Abstract
An annual seasonal cycle of composition of a bacterioplankton community in an oligotrophic coastal system was studied by denaturing gradient gel electrophoresis (DGGE) using five different primer sets. Analysis of DGGE fingerprints showed that primer set 357fGC-907rM grouped samples according to seasons. Additionally, we used the set of 16S rRNA genes archived in the RDPII database to check the percentage of perfect matches of each primer for the most abundant bacterial groups inhabiting coastal plankton communities. Overall, primer set 357fGC-907rM was the most suitable for the routine use of PCR-DGGE analyses in this environment.
Denaturing gradient gel electrophoresis (DGGE) (11, 12) has become one of the most frequently used methods for molecular fingerprinting. Although it is used widely in different areas of research (5, 7, 8, 10, 14), there seems to be little testing or justification for the correct choice of primers for each type of microbial community.
In the present work we have compared the PCR-DGGE profiles of a bacterioplankton community during a seasonal cycle in Blanes Bay Microbial Observatory by using five different primer sets. Seasonality of the bacterial assemblage in Blanes Bay has been studied previously by Schauer et al. (15) by use of DGGE with a single set of primers. That analysis showed that changes in the dominant bacterial members over the sampling period occurred at a gradual pace, without abrupt changes in composition. Alternative PCR-dependent (clone libraries) and PCR-independent (catalyzed reporter deposition-fluorescent in situ hybridization) studies of bacterioplankton seasonality have also been carried out in Blanes Bay (1), demonstrating that a detailed picture of seasonality can be obtained by combining several molecular approaches. Oceanic biogeochemical conditions have been shown to correctly predict the presence of annually recurring bacterial communities (6), thus suggesting that a link between these parameters exists.
A seasonal cycle provides a template for the temporal dynamics of the community, since samples from the same season are expected to have more similar fingerprints than samples that are distant in time. Thus, the most suitable set of primers will be the one that better reflects such seasonality. We optimized DGGE conditions for five primer sets commonly used in the literature and evaluated the community compositions inferred from the DGGE profiles. Additionally, we checked the number of perfect matches of each of the primers to 16S rRNA genes in the RDPII database (release 9.39). The results provide information about the most suitable DGGE conditions and primer sets for amplification of bacterioplankton communities.
Sampling.
Surface seawater samples (first 20 to 50 cm) were collected monthly during one seasonal cycle (from February 2003 to March 2004) from the Blanes Bay Microbial Observatory on the Catalan coast (northwest Mediterranean), 70 km north of Barcelona, Spain. Samples were taken 1 km offshore (41°40′N, 2°48′E), and seawater was kept in 25-liter polycarbonate carboys for less than 2 h until processing.
To collect bacterioplankton biomass, 5 liters of seawater was filtered by use of a peristaltic pump through a 3-μm-pore-size polycarbonate filter and a 0.2-μm Sterivex filter (Durapore; Millipore) in succession. The Sterivex unit was filled with 1.8 ml of lysis buffer and stored at −80°C.
PCR-DGGE fingerprinting.
Nucleic acid extraction was performed as described by Massana et al. (9). Five different sets of primers were tested in order to obtain fragments of the 16S rRNA gene suitable for DGGE analysis. Their sequences, as well as the annealing temperatures and DGGE conditions used in this study, are detailed in Table 1 (for PCR details, see the supplemental material).
TABLE 1.
