Abstract
Dissimilatory nitrate reduction is catalyzed by a membrane-bound and a periplasmic nitrate reductase. We set up a real-time PCR assay to quantify these two enzymes, using the narG and napA genes, encoding the catalytic subunits of the two types of nitrate reductases, as molecular markers. The narG and napA gene copy numbers in DNA extracted from 18 different environments showed high variations, with most numbers ranging from 2 × 102 to 6.8 × 104 copies per ng of DNA. This study provides evidence that, in soil samples, the number of proteobacteria carrying the napA gene is often as high as that of proteobacteria carrying the narG gene. The high correlation observed between narG and napA gene copy numbers in soils suggests that the ecological roles of the corresponding enzymes might be linked.
Nitrate in the environment can be either assimilated by plants and microorganisms or reduced to nitrite by one of two microbial dissimilatory processes: denitrification or dissimilatory reduction of nitrate to ammonium. Nitrate reduction by denitrification is of great importance since the produced nitrite is then reduced to N2O or N2 gases, which can lead to considerable nitrogen losses in agriculture and emissions of greenhouse gases (6, 13, 28). The reduction of nitrate present in contaminated water caused by the nitrate-reducing bacteria living in the human digestive tract is a potential health problem. As nitrite enters the bloodstream, it reacts with hemoglobin to form methemoglobin, blocking oxygen transport and causing a disease commonly called “blue baby syndrome” (26). Two types of dissimilatory nitrate reductase, differing in their locations, were characterized: a membrane-bound (Nar) and a periplasmic (Nap) nitrate reductase (2, 16, 29). The membrane-bound nitrate reductase is present in proteobacteria, firmicutes, actinobacteria, and even archaea, whereas the periplasmic nitrate reductase is present only in proteobacteria (21, 24). Nitrate-reducing proteobacteria can harbor Nar or Nap or both (18, 29). In contrast to that of Nar, the physiological role of Nap is still unclear and seems to differ between bacteria (11, 24). Thus, one proposed role for Nap is to support anaerobic metabolism as an alternative to Nar (1, 8). It has also been proposed that Nap facilitates the switch from aerobic respiration to denitrification (27) or scavenges nitrate in some pathogenic bacteria (23). The importance and diversity of the bacteria containing Nar have been extensively studied using both cultivation-based and direct molecular approaches (3, 5, 7, 17, 19, 22). However, only a few studies have focused on bacteria containing Nap (4, 9, 25). In this study, we investigated the relative abundances of the two types of nitrate reductases in various environments, using a real-time PCR-based assay.
Primer design, standard curves, and real-time PCR procedures.
In order to quantify the two types of nitrate reductases, a new real-time PCR assay was set up, using the narG and napA genes, encoding the catalytic subunits of the membrane-bound and periplasmic nitrate reductases, respectively, as molecular markers. All available sequences were aligned, and narG and napA primer sets specific to the proteobacteria were designed. The degenerated narG-f (5′-TCGCCSATYCCGGCSATGTC-3′), narG-r (5′-GAGTTGTACCAGTCRGCSGAYTCSG-3′), V17m (5′-TGGACVATGGGYTTYAAYC-3′; modified after reference 9), and napA4r (5′-ACYTCRCGHGCVGTRCCRCA-3′) primers were used to amplify fragments of 173 (for narG) and 152 (for napA) bp. Serial dilutions of linearized plasmids containing the narG and napA genes from Pseudomonas aeruginosa PAO1 were used to generate standard curves. The real-time PCR assays were carried out with a 20-μl reaction volume containing SYBR green PCR master mix (ABsoluteTM QPCR SYBR ROX Mix; Abgene, France), 2 μM of each primer, 100 ng of T4 gp32 (QBiogene, France), and 1.25 μl of template DNA (2 to 12.5 ng). Thermocycling conditions for narG were as follows: 15 min at 95°C; 6 cycles consisting of 30 s at 95°C and 30 s at 63°C, with a touchdown of −1°C by cycle; and 40 cycles consisting of 30 s at 95°C, 30 s at 58°C, 30 s at 72°C, and 30 s at 80°C. Conditions for napA were similar except that the annealing temperature was set at 61°C. All real-time PCRs were performed with an ABI Prism 7900 sequence detection system (Applied Biosystems). Quantification of the 16S rRNA gene was performed as described previously (15). DNAs extracted from triplicate samples from 18 different environments, such as soils (agricultural, industrial, or glacier), river sediments, waters, or biofilms, were used as templates.
