Abstract
Gliding arc (glidarc) discharge is a physicochemical technique for decontamination at atmospheric pressure and ambient temperature. It leads to the destruction of bacterial phytopathogens responsible for important losses in industrial agriculture, namely, Erwinia spp., without the formation of resistant forms. We investigated the effect of a novel optimized prototype allowing bacterial killing without lag time. This prototype also decreases the required duration of treatment by 50%. The study of the time course effect of the process on bacterial morphology suggests that glidarc induces major alterations of the bacterial membrane. We showed that glidarc causes the release of bacterial genomic DNA. By contrast, an apparent decrease in the level of extractible lipopolysaccharide was observed; however, no changes in the electrophoretic pattern and cytotoxic activity of the macromolecule were noted. Analysis of extractible proteins from the outer membrane of the bacteria revealed that glidarc discharge induces the release of these proteins from the lipid environment, but may also be responsible for protein dimerization and/or aggregation. This effect was not observed in secreted enzymatic proteins, such as pectate lyase. Analysis of the data supports the hypothesis that the plasma generated by glidarc discharge is acting essentially through oxidative mechanisms. Furthermore, these results indicate that, in addition to effectively destroying bacteria, glidarc discharge should be used to improve the extraction of bacterial molecules.
Industrial guidelines prescribing the use of nonchemical techniques of decontamination have evolved to avoid traces of antibacterial agents in the final product; consequently, this has encouraged research into physical techniques that kill bacteria, such as advanced oxidation processes.
Systems based on electric discharge, especially those that generate nonthermal plasma, appear to be very promising because of their low equipment and energy costs, their efficiency on diverse surfaces or media, and even their possible application to heat-sensitive materials (25). Producing nonthermal plasma requires specific conditions. Plasmas can be classified according to their energy level and proportion of light (electrons) and heavy species (e.g., molecules, atoms, radicals, and ions). Thermal plasmas, usually formed at high pressure (≥1 atm), are media in which the energies of the two species are similarly high. By contrast, the mean energy of the electrons at low pressure is much higher than that of the heavy species, and the resulting plasma is designated “cold plasma.” There are intermediate conditions at atmospheric pressure which allow for the formation of the nonthermal plasmas. Among the various techniques able to generate nonthermal plasma, we decided to study gliding arc, or “glidarc,” discharge. We chose glidarc particularly because of its high industrial potential and its efficiency for decontamination. This technique was previously used for treating Escherichia coli (35), Staphylococcus epidermidis (4), and various species of the potato pathogen, Erwinia (26). Glidarc has been shown to totally destroy Erwinia carotovora subsp. atroseptica, Erwinia chrysanthemi, and Erwinia carotovora subsp. carotovora cultures (26). Bacterial killing using glidarc was achieved directly in the liquid medium at room temperature and was not accompanied by the formation of viable but noncultivable forms. However, this process required a lag time poorly adapted to online industrial applications. Moreover, bacterial sensitivity to glidarc treatment was variable; E. carotovora subsp. atroseptica, the main potato pathogen in Europe and also responsible for crop losses amounting to millions of euros each year worldwide (34), was more resistant to the plasma treatment than the other Erwinia strains. Therefore, it was necessary to conceive a novel and more-efficient prototype for destroying these microorganisms. Additionally, identifying the major molecular and physiological targets for nonthermal plasma could lead to better control of the process of bacterial destruction by glidarc. This development will help in designing the next generation of equipment. The development also opens the way to new applications for glidarc, as shown by its use for the electrochemical conversion of pollutants.
In this study, we tested a novel glidarc prototype on the phytopathogen E. carotovora subsp. atroseptica. This bacterium was selected for two reasons, its resistance and its role in vegetal diseases (“black leg” of potato), as well as its relation to “quarantine microorganisms,” i.e., bacteria whose presence on fruits and tubers nowadays forces total destruction of the crops. We studied various molecular targets, including DNA, lipopolysaccharides (LPS), membrane proteins, and an exoenzyme, to determine the mechanism of bacterial killing by glidarc discharge. We also suggest possible new applications of glidarc for extracting membrane-linked bacterial molecules.
MATERIALS AND METHODS
Glidarc reactor.
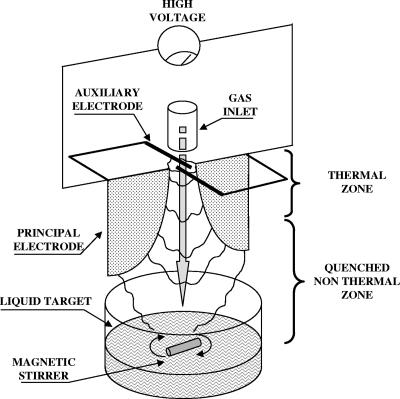
The reactor used for this study is based on the “glidarc” device previously described by Lesueur et al. (22) and developed by Czernichowski and coworkers (7, 8). This is a nonthermal quenched plasma system operated at atmospheric pressure and close to the ambient temperature. The main difference from our previous prototype (26) is the use of a four-electrode plasma generator. This new apparatus was designated “prototype 2” (Fig. 1); the discharge is produced by two principal, diverging, aluminum electrodes working at a high voltage fall (9 kV, 100 mA in open conditions). The two auxiliary electrodes, made of copper, are also connected to the generator initiating the divergent discharge. Gas is injected through a nozzle placed in the axis at the top of the reactor. An arc forms at the electrode minimum gap. It is pushed along the electrodes by the gas flow until it breaks into a plasma plume, while a new arc forms at the top of the electrodes. The plasma plume then makes contact with the liquid target, where the activated species can react at the liquid-gas interface.