Primers and PCR and DGGE conditions used in this study
| Primer | Sequence (5′-3′) | Annealing positions (nt)b | Target region | Annealing conditions | Amplicon length (bp)c | DGGE conditionsd | Total no. of matches (%)e | Reference |
|---|---|---|---|---|---|---|---|---|
| 63fa | GCC TAA CAC ATG CAA GTC | 46-63 | V1-V3 | 67-57°C, −1°C/cycle | 489 | 6%, 40-80%, 100 V, 17 h | 19,390 (21.5) | 2 |
| 518r | ATT ACC GCG GCT GCT GG | 518-534 | V1-V3 | 67-57°C, −1°C/cycle | 489 | 6%, 40-80%, 100 V, 17 h | 79,965 (88.6) | 2 |
| 357fa | CCT ACG GGA GGC AGC AG | 341-357 | V3-V5 | 65-55°C, −1°C/cycle | 586 | 6%, 40-80%, 100 V, 17 h | 83,279 (92.3) | 12 |
| 907rM | CCG TCA ATT CMT TTG AGT TT | 907-926 | V3-V5 | 65-55°C, −1°C/cycle | 586 | 6%, 40-80%, 100 V, 17 h | 81,609 (90.5) | 12 |
| 357fa | CCT ACG GGA GGC AGC AG | 341-357 | V3-V5 | 65-55°C, −1°C/cycle | 586 | 6%, 40-80%, 100 V, 17 h | 83,279 (92.3) | 13 |
| 907r | CCG TCA ATT CCT TTR AGT TT | 907-926 | V3-V5 | 65-55°C, −1°C/cycle | 586 | 6%, 40-80%, 100 V, 17 h | 71,026 (78,7) | 13 |
| 357fa | CCT ACG GGA GGC AGC AG | 341-357 | V3 | 65-55°C, −1°C/cycle | 194 | 8%, 40-80%, 75 V, 18 h | 83,279 (92.3) | 10 |
| 518r | ATT ACC GCG GCT GCT GG | 518-534 | V3 | 65-55°C, −1°C/cycle | 194 | 8%, 40-80%, 75 V, 18 h | 79,965 (88.6) | 10 |
| 968fa | AAC GCG AAG AAC CTT AC | 968-984 | V6-V8 | 63-53°C, −1°C/cycle | 434 | 6%, 40-80%, 100 V, 17 h | 52,285 (58.0) | 3 |
| 1401r | CGG TGT GTA CAA GAC CC | 1385-1401 | V6-V8 | 63-53°C, −1°C/cycle | 434 | 6%, 40-80%, 100 V, 17 h | 11,708 (13.0) | 3 |
| 1055f | ATG GCT GTC GTC AGC T | 1055-1070 | V8 | 65-55°C, −1°C/cycle | 352 | 8%, 40-80%, 75 V, 18 h | 46,218 (51.2) | 4 |
| 1392ra | ACG GGC GGT GTG TRC | 1392-1406 | V8 | 65-55°C, −1°C/cycle | 352 | 8%, 40-80%, 75 V, 18 h | 60,007 (66.5) | 4 |
Primer with a 40-bp clamp at the 5′ end.
Numbering according to the rrs gene of Escherichia coli. nt, nucleotides.
Calculated from the rrs gene of E. coli; the primers are included.
Conditions given are percent polyacrylamide, denaturing gradient, voltage, and running time, respectively.
The number of perfect matches for each primer to the RDPII database (release 9.39, May 2006) is given. The percentage is calculated from a total of 92,211 sequences with more than 1,200 bases.
DGGE analysis was run with a DCode (Bio-Rad) and a CBS system as previously described by Muyzer et al. (12). Nine hundred nanograms of PCR product was loaded for each sample.
Perpendicular DGGE analyses were carried out in order to select appropriate electrophoresis conditions for each set of primers. One thousand five hundred nanograms of PCR product from different cultures corresponding to Roseobacter sp. strain MED193, Polaribacter sp. strain MED152, and Dokdonia sp. strain MED134 obtained from the culture collection of the Blanes Bay Microbial Observatory was applied across the entire width of the gel and electrophoresed at 150 V for about 3 to 5 h. At a denaturant concentration range of 45 to 65% (63fGC-518r), 50 to 65% (357fGC-907rM), 45 to 65% (357fGC-518r), 45 to 60% (968fGC-1401r), or 50 to 60% (1055f-1392rGC), the three PCR products displayed reduced mobility. We thus confirmed that a gradient of 40 to 80% was adequate for all primers.
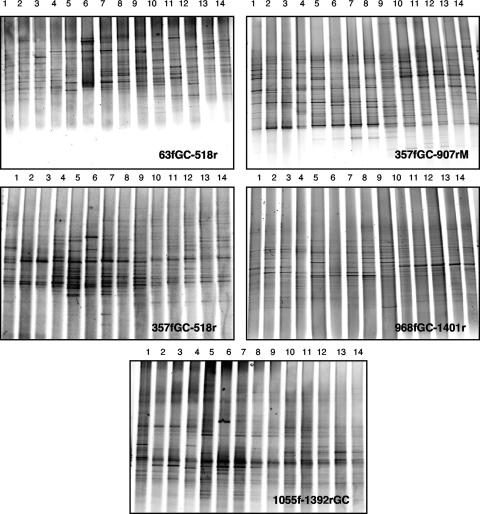
Using this 40 to 80% gradient, we ran five different DGGE gels including the 14 coastal samples from Blanes Bay obtained during a 1-year cycle (Fig. 1).