Evaluation of assay specificity and sensitivity.
A functional gene pipeline interactive tool (http://flyingcloud.cme.msu.edu/fungene/) was used for in silico evaluation of primer specificity. Searches for the narG and napA primer sequences among 36 and 40 narG and napA sequences from complete genomes of proteobacteria showed that 78, 70, 86, and 86% did not exhibit any mismatch with the narG-f, narG-r, V17m, and napA4r primers, respectively. Primer specificity was further confirmed experimentally using a collection of 19 strains (Table 1). Four firmicutes and one actinomycete nitrate-reducing strain were selected as negative controls, and 14 nitrate-reducing strains belonging to the alpha-, beta-, and gammaproteobacteria were selected as positive controls. None of the gram-positive nitrate reducers, which were used as negative controls, gave an amplicon. For Proteobacteria, an absence of PCR products with both narG and napA primers was recorded only with Alcaligenes faecalis ATCC 8750. Sequence analysis of 84 and 79 narG and napA real-time PCR products from agricultural soils (Côte Saint André and Yvetot), glacier soil (Rotmoosfermer), cave biofilm (Padirac), and river phototrophic biofilm (Garonne) revealed that all sequences were related to the narG or napA genes. A high level of diversity among the sequences of the real-time PCR products was observed, with identities as low as 67% for narG and 69% for napA to the sequences used for the designs of the primers (see Fig. S3 and S4 in the supplemental material). This indicates that our newly developed real-time PCR systems are suitable for general detection of proteobacterial nitrate reductase genes. However, 3 out of 84 narG sequences fell into a cluster containing only narG from actinobacteria, indicating that the designed narG primers were not entirely specific to proteobacteria.
TABLE 1.
Bacterial strains used in this study to test the specificities of the narG and napA primersa
| Strain or isolate | Phylum | Nitrate reductase activity | Nitrate reductase enzymes | Q-PCR result for:
|
|
|---|---|---|---|---|---|
| narG | napA | ||||
| Bacillus senegalensis M1518 | Firmicutes | + | ND, ND | − | − |
| Bacillus cereus M944 ND | Firmicutes | + | Nar, ND | − | − |
| Bacillus thuringiensis | Firmicutes | + | Nar, ND | − | − |
| Bacillus megaterium | Firmicutes | + | Nar, ND | − | − |
| Streptomyces bluensis | Actinobacteria | + | ND, ND | − | − |
| Agrobacterium tumefaciens | Alphaproteobacteria | + | −, Nap | − | + |
| Bradyrhizobium japonicum 562 | Alphaproteobacteria | + | ND, ND | − | + |
| Bradyrhizobium japonicum USDA 110 | Alphaproteobacteria | + | −, Nap | − | + |
| Hyphomicrobium denitrificans DSM 1869 | Alphaproteobacteria | + | ND, ND | + | − |
| Sinorhizobium morelense SN611 | Alphaproteobacteria | + | ND, ND | − | + |
| Sinorhizobium meliloti 1021 | Alphaproteobacteria | + | −, Nap | − | + |
| Rhizobium meliloti 50 | Alphaproteobacteria | + | ND, ND | − | + |
| Paracoccus denitrificans Pd1222 | Alphaproteobacteria | + | Nar, Nap | + | + |
| Alcaligenes faecalis ATCC 8750 | Betaproteobacteria | + | ND, ND | − | − |
| Alcaligenes eutrophus H16 | Betaproteobacteria | + | Nar, Nap | + | + |
| Achromobacter cycloclastes ATCC 21921 | Betaproteobacteria | + | ND, ND | − | + |
| Escherichia coli JM109 | Gammaproteobacteria | + | Nar, Nap | + | + |
| Pseudomonas denitrificans CCUG 2519 | Gammaproteobacteria | + | ND, ND | + | − |
| Pseudomonas fluorescens C7R12 | Gammaproteobacteria | + | Nar, − | + | − |
Q-PCR, quantitative PCR; ND, not determined; −, not present.
The detection limit of our assay was around 10 copies per ng of template DNA, and no signal was detected in the no-template control. The PCR efficiencies of the narG and napA real-time PCR assays were 86 and 83%, respectively. Genomic DNA from Pseudomonas aeruginosa PAO1, for which the theoretical 16S rRNA, narG, and napA gene copy numbers per ng of DNA were calculated, was used as an external control. The presence of PCR inhibitors coextracted with DNA was tested as described previously (12), and this test did not reveal any significant inhibition. Therefore, our assays based on novel primer sets narG-f-narG-r and V17m-napA4r provide an efficient and sensitive method for quantifying either Nar or Nap in environmental samples.
