% of fecal isolates (34). These bacteria play a variety of roles as members of the indigenous microbiota that contribute to normal gut physiology and function, whereas several Bacteroides species are important opportunistic pathogens. Bacteroides species are involved in hydrolysis and fermentation of exogenous polysaccharides and endogenous mucins, metabolism of xenobiotics, inactivation of trypsin activity, production of toxins, or modulation of host epithelial cell activities (16, 30, 34). Moreover, although cholesterol-reducing activity has never been reported in the genus Bacteroides, steroid metabolism, including bile salt hydrolase, sulfatase, and glucuronidase activities, has been detected in different Bacteroides species (25). Sequences from novel phylotypes of yet-to-be-cultured Bacteroides species have been detected in 16S rRNA gene clone libraries from human fecal samples (11, 37), and previously unknown Bacteroides species of human origin have recently been isolated and characterized (1, 2, 19). A comparison of 16S rRNA gene sequences revealed that the cholesterol-reducing strain D8 clustered with the two isolates described as B. dorei, displaying >99.5% sequence similarity. Although there is no precise correlation between 16S rRNA sequence divergence values and species delineation in some bacterial genera, all the Bacteroides species described so far have displayed divergence of >2%, suggesting that strain D8 constitutes a new isolate of the recently described species B. dorei (3). Phylogenetic tree construction also supports this assertion as the three isolates constitute an independent clade within the genus Bacteroides. Interestingly, all the bacterial strains belonging to this clade, including uncultured phylotypes, originated from mammalian, mostly human, digestive tracts. Biochemical properties determined with API 20A and API rapid ID 32A systems were found to be identical for Bacteroides sp. strain D8 and the B. dorei type strain. Nevertheless, no cholesterol-reducing activity could be detected in B. dorei type strain cultures, and no correlation was found between the incidence of this species and the cholesterol conversion pattern. These results indicate that not all the isolates belonging to this clade are able to reduce cholesterol to coprostanol.
Acknowledgments
We are grateful to D. Beitz for providing E. coprostanoligenes ATCC 51222T and for recommendations for preparing BC medium, to P. Lepage for TTGE advice, and to T. Meylheuc for scanning electron microscopy. We thank F. Béguet, V. Bourland, C. Bridonneau, J. Dabard, and J.-A. Martins for their technical assistance. We also thank G. Corthier, J. Doré, P. Langella, S. Rabot, J.-P. Furet, and O. Firmesse for useful discussions and V. Hawken for her careful reading of the English version of the manuscript.
Footnotes
Published ahead of print on 6 July 2007.
REFERENCES
- 1.Bakir, M. A., M. Kitahara, M. Sakamoto, M. Matsumoto, and Y. Benno. 2006. Bacteroides intestinalis sp. nov., isolated from human faeces. Int. J. Syst. Evol. Microbiol. 56:151-154. [DOI] [PubMed] [Google Scholar]
- 2.Bakir, M. A., M. Kitahara, M. Sakamoto, M. Matsumoto, and Y. Benno. 2006. Bacteroides finegoldii sp. nov., isolated from human faeces. Int. J. Syst. Evol. Microbiol. 56:931-935. [DOI] [PubMed] [Google Scholar]
- 3.Bakir, M. A., M. Sakamoto, M. Kitahara, M. Matsumoto, and Y. Benno. 2006. Bacteroides dorei sp. nov., isolated from human faeces. Int. J. Syst. Evol. Microbiol. 56:1639-1643. [DOI] [PubMed] [Google Scholar]
- 4.Björkhem, I., and J. Gustafsson. 1971. Mechanism of microbial transformation of cholesterol into coprostanol. Eur. J. Biochem. 21:428-432. [DOI] [PubMed] [Google Scholar]
- 5.Bligh, E. G., and W. J. Dyer. 1959. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37:911-917. [DOI] [PubMed] [Google Scholar]