FIG. 1.
Simplified scheme of the novel prototype for the glidarc reactor used in this study.
The gas used to create the plasma is ambient air, i.e., a mixture of N2 and O2 supplied by an air compressor in which the discharges allow the formation of highly reactive species, including NO· and OH·. The gas supplied is saturated with water by passing it through a flask filled with bubbling distilled water to create more reactive species. The UV content of this plasma remains negligible (18, 19). The reactor settings, such as the gas flow rate (liters · min−1), the electrode gap, and the distance between electrode tips and target, were kept constant during all experiments. The reactor was fitted with a circulating-water thermostatted jacket to limit the temperature increase during the experiments. The contact between plasma and liquid surface and the temperature exchanges were favored by continuous stirring.
Bacterial strain and culture conditions.
E. carotovora subsp. atroseptica strain 1526 used in this study was provided by the French Library of Phytopathogenic Bacteria (INRA, Anger, France). The strain was maintained as a glycerol stock culture and stored at −80°C until use. E. carotovora subsp. atroseptica cells were resuspended and incubated at 28°C for 24 h in sterile Luria Bertani (LB) broth. Then, 200 ml of sterile LB was inoculated with 1 ml of this preculture and grown for another 24 h at 28°C. Fractions of this culture (25 ml) were then exposed to glidarc discharge. For this study, we used bacteria that were in the late stationary phase. This choice was determined by two factors: (i) bacteria in the environment spend most of their life in this physiological state (late stationary phase) and (ii) bacteria in late stationary phase are particularly resistant to decontamination (29).
The classical plating technique on LB agar was used to plate cultures before and after treatment. Plates were incubated for 48 h at 28°C, after which viable colonies were counted.
Extraction of genomic DNA.
Genomic DNA was extracted according to Sambrook et al. (32). Briefly, 25-ml volumes of E. carotovora subsp. atroseptica cells (initial concentration, 1010 CFU · ml−1), exposed and not exposed to glidarc discharge, were centrifuged at 5,000 × g for 10 min at 20°C. The pellets were resuspended in 9.5 ml extraction buffer (1 M Tris, 0.5 M EDTA) and mixed with 0.5 ml of 1% sodium dodecyl sulfate (SDS) and 50 μl of 20 mg · ml−1 proteinase K. After shaking, the suspension was incubated for 1 h at 37°C, and then 1.8 ml of 5 M NaCl and 1.5 ml CTAB buffer (10% hexadecyltrimethyl-ammonium bromide, 0.7 M NaCl in water) were added. The samples were incubated for another hour at 65°C.
We added 13 ml of chloroform-isoamyl alcohol (24/1; vol/vol) to the suspensions, and they were then centrifuged at 5,000 × g for 10 min. Isopropanol was added to the supernatant in a proportion of 0.6 per volume.
The suspensions were gently shaken to release DNA. The suspensions were centrifuged, and the precipitant was then mixed with 1 ml of 70% ethanol and centrifuged at 10,000 × g for a few seconds. The ethanol was removed, and the DNA was dried overnight at room temperature and resuspended in distilled water.
Extracted genomic DNA was run on a 1% agarose gel in 1× TAE buffer (Tris-acetate-EDTA). The gels were then stained with ethidium bromide and visualized under UV light.
DNA assay.
We determined the concentrations of DNA released from bacteria after they were exposed to glidarc discharge. The DNA concentrations were determined by measuring the optical density at 260 nm, assuming that 1 absorbance unit corresponds to 50 μg · ml−1 nucleic acid. The degree of purity of the DNA was checked by measuring the ratio between the absorbance of the samples at 260 nm and at 280 nm (absorbance wavelength of proteins). Only samples presenting a ratio of >1.5 were used.
Scanning electron microscopy.
E. carotovora subsp. atroseptica cells were fixed with 3% glutaraldehyde overnight. The cultures were then centrifuged (7,000 × g, 10 min), and the pellets were rinsed twice in phosphate buffer. The samples were dehydrated by successive treatments of ethanol in water; the ethanol concentration for each treatment was increased in a step-wise manner with concentrations from 50% to 100%. The samples were finally dried at ambient temperature. The samples were coated with gold in a sputter coater (Jeol JFC 1100 E; Jeol, Japan) and were examined with a scanning electron microscope (SEM S4500; Hitachi, Japan).
Extraction and purification of the LPS.
After exposure to glidarc discharge for various times, 25 ml cultures of E. carotovora subsp. atroseptica were centrifuged for 10 min at 5,000 × g at 20°C. The pellets were then suspended in PBS buffer (0.8% NaCl, 0.02% KCl, 0.17% Na2HPO4, 0.8% KH2PO4) and centrifuged again for 10 min. This washing step was repeated three times to eliminate all bacterial mucus that could interfere with the extraction of LPS. LPS was then purified as previously described (9).
Assay and analysis of LPS.
The apparent concentration of LPS in the extracts was determined by the measurement of 2-keto-3-deoxyoctunolic acid (KDO), using the technique described by Karkhanis et al. (15).
A standard curve from 0 to 200 μg · ml−1 KDO was extemporaneously established using purified KDO from Escherichia coli (Sigma) dissolved in 0.2 N H2SO4.
LPS extracted from treated (cells had undergone glidarc discharge) and untreated E. carotovora subsp. atroseptica cells were analyzed by SDS-polyacrylamide gel electrophoresis (PAGE) (16). The gel was stained with ethidium bromide (30 μg · ml−1; 10 s) after electrophoresis, and the LPS bands were visualized by transillumination.
Determination of the cytotoxic effect of the LPS.