FIG. 1.
DGGE fingerprints with different primer sets of bacterial assemblages obtained from Blanes Bay during a seasonal cycle (lane 1, February 2003; lane 2, March 2003; lane 3, April 2003; lane 4, May 2003; lane 5, June 2003; lane 6, July 2003; lane 7, August 2003; lane 8, September 2003; lane 9, October 2003; lane 10, November 2003; lane 11, December 2003; lane 12, January 2004; lane 13, February 2004; lane 14, March 2004).
Quantitative analyses.
Digitized DGGE images were analyzed with Quantity One software (Bio-Rad) (for details, see the supplemental material). Bands occupying the same position in the different lanes of the gels were identified. Primer set 357fGC-907rM retrieved a larger number of bands both globally (462 bands) and in each sample (average, 33.0 bands).
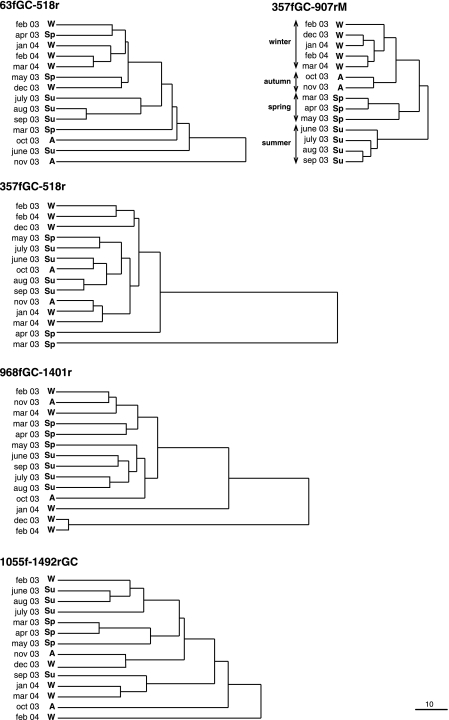
A matrix was constructed for all lanes, taking into account the presence or absence of the individual bands and the relative contribution of each band (by percentage) to the total intensity of the lane. This matrix was used to calculate a distance matrix using normalized Euclidean distances (root mean square differences) with the software STATISTICA. Finally, a dendrogram comparing samples for each set of primers was obtained by use of the unweighted-pair group method using average linkages and STATISTICA (Fig. 2). Primer set 357fGC-907rM clustered the samples according to different seasons. In contrast, other primers did not show a completely coherent seasonal clustering.
FIG. 2.
Euclidean-distance dendrograms generated from the DGGE profiles of the 14 samples analyzed for each primer set, determined by the unweighted-pair group method using average linkages. The scale bar is linkage distance and applies to all dendrograms. To clarify interpretation, the season of each sample is indicated (W, winter; Sp, spring; Su, summer; A, autumn). Dates during which samples were obtained are indicated by month and year.
Statistical analyses.
We used ancillary physical and ecological data from the Blanes Bay Observatory to run one-way analyses of variance (ANOVA) according to the classification based on DGGE clusters. For details of statistical analyses, see the supplemental material.
As can be seen in Table 2, most variables, particularly the phytoplankton-related variables, nutrients, and other physical variables, showed that the seasonal classification carried out by DGGE with primer set 357fGC-907rM explained the variability in the data. Significant exceptions were NH4 and bacterial abundance, two variables that seemed to be particularly well buffered in this and in most other systems (L. Alonso-Sáez, E. Vázquez-Domínguez, J. Pinhassi, C. Cardelús, M. M. Sala, I. Lekunberri, M. Vila-Costa, F. Unrein, R. Massana, R. Simó, and J. M. Gasol, submitted for publication). A post hoc Tukey-Kramer test classified summer as the most distinct season of the year (Table 2), in the same way as the DGGE dendrogram based on primer set 357fGC-907rM separated summer from the other seasons (Fig. 2).