- 6.Brinkley, A. W., A. R. Gottesman, and G. E. Mott. 1980. Growth of cholesterol-reducing Eubacterium on cholesterol-brain agar. Appl. Environ. Microbiol. 40:1130-1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brinkley, A. W., A. R. Gottesman, and G. E. Mott. 1982. Isolation and characterization of new strains of cholesterol-reducing bacteria from baboons. Appl. Environ. Microbiol. 43:86-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dam, H. 1934. The formation of coprosterol in the intestine: the action of intestinal bacteria on cholesterol. Biochem. J. 28:820-825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.De Ley, J., H. Cattoir, and A. Reynaerts. 1970. The quantitative measurement of DNA hybridization from renaturation rates. Eur. J. Biochem. 12:133-142. [DOI] [PubMed] [Google Scholar]
- 10.Doré, J., A. Sghir, G. Hannequart-Gramet, G. Corthier, and P. Pochart. 1998. Design and evaluation of a 16S rRNA-targeted oligonucleotide probe for specific detection and quantitation of human faecal Bacteroides populations. Syst. Appl. Microbiol. 21:65-71. [DOI] [PubMed] [Google Scholar]
- 11.Eckburg, P. B., E. M. Bik, C. N. Bernstein, E. Purdom, L. Dethlefsen, M. Sargent, S. R. Gill, K. E. Nelson, and D. A. Relman. 2005. Diversity of the human intestinal microbial flora. Science 308:1635-1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eyssen, H. J., G. G. Parmentier, F. C. Compernolle, G. De Pauw, and M. Piessens-Denef. 1973. Biohydrogenation of sterols by Eubacterium ATCC 21408 nova species. Eur. J. Biochem. 36:411-421. [DOI] [PubMed] [Google Scholar]
- 13.Freier, T. A., D. C. Beitz, L. Li, and P. A. Hartman. 1994. Characterization of Eubacterium coprostanoligenes sp. nov., a cholesterol-reducing anaerobe. Int. J. Syst. Bacteriol. 44:137-142. [DOI] [PubMed] [Google Scholar]
- 14.Gérard, P., F. Béguet, P. Lepercq, L. Rigottier-Gois, V. Rochet, C. Andrieux, and C. Juste. 2004. Gnotobiotic rats harboring human intestinal microbiota as a model for studying cholesterol-to-coprostanol conversion. FEMS Microbiol. Ecol. 47:337-343. [DOI] [PubMed] [Google Scholar]
- 15.Godon, J.-J., E. Zumstein, P. Dabert, F. Habouzit, and R. Moletta. 1997. Molecular microbial diversity of an anaerobic digestor as determined by small-subunit rDNA sequence analysis. Appl. Environ. Microbiol. 63:2802-2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hooper, L. V., M. H. Wong, A. Thelin, L. Hansson, P. G. Falk, and J. I. Gordon. 2001. Molecular analysis of commensal host-microbial relationships in the intestine. Science 291:881-884. [DOI] [PubMed] [Google Scholar]
- 17.Hopkins, M. J., G. T. Macfarlane, E. Furrie, A. Fite, and S. Macfarlane. 2005. Characterisation of intestinal bacteria in infant stools using real-time PCR and northern hybridisation analyses. FEMS Microbiol. Ecol. 54:77-85. [DOI] [PubMed] [Google Scholar]
- 18.Kaneuchi, C., K. Watanabe, A. Terada, Y. Benno, and T. Mitsuoka. 1976. Taxonomic study of Bacteroides clostridiiformis subsp. clostridiiformis (Burri and Ankersmith) Holdeman and Moore and of related organisms: proposal of Clostridium clostridiiformis (Burri and Ankersmith) comb. nov. and Clostridium symbosium (Stevens) comb. nov. Int. J. Syst. Bacteriol. 26:195-204. [Google Scholar]
- 19.Kitahara, M., M. Sakamoto, M. Ike, S. Sakata, and Y. Benno. 2005. Bacteroides plebeius sp. nov. and Bacteroides coprocola sp. nov., isolated from human faeces. Int. J. Syst. Evol. Microbiol. 55:2143-2147. [DOI] [PubMed] [Google Scholar]
- 20.Korpela, J. T., and H. Adlerkreutz. 1985. Fecal neutral sterols in omnivorous and vegetarian women. Scand. J. Gastroenterol. 20:1180-1184. [DOI] [PubMed] [Google Scholar]
- 21.Lepage, P., P. Seksik, M. Sutren, M. F. de la Cochetiere, R. Jian, P. Marteau, and J. Doré. 2005. Biodiversity of the mucosa-associated microbiota is stable along the distal digestive tract in healthy individuals and patients with IBD. Inflamm. Bowel Dis. 11:473-480. [DOI] [PubMed] [Google Scholar]
- 22.Li, L., T. A. Freier, P. A. Hartman, J. W. Young, and D. C. Beitz. 1995. A resting-cell assay for cholesterol reductase activity in Eubacterium coprostanoligenes ATCC 51222. Appl. Microbiol. Biotechnol. 43:887-892. [Google Scholar]
- 23.Lichtenstein, A. H. 1990. Intestinal cholesterol metabolism. Ann. Med. 22:49-52. [DOI] [PubMed] [Google Scholar]