The cytotoxic effects of LPS extracted from glidarc-treated and untreated E. carotovora subsp. atroseptica cells were investigated in primary cultures of glial cells using biochemical indicators of apoptosis and necrosis as previously described (28).
Rat glial cells, obtained from newborn rats (24 to 48 h), were grown in Dulbecco's modified Eagle's medium-Ham's medium (2/1) supplemented with 10% fetal calf serum, 2 mM glutamine, 0.001% insulin, 5 mM HEPES, 0.3% glucose, and 1% antibiotic-antimycotic solution. The cells were layered at a concentration of 105 cells/slide on glass slides coated with poly-l-lysine (50 μg · ml−1) and were incubated at 37°C in a 5% CO2 humidified atmosphere. The culture medium was changed the first day after plating and then every 2 days. The cells were allowed to grow for 7 days before use.
Apoptosis was measured by the accumulation of nitrite ions (NO2−) in the incubation medium; nitrite ions result from the spontaneous conversion of nitric oxide (NO) following the activation and induction of apoptosis-dependent NO synthase activities (23). The concentration of NO2− was determined by using a technique derived from the Griess colorimetric reaction. This technique has been previously validated by Picot et al. (28).
Necrosis was quantified by measuring the amount of lactate dehydrogenase (LDH)—a stable cytosolic eukaryotic enzyme—released in the culture medium following disruption of the cytoplasmic membrane of the cells. The concentration of LDH was determined by using a Cytotox 96 enzymatic assay (Promega, Charbonnieres, France). This technique has been previously used as an indicator for necrosis in eukaryotic cells (11, 28).
Extraction and determination of the concentration of outer membrane proteins.
Outer membrane proteins were extracted by the spheroplast procedure (24) as modified by Dé et al. (10). The total protein content was determined by two complementary methods, the Bradford (3) and the biuret (13) assays; this was because of the intrinsic variability of the protein assay techniques (33) and their possible sensitivity to active chemical species generated during glidarc treatment. The Bradford protein assay depends on the change of absorbance of Coomassie blue G-250 on the binding protein. By contrast, the biuret assay is based on the formation of a purple complex between biuret (NH2-CO-NH-CO-NH2) and consecutive peptic bonds of the protein in alkaline medium.
The outer membrane protein fractions were analyzed by SDS-PAGE in a 7 to 15% discontinuous gel system. The proteins were visualized by Coomassie blue staining.
Assay of the PEL activity.
The PEL activity was determined by measuring the formation of C4 and C5 unsaturated oliguronides at 235 nm, according to the technique reported by Collmer et al. (6) and modified by Laurent et al. (20).
Statistical analysis.
Each value in the assay was determined over a minimum of three independent measures. Student's t test was used to compare the means of various values within the same set of experiments.
RESULTS
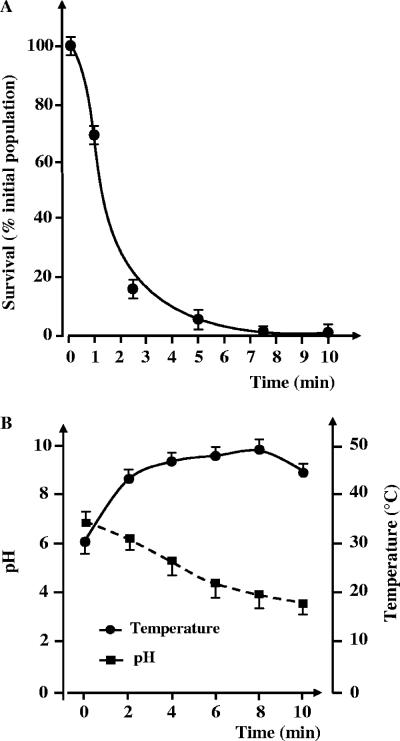
We studied the effect of glidarc discharge on Erwinia carotovora subsp. atroseptica strain 1526 over a period of 10 min (Fig. 2). The surviving bacterial population growth as a function of the treatment time, i.e., the percentage of the initial population versus time, showed two steps: (i) a rapid decrease of up to 9 logarithmic units of the initial bacterial population within 2.5 min and (ii) a second step with slower kinetics, leading to a total reduction of 10 logarithmic units within 5 min.
FIG. 2.
(A) Destruction curve of Erwinia carotovora subsp. atroseptica 1526 by glidarc discharge. The values are the means (± SEM) of the results of a minimum of six independent experiments. (B) Graph illustrating the pH and the temperature in a solution exposed to glidarc discharge over the course of a 10-min experiment. The values are the means (± SEM) of the results of a minimum of three independent experiments.
We monitored the temperature and pH of the cultures exposed to glidarc discharge in parallel during the same time. The pH decreased from 7.19 ± 0.09 (mean ± standard error of the mean [SEM]) to 3.7 ± 0.05 after 10 min of treatment (Fig. 2). The pH appeared to stabilize around pH 3.7, and no significant change was observed for the longer treatment period (data not shown). We also observed slight changes in the temperature of the culture during treatment (Fig. 2). The temperature increased slowly from 30.8 ± 0.2°C (initial temperature) to 46.0 ± 0.1°C after 2.5 min of exposure to glidarc discharge. The temperature value (46.0 ± 0.1°C) tended to stabilize between 2.5 and 10 min, as is visible on the graph (Fig. 2B). E. carotovora subsp. atroseptica cells were exposed to the same temperature (46°C) and pH (3.7) in control experiments; however, these conditions did not result in bacterial death as observed in the glidarc reactor (data not shown).