TABLE 2.
Results of ANOVA done with the Blanes Bay Microbial Observatory physical and microbiological dataa
| Variable | No. of samples tested | F-value | P value | Tukey-Kramer post hoc test result
|
|||
|---|---|---|---|---|---|---|---|
| Summer | Autumn | Winter | Spring | ||||
| Temp | 101 | 165.0 | <0.001 | A | B | C | C |
| Secchi depth | 74 | 9.0 | <0.001 | A | B | B | B |
| Chlorophyll a | 102 | 14.0 | <0.001 | A | AB | C | BC |
| % Chlorophyll <3 μm | 81 | 3.6 | 0.018 | A | AB | B | AB |
| NH4 | 83 | 0.9 | NSb | ||||
| NO2 | 83 | 5.3 | 0.002 | A | AB | B | B |
| NO3 | 83 | 6.1 | 0.001 | A | AB | B | B |
| SiO2 | 83 | 4.0 | 0.011 | A | AB | B | B |
| Bacterial production | 56 | 2.6 | 0.065 | A | AB | B | AB |
| Leucine:thymidine ratio | 24 | 3.2 | 0.048 | A | A | A | A |
| Bacterial abundance | 50 | 1.5 | NS | ||||
| Heterotrophic nanoflagellates | 45 | 4.3 | 0.010 | A | AB | B | AB |
| Autotrophic nanoflagellates | 45 | 11.1 | <0.001 | A | B | B | B |
| Prochlorococcus | 89 | 16.6 | <0.001 | AB | C | A | B |
| Synechococcus | 89 | 14.8 | <0.001 | A | AB | C | B |
| Picoeukaryotes | 88 | 6.2 | 0.001 | A | A | B | AB |
The samples collected from 2000 to 2006 were classified in four categories as determined by the DGGE analysis (with primer set 357fGC-907rM), with the categories labeled according to season. A positive ANOVA result means that the classification of a variable into these groups significantly explained the variability in the data. The post hoc Tukey-Kramer test results indicate which groups are significantly different from the others, represented by different letters (A, B, and C).
NS, not significant.
We also tested whether the clustering generated by the other primer sets explained the seasonality of the variables. In most cases (70%), the F-value of ANOVA (used as an indicator of the ability of the clustering to organize the ecological data) was higher with primer set 357fGC-907rM than with any of the other significant ordinations with the other primers. In 20% of the cases, primer set 63fGC-518r organized better than the other sets of primers, and in 10% of the cases the best was primer set 357fGC-518r. Therefore, the first set (357fGC-907rM) ordered the environmental variables in an ecologically coherent fashion.
Probe match with RDPII database.
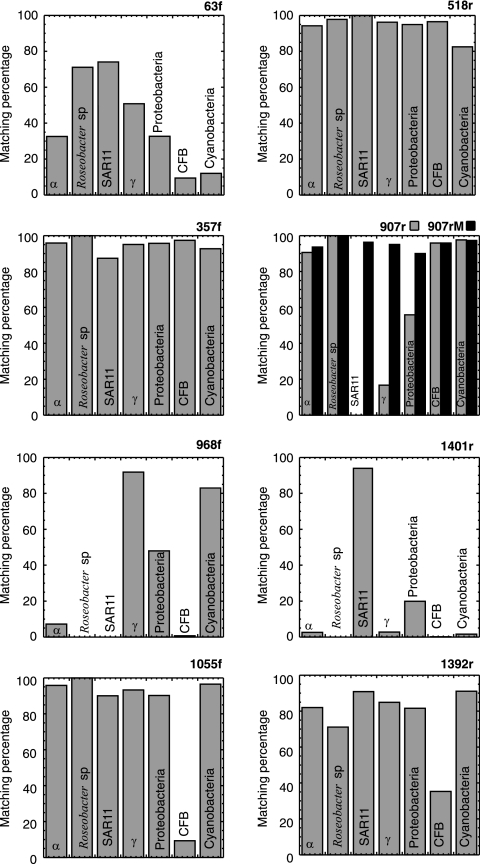
Additionally, the specificities of the primers selected for our study were analyzed against the RDPII database (release 9.39, 2006 [http://rdp.cme.msu.edu/]), which contained 90,211 bacterial sequences with almost the complete 16S rRNA gene sequence (≥1,200 bp). The numbers of sequences in the complete database with no mismatches varied widely. Three primers matched more than 75% of the sequences from the whole database (357f, 518r, and 907rM).