Quantification of narG and napA genes.
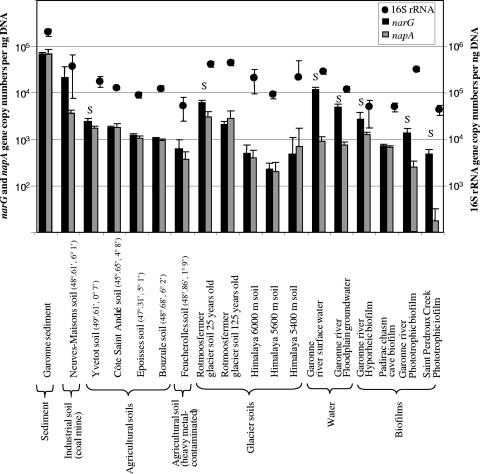
To compare with accuracy the numbers of genes in the different environments, results were expressed as gene copy numbers per ng of extracted DNA. Quantification of the narG and napA genes revealed high variations between environments, with most numbers ranging from 2 × 102 to 6.8 × 104 copies per ng of DNA (Fig. 3). The highest copy numbers for both narG and napA genes were observed in the river sediment samples. The napA gene copy numbers were lower than those of narG in most of the freshwater samples, whereas similar narG and napA gene copy numbers were observed in all soil samples except the Yvetot soil and the Rotmoosfermer glacier soil (Fig. 1). The numbers of 16S rRNA genes were between 1 and 3 logs higher than those of narG or napA genes. However, up to 12 copies of 16S rRNA may be found in the same bacterial genome (10), while only 1 napA copy and a maximum of 3 narG copies have been identified in the same strain (18). Hence, our study provides evidence that in soils, the numbers of proteobacteria containing periplasmic nitrate reductase are similar to the numbers of proteobacteria with the membrane-bound nitrate reductase. This is consistent with the results from Roussel-Delif et al. (25) and Carter et al. (4), which showed that a large proportion of nitrate-reducing, gram-negative isolates contain Nap. Unfortunately, it is not possible to conclude from our study whether the similar narG and napA copy numbers in many samples resulted from a majority of nitrate reducers possessing both types of nitrate reductase or from similar numbers of bacteria possessing either Nar or Nap. The narG genes from proteobacteria were mainly targeted in our assay, whereas these are also present in gram-positive bacteria and archaea, in contrast to napA. Therefore, the fact that the narG gene copy numbers were either similar to or higher than the napA gene copy numbers indicates that Nar is probably predominant in the environment.
FIG. 1.
Abundance of 16S rRNA, narG, and napA genes expressed as gene copy numbers per ng of extracted DNA, analyzed from three independent replicates per site. Error bars indicate standard deviations. S indicates significant differences (P < 0.05) between the narG and napA gene copy numbers (Student's t test) for each site.
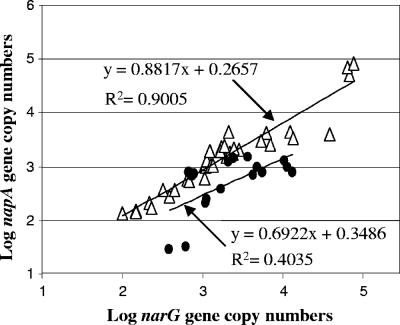
Interestingly, a high correlation coefficient of 0.9 between narG and napA gene copy numbers was calculated for the soil and sediment samples, whereas this coefficient was only 0.4 for samples from river biofilm and water samples (Fig. 2). In contrast, the correlation between narG or napA and 16S rRNA gene copy numbers did not exceed 0.5. The physiological role of Nap is still unclear and probably differs between strains (11, 21, 24). The high correlation observed between the numbers of narG and napA genes but not with 16S rRNA suggests that the ecological role of Nap in the majority of soil proteobacteria might be similar or complementary to that of Nar. Our results also showed that the abundances of the two types of nitrate reductases differed between environments, which could be due to the selection of nitrate reducers in some habitats. Since detection of functional genes is only a weak hint of the presence of the corresponding activity (20), investigation of the relative contributions of the two types of nitrate reductases to the total nitrate reduction activity in the different environments is of interest. Unfortunately, the activities of the two types of nitrate reductase are simultaneously monitored by the nitrate reduction assay developed by Kandeler (14), and the activities of Nar and Nap can be distinguished only on bacterial isolates (4). In the future, integrated studies are needed to compile further information on the physiology, diversity, and distribution of nitrate reducers for a more comprehensive understanding of nitrate reduction in the environment.