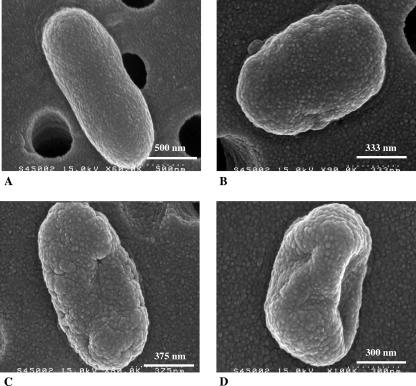
Scanning electron microscope observations of the bacteria submitted to glidarc discharge indicated that the overall structure of the microorganism was significantly affected (Fig. 3). The percentage of bacterial survival remained relatively high after 1 min of treatment (Fig. 3A), and bacterial structural changes were limited; however, in some microorganisms, the surfaces appeared more rugged and their shape appeared less regular (Fig. 3B). Bacteria exposed for 2.5 min appeared to lose their elongated shape and their surfaces became irregular (Fig. 3C). The structural change was even more dramatic after 5 min of treatment. These bacteria showed deep surface cracks and a complete decay of any organized structure (Fig. 3D).
FIG. 3.
Scanning electron microscope photomicrographs illustrating the effects of glidarc discharge on the morphology of Erwinia carotovora subsp. atroseptica strain 1526. (A) Typical aspect of nontreated E. carotovora. (B) Morphology of E. carotovora after 1 min of glidarc discharge treatment. The surface of the bacterium is less regular. (C) E. carotovora cells treated by glidarc for 2 min show the beginning of the formation of surface cracks. (D) Bacterium exposed for 5 min to glidarc discharge illustrates the effects of the process on the shape and surface of the microorganism. Holes in the background correspond to the pores of the filters on which bacteria were collected.
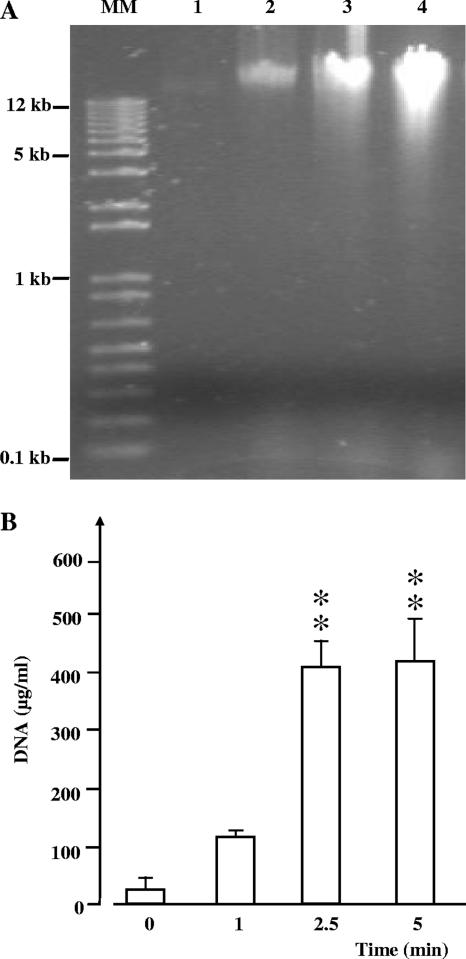
The influence of glidarc discharge on the genomic DNA of E. carotovora subsp. atroseptica was investigated by electrophoresis and direct assay. Genomic DNA extracted from E. carotovora subsp. atroseptica cells treated with glidarc discharge for 1, 2.5, and 5 min revealed a marked increase in the spot intensity on the SDS-PAGE gel and a progressive spreading of the signal with the appearance of a smear (Fig. 4A). Spectrophotometric DNA assay showed that when E. carotovora subsp. atroseptica cultures were exposed to glidarc discharge, the concentration of extractible genomic DNA increased greatly. The concentration of extractible genomic DNA was 37.5 ± 1 μg · ml−1 for untreated cultures, but it was 110 ± 32 μg · ml−1 for a culture treated for 1 min. For longer exposures, the total amount of extractible DNA increased significantly (P > 0.05) and reached 412 ± 114 μg · ml−1 after a 5-min treatment (Fig. 4B).
FIG. 4.
Effect of glidarc treatment on the genomic DNA of Erwinia carotovora subsp. atroseptica strain 1526. (A) Visualization of the effect of glidarc discharge on the electrophoretic pattern of DNA extracted from nontreated bacteria (lane 1) and microorganisms submitted to treatments of 1 min (lane 2), 2.5 min (lane 3), and 5 min (lane 4). MM, molecular mass markers. (B) Spectrophotometric quantification of the genomic DNA extracted from cultures of E. carotovora after exposure to glidarc discharge from 0 to 5 min. The values are the means (± SEM) of the results of a minimum of eight independent experiments. **, significant difference with regard to the control (0). Level of confidence, >99%.
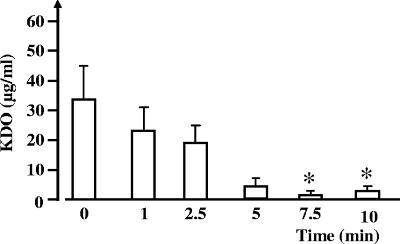
The effect of glidarc discharge on E. carotovora subsp. atroseptica LPS was studied by direct measurement of KDO and SDS-PAGE. The apparent quantities of KDO, measured in cultures of E. carotovora subsp. atroseptica exposed to glidarc, decreased during treatment (Fig. 5). However, in control cultures, the concentration of detectable KDO was 32 ± 11 μg · ml−1; it decreased significantly (P > 0.01) to 1 ± 0.5 μg · ml−1 in cultures that were treated for 10 min. These results were not consistent with visible changes in the electrophoretic properties of LPS extracted from E. carotovora subsp. atroseptica cells, even after a 10-min exposure to glidarc discharge (data not shown). The apparent molecular mass of LPS remained unchanged, and no smear or spreading of the bands was observed.