The percentages of sequences for the most representative groups of bacteria common in coastal bacterioplankton communities (Alphaproteobacteria [mostly from the SAR11 and Roseobacter clades], Gammaproteobacteria, Bacteroidetes [Cytophaga-Flavobacterium-Bacteroides, or CFB], and cyanobacteria) targeted by each primer were also different (Fig. 3). Some primers (518r, 357f, 907rM, and 1392r) matched these abundant groups of bacteria in Blanes Bay, while other primers (63f, 968f, 1055f, and 1401r) missed some of these groups, such as members of the Roseobacter (968f and 1401r), SAR11 (968f), and CFB (63f, 968f, 1401r, and 1055f) groups. However, it has to be taken into account that primers 63f and 1392r target the initial and final regions of the rrs gene, respectively, and that, although we have considered for the analysis of probe matching only sequences of more than 1,200 bp, the matching percentages for these primers are likely underestimates.
FIG. 3.
Histograms of matching percentages for the most abundant phylogenetic groups in coastal bacterioplankton obtained from data in the RDPII database, release 9.39 (sequences with ≥1,200 bases, 0 mismatches), for each primer used in this study (α, Alphaproteobacteria; γ, Gammaproteobacteria). Percentages have been calculated on the basis of 8,651 sequences of Alphaproteobacteria, 45 sequences of Roseobacter sp., 54 sequences of the SAR11 cluster, 13,993 sequences of Gammaproteobacteria, 33,203 sequences of proteobacteria, 12,021 sequences of CFB, and 1,812 sequences of cyanobacteria.
We also included primer 907r used by Schauer et al. (15) in the comparison. This primer differs by only 1 base from primer 907rM and does not perfectly match either the SAR11 cluster or some Gammaproteobacteria.
Although primer pair 357fGC-518r should be suitable to describe marine bacterioplankton, given that it matches with almost all sequences in the RDPII database, the quantitative analysis of the DGGE fingerprint showed that this set was not the best combination for Blanes Bay, since it did not reflect the seasonality of bacterial assemblage composition and the short length of the amplicon obtained limits the phylogenetic information contained in the sequenced bands.
Primer set 357fGC-907rM matched most of the groups present in Blanes Bay and seemed a priori a good choice for describing the bacterial community. Analysis of the DGGE fingerprints showed that primer set 357fGC-907rM was also the combination that better reflected the seasonal changes in the plankton of Blanes Bay. These primers generated clusters that were consistent with seasons (Fig. 2) and that corresponded to changes in the physicochemical and phytoplankton variables (Table 2).
In conclusion, the primer set 357fGC-907rM, which amplifies the V3 to V5 region of rrs genes, is recommended for the routine use of PCR-DGGE analyses of bacterioplankton samples at least from coastal Mediterranean waters but probably also from other coastal and open sea environments since the bacterial compositions in all of these systems are rather similar (1, 15). However, determination of the most suitable set of primers should be carried out for every habitat. Once appropriate fingerprints have been obtained, the most interesting samples can be selected with confidence for in depth analysis by other techniques, such as fluorescent in situ hybridization or clone libraries.
Supplementary Material
Acknowledgments
This work was supported by the Spanish projects MicroDiff (REN2001-2110/MAR), ESTRAMAR (CTM2004-12631/MAR), GENMμMAR (CTM2004-02586/MAR), TRAGUA (CSD2006-00044), TEC2006-13109-C03-02/MIC, MODIVUS (CTM2005-04975/MAR), and NoEs MARBEF and Marine Genomics Europe.
We thank E. L. Sà for her help in the lab, V. Balagué and I. Forn for their help with field sampling and lab support, K. Jürgens for his useful comments, L. Alonso-Sáez for access to her unpublished data, and all coworkers at Blanes for sharing the data.