FIG. 2.
Correlation between narG and napA gene copy numbers in the soil and sediment samples (white triangles) and in water and biofilm samples (black circles). Each replicate of the 18 selected environments is plotted.
Nucleotide sequence accession numbers.
The GenBank accession numbers for the sequences reported here are EF217059 to EF217221.
Supplementary Material
Acknowledgments
We are grateful to A. Brauman, G. Braker, F. Garabetian, S. Hallin, E. Kandeler, S. Nazareth, and B. Stres for providing reference strains or environmental samples. We also thank S. Henry and S. Hallet for their assistance.
This work was supported by the French Ministry of Research (ACI PNBC MUTEN).
This work is dedicated to the memory of Frédéric Marmont and Ivan Mahne.
Footnotes
Published ahead of print on 13 July 2007.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Bedzyk, L., T. Wang, and R. W. Ye. 1999. The periplasmic nitrate reductase in Pseudomonas sp. strain G-179 catalyzes the first step of denitrification. J. Bacteriol. 181:2802-2806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berks, B. C., S. J. Ferguson, J. W. B. Moir, and D. J. D. Richardson. 1995. Enzymes and associated electron transports systems that catalyse the respiratory reduction of nitrogen oxides and oxyanions. Biochim. Biophys. Acta 1232:97-173. [DOI] [PubMed] [Google Scholar]
- 3.Brunel, B., J. D. Janse, H. J. Laanbroek, and J. D. Woldendorp. 1992. Effect of transient oxic conditions on the composition of the nitrate-reducing community from the rhizosphere of Typha angustipholia. Microb. Ecol. 24:51-61. [DOI] [PubMed] [Google Scholar]
- 4.Carter, J. P., Y. H. Hsaio, S. Spiro, and D. J. Richardson. 1995. Soil and sediment bacteria capable of aerobic nitrate respiration. Appl. Environ. Microbiol. 61:2852-2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chèneby, D., S. Hallet, M. Mondon, F. Martin-Laurent, J. C. Germon, and L. Philippot. 2003. Genetic characterization of the nitrate reducing community based on narG nucleotide sequence analysis. Microb. Ecol. 46:113-121. [DOI] [PubMed] [Google Scholar]
- 6.Conrad, R. 1996. Soil microorganisms as controllers of atmospheric trace gases (H2, CO, CH4, OCS, N2O, and NO). Microbiol. Rev. 60:609-640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deiglmayr, K., L. Philippot, U. A. Hartwig, and E. Kandeler. 2004. Structure and activity of the nitrate-reducing community in the rhizosphere of Lolium perenne and Trifolium repens under long-term elevated atmospheric pCO2. FEMS Microbiol. Ecol. 49:445-454. [DOI] [PubMed] [Google Scholar]
- 8.Delgado, M. J., N. Bonnard, A. Tressierra-Ayala, E. J. Bedmar, and P. Müller. 2003. The Bradyrhizobium japonicum napEDABC genes encoding the periplasmic nitrate reductase are essential for nitrate respiration. Microbiology 149:3395-3403. [DOI] [PubMed] [Google Scholar]
- 9.Flanagan, D. A., L. G. Gregory, J. P. Carter, A. Karakas-Sen, D. J. Richardson, and S. Spiro. 1999. Detection of genes for periplasmic nitrate reductase in nitrate respiring bacteria and in community DNA. FEMS Microbiol. Lett. 177:263-270. [DOI] [PubMed] [Google Scholar]
- 10.Fogel, G. B., C. R. Collins, J. Li, and C. F. Brunk. 1999. Prokaryotic genome size and SSU rDNA copy number: estimation of microbial relative abundance from a mixed population. Microb. Ecol. 38:93-113. [DOI] [PubMed] [Google Scholar]
- 11.Gonzalez, P. J., C. C. I. Moura, C. D. Brondino, and J. J. G. Moura. 2006. Bacterial nitrate reductases: molecular and biological aspects of nitrate reduction. J. Inorg. Biochem. 100:1015-1023. [DOI] [PubMed] [Google Scholar]
- 12.Henry, S., D. Bru, B. Stres, S. Hallet, and L. Philippot. 2006. Quantitative dectection of the nosZ gene, encoding nitrous oxide reductase, and comparison of the abundance of 16S rRNA, narG, nirK, and nosZ genes in soils. Appl. Environ. Microbiol. 72:5181-5189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hofstra, N., and A. F. Bouwman. 2005. Denitrification in agricultural soils: summarizing published data and estimating global annual rates. Nutr. Cycl. Agroecosyst. 72:267-278. [Google Scholar]
- 14.Kandeler, E. 1995. Nitrate reductase activity, p. 176-179. In F. Schinner, R. Öhlinger, E. Kandeler, and R. Margesin (ed.), Methods in soil biology. Springer, Berlin, Germany.