FIG. 5.
Quantification of the KDO extracted from cultures of E. carotovora after exposure to glidarc discharge for 0 to 10 min. The values are the means (± SEM) of the results of a minimum of three independent experiments. *, significant difference with regard to the control (0). Level of confidence, >95%.
We investigated the effect of glidarc discharge on the cytotoxic activity of E. carotovora subsp. atroseptica LPS by using biochemical indicators of apoptosis and necrosis. Nitrites (NO2−) are representative of the activation of inducible nitric oxide synthase associated with the apoptotic process; thus, we assayed the concentration of nitrites using a technique derived from the Griess colorimetric reaction. The control cell culture nitrite concentration was very limited (1.3 ± 0.7 μM). Incubation with LPS extracted from E. carotovora subsp. atroseptica cells (500 ng · ml−1) not previously exposed to glidarc discharge significantly stimulated inducible nitric oxide synthase activity: the concentration of NO2− in the medium reached 14 ± 3 μM. The NO2− levels measured using LPS extracted from E. carotovora subsp. atroseptica cells exposed to glidarc for 1 min (15 ± 2 μM) to 10 min (15 ± 0.1 μM) did not significantly differ from that of the control (data not shown).
LDH is a stable cytosolic enzyme released in the medium following destabilization of the plasma membrane, and its accumulation in the extracellular compartment is considered a necrosis marker (11). The 0% LDH release was determined by using culture medium incubated with control extraction buffer in the absence of the cells. The 100% LDH release was defined as the total amount of LDH released in the culture medium after total necrosis of the cell population, provoked by adding a lysis buffer. We found only a low concentration of LDH, 20% ± 2% of the total amount of enzyme potentially released after necrosis of the whole cell population, in the medium after 24 h of incubation in the control cells. This low level is indicative of the good health of the cultures. The percentage of LDH release in cells incubated with LPS purified from untreated E. carotovora subsp. atroseptica cells (500 ng · ml−1) was 51% ± 2.5%. The percentage of LDH recovered from cultures exposed to LPS from E. carotovora subsp. atroseptica cells treated by glidarc was between 57% ± 3% and 71% ± 8%, but was never significantly different from the control values (data not shown).
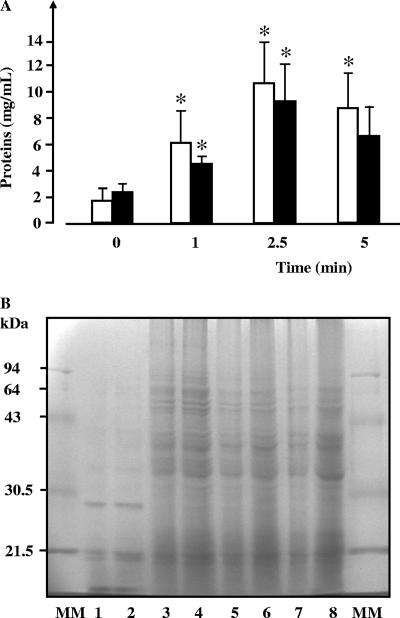
The effect of glidarc discharge on membrane proteins in E. carotovora subsp. atroseptica cells was investigated by measuring the total concentration of extractible outer membrane proteins; we used two biochemical assays based on different principles, i.e., the Bradford and biuret assays. Using the Bradford technique, the apparent concentration of outer membrane proteins extracted from E. carotovora subsp. atroseptica cells increased when the cultures were exposed to glidarc discharge (Fig. 6A). Similar results were obtained using the biuret assay, i.e., a significant increase in the apparent concentration of extractible outer membrane proteins after the cultures were exposed to glidarc discharge.
FIG. 6.
Evaluating the effect of glidarc treatment on the extractability and structure of Erwinia carotovora subsp. atroseptica strain 1526 outer membrane proteins. (A) Quantification by the Bradford (open bars) and biuret (black bars) assays of outer membrane proteins extracted from cultures of E. carotovora after exposure to glidarc discharge for 0 to 5 min. *, significant difference with regard to the control (0). Level of confidence, >95%. (B) SDS-PAGE analysis of outer-membrane proteins extracted from E. carotovora cells before (lanes 1 and 2) and after treatment by glidarc discharge for 1 min (lanes 3 and 4), 2 min (lanes 5 and 6), and 5 min (lanes 7 and 8). The values are the means (± SEM) of the results of a minimum of five independent experiments. MM, molecular mass markers.
Outer membrane proteins extracted after exposing the bacteria to glidarc were analyzed by SDS-PAGE. There were major differences between the electrophoretic profiles of outer membrane proteins in cultures exposed to glidarc discharge and those not exposed (Fig. 6B). For instance, a protein band of an apparent molecular mass close to 30 kDa was visualized in extracts from untreated cultures (Fig. 6B, lanes 1 and 2), but was not present in samples from bacteria exposed to glidarc (Fig. 6B, lanes 3 to 8). By contrast, various bands corresponding to high-molecular-mass proteins were observed in extracts from cultures exposed to glidarc (Fig. 6B, lanes 3 to 8), whereas these bands were undetectable in extracts from control cultures (Fig. 6B, lanes 1 and 2).