- 24.Ludwig, W., O. Strunk, R. Westram, L. Richter, H. Meier, Yadhukumar, A. Buchner, T. Lai, S. Steppi, G. Jobb, W. Förster, I. Brettske, S. Gerber, A. W. Ginhart, O. Gross, S. Grumann, S. Hermann, R. Jost, A. König, T. Liss, R. Lüβmann, M. May, B. Nonhoff, B. Reichel, R. Strehlow, A. Stamatakis, N. Stuckmann, A. Vilbig, M. Lenke, T. Ludwig, A. Bode, and K.-H. Schleifer. 2004. ARB: a software environment for sequence data. Nucleic Acids Res. 32:1363-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Macdonald, I. A., V. D. Bokkenheuser, J. Winter, A. M. McLernon, and E. H. Mosbach. 1983. Degradation of steroids in the human gut. J. Lipid Res. 24:675-700. [PubMed] [Google Scholar]
- 26.Marmur, J., and P. Doty. 1962. Determination of base composition of deoxyribonucleic acid from its thermal denaturation temperature. J. Mol. Biol. 5:109-118. [DOI] [PubMed] [Google Scholar]
- 27.Midtvedt, T., E. Lingaas, B. Carlstedt-Duke, T. Höverstad, A.-C. Midtvedt, H. Saxerholt, M. Steinbakk, and K. E. Norin. 1990. Intestinal microbial conversion of cholesterol to coprostanol in man. APMIS 98:839-844. [DOI] [PubMed] [Google Scholar]
- 28.Panda, S. K., S. C. Chattoraj, and S. A. Broitman. 1999. Correlation of neomycin, faecal neutral and acid sterols with colon carcinogenesis in rats. Br. J. Cancer 80:1132-1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Peuchant, E., C. Salles, and R. Jensen. 1987. Relationship between fecal neutral steroid concentrations and malignancy in colon cells. Cancer 60:994-999. [DOI] [PubMed] [Google Scholar]
- 30.Ramare, F., I. Hautefort, F. Verhe, P. Raibaud, and J. Iovanna. 1996. Inactivation of tryptic activity by a human-derived strain of Bacteroides distasonis in the large intestines of gnotobiotic rats and mice. Appl. Environ. Microbiol. 62:1434-1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ren, D., L. Li, S. W. Schwabacher, J. W. Young, and D. C. Beitz. 1996. Mechanism of cholesterol reduction to coprostanol by Eubacterium coprostanoligenes ATCC 51222. Steroids 61:33-40. [DOI] [PubMed] [Google Scholar]
- 32.Riottot, M., P. Olivier, A. Huet, J. J. Caboche, M. Parquet, J. Khallou, and C. Lutton. 1993. Hypolipidemic effects of β-cyclodextrin in the hamster and in the genetically hypercholesterolemic Rico rat. Lipids 28:181-188. [DOI] [PubMed] [Google Scholar]
- 33.Rosenfeld, R. S., and T. F. Gallagher. 1964. Further studies of the biotransformation of cholesterol to coprostanol. Steroids 4:515-520. [Google Scholar]
- 34.Salyers, A. A. 1984. Bacteroides of the human lower intestinal tract. Annu. Rev. Microbiol. 38:293-313. [DOI] [PubMed] [Google Scholar]
- 35.Sekimoto, H., O. Shimada, M. Makanishi, T. Nakano, and O. Katayama. 1983. Interrelationship between serum and fecal sterols. Jpn. J. Med. 22:14-20. [DOI] [PubMed] [Google Scholar]
- 36.Smibert, R. M., and N. R. Krieg. 1994. Phenotypic characterization, p. 607-654. In P. Gerhardt, R. G. E. Murray, W. A. Wood, and N. R. Krieg (ed.), Methods for general and molecular bacteriology. American Society for Microbiology, Washington, DC.
- 37.Suau, A., R. Bonnet, M. Sutren, J. J. Godon, G. R. Gibson, M. D. Collins, and J. Doré. 1999. Direct analysis of genes encoding 16S rRNA from complex communities reveals many novel molecular species within the human gut. Appl. Environ. Microbiol. 65:4799-4807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Veiga, P., C. Juste, P. Lepercq, K. Saunier, F. Béguet, and P. Gérard. 2005. Correlation between faecal microbial community structure and cholesterol-to-coprostanol conversion in the human gut. FEMS Microbiol. Lett. 242:81-86. [DOI] [PubMed] [Google Scholar]
- 39.Weisburg, W. G., S. M. Barns, D. A. Pelletier, and D. J. Lane. 1991. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 173:697-703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wilkins, T. D., and A. S. Hackman. 1974. Two patterns of neutral sterol conversion in the feces of normal North Americans. Cancer Res. 34:2250-2254. [PubMed] [Google Scholar]
- 41.Zoetendal, E. G., A. D. Akkermans, and W. M. De Vos. 1998. Temperature gradient gel electrophoresis analysis of 16S rRNA from human fecal samples reveals stable and host-specific communities of active bacteria. Appl. Environ. Microbiol. 64:3854-3859. [DOI] [PMC free article] [PubMed] [Google Scholar]