- 15.Lopez-Gutierrez, J. C., S. Henry, S. Hallet, F. Martin-Laurent, G. Catroux, and L. Philippot. 2004. Quantification of a novel group of nitrate-reducing bacteria in the environment by real-time PCR. J. Microbiol. Methods 57:399-407. [DOI] [PubMed] [Google Scholar]
- 16.Moreno-Vivian, C., P. Cabello, M. Martinez-Luque, R. Blasco, and F. Castillo. 1999. Prokaryotic nitrate reduction: molecular properties and functional distinction among bacterial nitrate reductases. J. Bacteriol. 181:6573-6584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nijburg, J., M. J. L. Coolen, S. Gerards, P. J. A. K. Gunnewiek, and H. J. Lannbroek. 1997. Effects of nitrate availability and the presence of Glyceria maxima on the composition and activity of the dissimilatory nitrate-reducing bacteria community. Appl. Environ. Microbiol. 63:931-937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Philippot, L. 2002. Denitrifying genes in bacterial and Archaeal genomes. Biochim. Biophys. Acta 1577:355-376. [DOI] [PubMed] [Google Scholar]
- 19.Philippot, L. 2005. Tracking nitrate reducers and denitrifiers in the environment. Biochem. Soc. Trans. 33:204-207. [DOI] [PubMed] [Google Scholar]
- 20.Philippot, L., and S. Hallin. 2005. Finding the missing link between diversity and activity using denitrifying bacteria as a model functional community. Curr. Opin. Microbiol. 8:234-239. [DOI] [PubMed] [Google Scholar]
- 21.Philippot, L., and O. Hojberg. 1999. Dissimilatory nitrate reductases in bacteria. Biochim. Biophys. Acta 1446:1-23. [DOI] [PubMed] [Google Scholar]
- 22.Philippot, L., S. Piutti, F. Martin-Laurent, S. Hallet, and J. C. Germon. 2002. Molecular analysis of the nitrate-reducing community from unplanted and maize-planted soil. Appl. Environ. Microbiol. 68:6121-6128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Potter, L., H. Angove, D. Richardson, and J. Cole. 2001. Nitrate reduction in the periplasm of Gram-negative bacteria. Adv. Microb. Physiol. 45:51-112. [DOI] [PubMed] [Google Scholar]
- 24.Richardson, D. J., B. C. Berks, D. A. Russel, S. Spiro, and C. J. Taylor. 2001. Functional, biochemical and genetic diversity of prokaryotic nitrate reductase. Cell. Mol. Life Sci. 58:165-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Roussel-Delif, L., S. Tarnawski, J. Hamelin, L. Philippot, M. Aragno, and N. Fromin. 2005. Frequency and diversity of nitrate reductase genes among nitrate-dissimilating Pseudomonas in the rhizosphere of perennial grasses grown in field conditions. Microb. Ecol. 49:63-72. [DOI] [PubMed] [Google Scholar]
- 26.Shearer, L. A., J. R. Goldsmith, C. Young, O. A. Kearns, and B. R. Tamplin. 1972. Methemoglobin levels in infants, in an area with high nitrate water supply. Am. J. Public Health 62:1174-1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Siddiqui, R. A., U. Warnecke-Eberz, A. Hengsberger, B. Schneider, and B. Friedrich. 1993. Structure and function of a periplasmic nitrate reductase in Alcaligenes eutrophus H16. J. Bacteriol. 175:5867-5876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tiedje, J. M. 1988. Ecology of denitrification and dissimilatory nitrate reduction to ammonium, p. 179-244. In A. J. B. Zendher. (ed.), Biology of anaerobic microorganisms. John Wiley & Sons, Inc., New York, NY.
- 29.Zumft, W. G. 1997. Cell biology and molecular basis of denitrification. Microbiol. Mol. Biol. Rev. 61:533-536. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.