PEL was selected as a representative indicator of secreted bacterial proteins. The PEL activities detected in the media from control and glidarc-treated E. carotovora subsp. atroseptica cells were not significantly different. The PEL activity in medium from control cultures was 1.1 ± 0.1 U · ml−1, whereas the PEL activity for bacteria exposed to glidarc discharge was between 0.9 ± 0.1 U · ml−1 and 1.4 ± 0.9 U · ml−1 (data not shown).
DISCUSSION
In this study, we developed a new glidarc discharge reactor. The main characteristic of this new prototype is that it has four electrodes instead of two, as in the older one. This modification generates a greater volume of plasma, a more-stable discharge, and a greater increase in the energy released in the discharge than those of the initial model. Thus, we obtained a more-effective prototype, which was confirmed by comparing the kinetics of E. carotovora subsp. atroseptica strain 1526 cell death in the two systems. The treatment time required to reduce an E. carotovora subsp. atroseptica population by 10 logarithmic units was about 5 min, whereas previously it was double that (10 min) (26). Moreover, with this new prototype, bacterial killing appears to be immediate. The lag phase of the destruction kinetics observed with the first prototype was totally absent with the new prototype. The absence of this lag phase results from suitable modifications of the reactor, which release more energy and create more active species, such as OH· and NO·, in a very short time.
We also observed a rapid fall in the pH of the medium during exposure to glidarc discharge, as has been previously reported (26). This decrease in pH can be explained by the creation of NO· radicals that form acid species in solution, such as HNO2/HNO3. The variation in pH was in the same range as previously noted and, therefore, is not responsible for the efficiency differences between the two systems.
We monitored the temperature during a 10-min decontamination run; we observed a mean increase of 16°C in the target medium, whereas no significant variation in the temperature was noted for the first prototype. This increase in temperature was certainly due to the plasma generated by the new reactor being more energetic. The temperature change and the pH fall probably contribute to bacterial stress. However, their role in bacterial death is apparently limited, as indicated by the results of our control experiments with E. carotovora subsp. atroseptica cells exposed to a similar temperature (46°C) and pH (3.7): these simple environmental variations did not provoke any significant bacterial death. These data suggest that temperature and pH combine with at least another factor, presumably a highly reactive chemical species, to generate the kinetics of bacterial killing observed using glidarc. Few hypotheses explain the effects of electrical discharge, particularly those of glidarc, on bacteria. Although Pelletier (27) and Lerouge et al. (21) did not use the same type of plasma, their studies suggest that bacterial death could result from alteration of the integrity of the membrane followed by irreparable damage to nucleic acids. NO· and OH· are the main species formed in the plasma during glidarc discharge in humid air. NO· is responsible for acid effects, and OH·, produced in high quantities, is responsible for a strong oxidizing effect (2, 19). Both acid and oxidative stresses strongly affect the permeability of the bacterial membrane, transmembrane potential, and intracellular pH (1, 31). Glidarc provokes major structural alterations in the bacterial membrane, as shown by our scanning electron microscope observations. These structural changes can have multiple origins. Indeed, oxidative reactions can destabilize the bacterial chromosomal DNA-membrane interactions (30). Similarly, it is well known that a mild acidic attack causes the cleavage of the O antigen of the LPS of the outer membrane of gram-negative bacteria (5) and that highly oxidative chemical species can induce changes in the protein content of the bacterial membrane (14). Therefore, we investigated the effects of glidarc discharge on these various molecular targets.
SDS-PAGE analysis of the chromosomal DNA extracted from E. carotovora subsp. atroseptica cells treated by glidarc discharge showed the rapid appearance of a smear. Smears generally indicate partial fragmentation of the nucleic acid. Oxidative damage to DNA is well documented in E. coli, in which even low doses of hydrogen peroxide generate breaks in the DNA sequence (17). Our results are consistent with oxidative mechanisms being involved in the antibacterial effect of glidarc. However, the intensity of the spots on the electrophoresis gel increased with the duration of exposure to glidarc. This observation was confirmed by measuring the amount of DNA that was extractible from E. carotovora subsp. atroseptica cells submitted to various treatment times. The bacterial concentration was identical in all the samples; therefore, the only possible interpretation is that exposure to glidarc causes the release of chromosomal DNA from its membrane complex where it is normally anchored (12), resulting in a better extraction yield of the macromolecule.
LPS, the major component of the outer membrane of gram-negative bacteria, is sensitive to low pH (5), but it appears poorly sensitive to oxidative degradation. We determined the LPS concentration in extracts from E. carotovora subsp. atroseptica cells exposed to glidarc: it was shown to decrease during treatment. This result was unexpected since KDO, the marker in the LPS assay, is located in the core of the macromolecule and should not be cleaved in the case of acidic attack on the O antigen. In addition, no change in the electrophoretic properties of the LPS were observed, suggesting that, in fact, the KDO assay was biased by the oxidative species generated by the glidarc system in the medium. The absence of structural changes in the LPS was confirmed because the cytotoxic activity of the molecule remained unchanged, even after 10 min of glidarc treatment. A direct correlation between changes in the structure of the LPS and variations in the apoptotic and necrotic activities of the endotoxin has been previously demonstrated (28). These results also support the hypothesis that, at least in the case of LPS, the role of the pH fall in bacterial death is limited.