Footnotes
Published ahead of print on 27 July 2007.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Alonso-Sáez, L., V. Balagué, E. L. Sà, O. Sánchez, J. M. González, J. Pinhassi, R. Massana, J. Pernthaler, C. Pedrós-Alió, and J. M. Gasol. 2007. Seasonality in bacterial diversity in north-west Mediterranean coastal waters: assessment through clone libraries, fingerprinting and FISH. FEMS Microbiol. Ecol. 60:98-112. [DOI] [PubMed] [Google Scholar]
- 2.El Fantroussi, S., L. Verschuere, W. Verstraete, and E. M. Top. 1999. Effect of phenylurea herbicides on soil microbial communities estimated by analysis of 16S rRNA gene fingerprints and community-level physiological profiles. Appl. Environ. Microbiol. 65:982-988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Felske, A., B. Engelen, U. Nübel, and H. Backhaus. 1996. Direct ribosome isolation from soil to extract bacterial rRNA for community analysis. Appl. Environ. Microbiol. 62:4162-4167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ferris, M. J., G. Muyzer, and D. M. Ward. 1996. Denaturing gradient gel electrophoresis profiles of 16S rRNA-defined populations inhabiting a hot spring microbial mat community. Appl. Environ. Microbiol. 62:340-346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fontana, C., G. Vignolo, and P. S. Cocconcelli. 2005. PCR-DGGE analysis for the identification of microbial populations from Argentinean dry fermented sausages. J. Microbiol. Methods 63:254-263. [DOI] [PubMed] [Google Scholar]
- 6.Fuhrman, J. A., I. Hewson, M. S. Swalbach, J. A. Steele, M. V. Brown, and S. Naeem. 2006. Annually reoccurring bacterial communities are predictable from ocean conditions. Proc. Natl. Acad. Sci. USA 103:13104-13109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lindström, E. S., and E. Leskinen. 2002. Do neighboring lakes share common taxa of bacterioplankton? Comparison of 16S rDNA fingerprints and sequences from three geographic regions. Microb. Ecol. 44:1-9. [DOI] [PubMed] [Google Scholar]
- 8.Martínez-Alonso, M., J. Bleijsjwijk, N. Gaju, and G. Muyzer. 2005. Diversity of anoxygenic phototrophic sulfur bacteria in the microbial mats of the Ebro Delta: a combined morphological and molecular approach. FEMS Microbiol. Ecol. 52:339-350. [DOI] [PubMed] [Google Scholar]
- 9.Massana, R., A. E. Murray, C. M. Preston, and E. F. DeLong. 1997. Vertical distribution and phylogenetic characterization of marine planktonic Archaea in the Santa Barbara Channel. Appl. Environ. Microbiol. 63:50-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Murray, A. E., J. T. Hollibaugh, and C. Orrego. 1996. Phylogenetic compositions of bacterioplankton from two California estuaries compared by denaturing gradient gel electrophoresis of 16S rDNA fragments. Appl. Environ. Microbiol. 62:2676-2680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Muyzer, G., E. de Waal, and A. G. Uitterlinden. 1993. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 59:695-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Muyzer, G., T. Brinkhoff, U. Nübel, C. Santegoeds, H. Schäfer, and C. Wawer. 1998. Denaturing gradient gel electrophoresis (DGGE) in microbial ecology, p. 3.4.4/1-3.4.4/27. In A. D. L. Akkermans, J. D. van Elsas, and F. J. Bruijn (ed.), Molecular microbial ecology manual. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 13.Muyzer, G., A. Teske, C. O. Wirsen, and H. W. Jannasch. 1995. Phylogenetic relationships of Thiomicrospira species and their identification in deep-sea hydrothermal vent samples by denaturing gradient gel electrophoresis of 16S rDNA fragments. Arch. Microbiol. 164:165-172. [DOI] [PubMed] [Google Scholar]
- 14.Schabereiter-Gurtner, C., C. Saiz-Jiménez, G. Pinar, W. Lubitz, and S. Rolleke. 2002. Phylogenetic 16S rRNA analysis reveals the presence of complex and partly unknown bacterial communities in Tito Bustillo cave, Spain, and on its Palaeolithic paintings. Environ. Microbiol. 4:392-400. [DOI] [PubMed] [Google Scholar]
- 15.Schauer, M., V. Balagué, C. Pedrós-Alió, and R. Massana. 2003. Seasonal changes in the taxonomic composition of bacterioplankton in a coastal oligotrophic system. Aquat. Microb. Ecol. 31:163-174. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.