The effect of glidarc discharge on outer membrane proteins, the third possible molecular target of the bacterial surface, was investigated by using two biochemical assays. Two assays were used to avoid any possible interference with the highly reactive chemical species formed during treatment, as observed in the KDO assay. The biuret and Bradford assays both indicated that, for treatments of 2.5 min or longer, the extraction yield of outer membrane proteins increased, presumably as a result of membrane destabilization. We also observed significant changes in the SDS-PAGE electrophoretic pattern of outer membrane proteins before and after exposure to glidarc discharge. We noted the disappearance of low-molecular-mass proteins of close to 30 kDa, but observed a greater proportion of high-molecular-mass forms of between 35 and 90 kDa. These results are consistent with the observations of Igumenov et al. (14), who demonstrated that oxidation can initiate the formation of the protein complex in the E. coli membrane. However, no variation was observed in the PEL activity, which is used as an indicator for secreted protein. Therefore, it appears that oxidation specifically affects membrane-linked proteins, presumably because of their close contacts within the membrane. These results are consistent with the hypothesis that glidarc discharge acts on the bacterial structure essentially through oxidative mechanisms. The role of oxidation in glidarc discharge indicates that postdischarge processes take place in the sample after the end of the treatment. This hypothesis is worth investigating further.
Conclusion.
This study involved the use of a novel prototype for a glidarc reactor. Changes in the configuration of electrodes allow an increase in the performance of plasma, resulting in a twofold reduction of the time required for bacterial killing. Detailed analysis of the potential molecular targets in this process reveals that nonthermal plasma acts under various mechanisms driven essentially by oxidation and mediated by destabilization of the interaction of DNA and proteins with the plasma membrane. The unexpected consequences of this effect, characterized by an increase in the extraction yield of DNA and proteins, suggest that glidarc discharge should be used for extracting bacterial molecules. Together, these results allow the development of glidarc decontamination systems at an industrial level. The potential applications of this technology are widespread, including the rapid decontamination of laboratory surfaces, the treatment of liquid waste, and the recycling of water in agriculture. Today, various collaborative industrial projects based on glidarc technology are being carried out by the Technological Platform of Evreux (PFT) and other European partners.
Acknowledgments
We thank the GIE Comité Nord and ANRT for supporting this work and for funding a grant for M. Moreau.
Footnotes
Published ahead of print on 20 July 2007.
REFERENCES
- 1.Baatout, S., P. De Boever, and M. Mergeay. 2006. Physiological changes induced in four bacterial strains following oxidative stress. Prikl. Biokhim. Mikrobiol. 42:418-427. [PubMed] [Google Scholar]
- 2.Benstaali, B., D. Moussa, A. Addou, and J. L. Brisset. 1998. Plasma treatment of aqueous solutes: some chemical properties of gliding arc in humid air. Eur. J. Appl. Phys. 4:171-179. [Google Scholar]
- 3.Bradford, M. M. 1976. A rapid and sensitive method for the quantification of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 4.Briandet, R., J. O. Kamgang, Y. Rutter, J. M. Herry, M. N. Bellon-Fontaine, and J. L. Brisset. 2003. Potential of a non thermal gliding arc plasma device for the control of Staphylococcus epidermidis contamination on solid surfaces. Proc. Biofilms Med. (Elsinor, Denmark)
- 5.Bystrova, O. V., Y. A. Knirel, B. Lindner, N. A. Kocharova, A. N. Kondakova, U. Zahringer, and G. B. Pier. 2006. Structure of the core oligosaccharide and O-units in the R- and SR-type lipopolysaccharides of reference strains of Pseudomonas aeruginosa O-serogroups. FEMS Immunol. Med. Microbiol. 46:85-99. [DOI] [PubMed] [Google Scholar]
- 6.Collmer, A., J. L. Ried, and M. S. Mount. 1988. Assay methods for pectic enzymes. Methods Enzymol. 161:329-399. [Google Scholar]
- 7.Czernichowski, A., B. Hnatiuc, P. Pastva, and A. Ranaivosoloarimanan. 2000. Générateurs et circuits électriques pour alimenter des décharges instables de hautes tension. French patent 2817 444.
- 8.Czernichowski, A., and M. Czernichowski. 2002. Dispositif modulaire pour générer de multiples décharges électriques de haute tension. French patent 2842 389.
- 9.Darveau, R. P., and R. E. Hancock. 1983. Procedure for isolation of bacterial lipopolysaccharide from both smooth and rough Pseudomonas aeruginosa and Salmonella Typhimurium strains. J. Bacteriol. 155:831-838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dé, E. R., N. De Mot, N. Orange, N. Saint, and G. Molle. 1995. Channel-forming properties and structural homology of major outer membrane proteins from Pseudomonas fluorescens MF0 and OE 28-3. FEMS. Microbiol. Lett. 127:267-272. [DOI] [PubMed] [Google Scholar]
- 11.Decker, T., and M. L. Lohmann-Matthes. 1988. A quick and simple method for the quantification of lactate dehydrogenase release in measurements of cellular cytotoxicity and tumor necrosis factor (TNF) activity. J. Immunol. Methods 115:61-69. [DOI] [PubMed] [Google Scholar]
- 12.Firshein, W. 1989. Role of the DNA-membrane complex in prokaryotic DNA replication. Annu. Rev. Microbiol. 43:89-120. [DOI] [PubMed] [Google Scholar]
- 13.Gornall, A. G., C. J. Bardawill, and M. M. David. 1949. Determination of serum proteins by means of the biuret reaction. J. Biol. Chem. 177:751-766. [PubMed] [Google Scholar]
- 14.Igumenov, V. L., B. P. Sharonov, and V. A. Pasechnik. 1988. Aggregation of membrane proteins of E. coli cells after treatment with singlet oxygen. Biokhimiia 53:925-930. [PubMed] [Google Scholar]
- 15.Karkhanis, Y. D., J. Y. Zeltner, J. J. Jackson, and D. J. Carlo. 1978. A new improved microassay to determine 2-keto-3-deoxyoctonate in lipopolysaccharide of gram negative bacteria. Anal. Biochem. 85:595-601. [DOI] [PubMed] [Google Scholar]
- 16.Kido, N., M. Ohta, and N. Kato. 1990. Detection of lipopolysaccharide by ethidium bromide staining after sodium dodecyl sulfate-polyacrylamide gel electrophoresis. J. Bacteriol. 172:1145-1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Konola, J. T., K. E. Sargent, and J. B. Gow. 2000. Efficient repair of hydrogen peroxide-induced DNA damage by Escherichia coli requires SOS induction of RecA and RuvA proteins. Mutat. Res. 459:187-194. [DOI] [PubMed] [Google Scholar]
- 18.Kuzmichev, A. I., I. A. Soloshenko, V. V. Tsiolko, V. I. Kryzhanovsky, V. Y. Bazhenov, I. L. Mikhno, and V. A. Khomich. 2000. Features of sterilization by different types of atmospheric pressure discharges, p. 402-406. Proc. 7th Int. Symp. High Press. Low Temp. Plasma Chem., Hakone VII, Greifswald, Germany.
- 19.Laroussi, M., and F. Leipold. 2004. Evaluation of the roles of reactive species, heat, and UV radiation in the inactivation of bacterial cells by air plasma at atmospheric pressure. Int. J. Mass Spectrom. 223:81-86. [Google Scholar]
- 20.Laurent, P., L. Buchon, J. F. Guespin-Michel, and N. Orange. 2000. Production of pectate lyases and cellulases by Chryseomonas luteola strain MFCL0 depends on the growth temperature and the nature of the culture medium: evidence for two critical temperatures. Appl. Environ. Microbiol. 66:1538-1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lerouge, S., M. R. Wertheimer, R. Marchand, M. Tabrizian, and L. Yahia. 2000. Effect of gas composition on spore mortality and etching during low pressure plasma sterilization. J. Biomed. Mater. Res. 51:128-135. [DOI] [PubMed] [Google Scholar]
- 22.Lesueur, H., A. Czernichowski, and J. Chapelle. 1988. Dispositif de génération de plasmas basse température par formation de décharges électriques glissantes. French patent 2639 172.
- 23.Mezghani-Abdelmoula, S., A. Khemiri, O. Lesouhaitier, S. Chevalier, N. Orange, L. Cazin, and M. G. J. Feuilloley. 2004. Sequential activation of constitutive and inducible nitric oxide synthase (NOS) in rat cerebellar granule neurons by Pseudomonas fluorescens and invasive behavior of the bacteria. Microbiol. Res. 159:355-363. [DOI] [PubMed] [Google Scholar]
- 24.Mizuno, T., and M. Kageyama. 1978. Separation and characterization of the outer membrane of Pseudomonas aeruginosa. J. Biochem. 84:179-191. [DOI] [PubMed] [Google Scholar]
- 25.Moisan, M., J. Barbeau, S. Moreau, J. Pelletier, M. Tabrizian, and L. Yahia. 2001. Low-temperature sterilization using plasmas: a review of the experiments and analysis of the inactivation mechanisms. Int. J. Pharm. 226:1-21. [DOI] [PubMed] [Google Scholar]
- 26.Moreau, M., M. G. J. Feuilloley, N. Orange, and J. L. Brisset. 2005. Lethal effect of the gliding arc discharges on Erwinia spp. J. Appl. Microbiol. 98:1039-1046. [DOI] [PubMed] [Google Scholar]
- 27.Pelletier, J. 1993. La stérilisation par le procédé plasma. Agressologie 33:105-110. [PubMed] [Google Scholar]
- 28.Picot, L., S. Chevalier, S. Mezghani-Abdelmoula, A. Meriau, O. Lesouhaitier, P. Leroux, L. Cazin, N. Orange, and M. G. J. Feuilloley. 2003. Cytotoxic effects of the lipopolysaccharide from Pseudomonas fluorescens on neurons and glial cells. Microb. Pathog. 35:95-105. [DOI] [PubMed] [Google Scholar]
- 29.Ponniah, G., H. Chen, R. Michielutti, N. Salonen, and P. Blum. 2003. Single-cell protein profiling of wastewater enterobacterial communities predicts disinfection efficiency. Appl. Environ. Microbiol. 69:4227-4235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rosen, H., J. Orman, R. M. Rakita, B. R. Michel, and D. R. VanDevanter. 1990. Loss of DNA-membrane interactions and cessation of DNA synthesis in myeloperoxidase-treated Escherichia coli. Proc. Natl. Acad. Sci. USA 87:10048-10052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ruiz, N., and T. J. Silhavy. 2005. Sensing external stress: watchdogs of the Escherichia coli cell envelope. Curr. Opin. Microbiol. 8:122-126. [DOI] [PubMed] [Google Scholar]
- 32.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed., p. 260-280. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 33.Sapan, C. V., R. L. Lundblad, and N. C. Price. 1999. Colorimetric protein assay techniques. Biotechnol. Appl. Biochem. 29:99-108. [PubMed] [Google Scholar]
- 34.Toth, I. K., L. Pritchard, and P. R. Birch. 2006. Comparative genomics reveals what makes an enterobacterial plant pathogen. Annu. Rev. Phytopathol. 44:305-336. [DOI] [PubMed] [Google Scholar]
- 35.Vitrac, H., J. F. Guespin-Michel, and J. L. Brisset. 2000. A microbiological investigation of the gliding arc treatment of aqueous media, p. 393-397. Proc. 7th Int. Symp. High Press. Low Temp. Plasma Chem., Hakone VII, Greifswald, Germany.