Abstract
In a previous column study, we investigated the long-term impact of ethanol additions on U and Tc mobility in groundwater (M. M. Michalsen et al., Environ. Sci. Technol. 40:7048-7053, 2006). Ethanol additions stimulated iron- and sulfate-reducing conditions and significantly enhanced U and Tc removal from groundwater compared to an identical column that received no ethanol additions (control). Here we present the results of a combined signature lipid and nucleic acid-based microbial community characterization in sediments collected from along the ethanol-stimulated and control column flow paths. Phospholipid fatty acid analysis showed both an increase in microbial biomass (∼2 orders of magnitude) and decreased ratios of cyclopropane to monoenoic precursor fatty acids in the stimulated column compared to the control, which is consistent with electron donor limitation in the control. Spatial shifts in microbial community composition were identified by PCR-denaturing gradient gel electrophoresis analysis as well as by quantitative PCR, which showed that Geobacteraceae increased significantly near the stimulated-column outlet, where soluble electron acceptors were largely depleted. Clone libraries of 16S rRNA genes from selected flow path locations in the stimulated column showed that Proteobacteria were dominant near the inlet (46 to 52%), while members of candidate division OP11 were dominant near the outlet (67%). Redundancy analysis revealed a highly significant difference (P = 0.0003) between microbial community compositions within stimulated and control sediments, with geochemical variables explaining 68% of the variance in community composition on the first two canonical axes.
In situ bioimmobilization has recently gained attention as a potentially effective remediation strategy for metal- or radionuclide-contaminated groundwater (4, 29, 39, 65). During in situ bioimmobilization, electron donor additions are used to stimulate iron- and sulfate-reducing conditions, which promote the reductive precipitation of redox-sensitive metals and radionuclides from groundwater. Diverse or extreme geochemical conditions common to radionuclide-contaminated sites present unique challenges to successful implementation of bioimmobilization. One such site, located in Oak Ridge, TN, was established by the U.S. Department of Energy as a field research center (FRC). Groundwater at the FRC has a wide concentration range of U (up to 210 μM), Tc (up to 24 nM), and nitrate (up to 168 mM), with pH varying from 3 to 7 (Environmental Remediation Sciences Program, Oak Ridge Field Research Center site descriptions [http://public.ornl.gov/nabirfrc/sitenarrative.cfm]).
Several batch studies have been conducted to characterize the subsurface microbial community at the FRC and to evaluate its bioimmobilization potential with varied electron donors, geochemical conditions, and microbiological methods. In one study, contaminated FRC sediments were incubated with ethanol-amended, pH 4 site groundwater (53). Clone libraries of 16S rRNA genes indicated that Firmicutes were initially dominant but that Betaproteobacteria sequences were dominant after 78 days. Though 12 μM U was removed from solution, 46 mM nitrate remained in solution and U removal was not attributed to reduction. Such shifts have also been observed in 16S rRNA gene clone libraries from iron-reducing enrichment cultures prepared using FRC site sediment with acetate, lactate, or glycerol as the electron donor (50). Geobacter and Pelobacter were dominant in cultures prepared using uncontaminated, pH 6 sediment, while Anaeromyxobacter and Anaerovibrio were mostly dominant in cultures prepared using contaminated, pH 4 sediments. In a separate study conducted using FRC sediments that were not electron donor stimulated, composition of the metabolically active microbial community was shown to be different from that of the community overall (2). For example, in pH 6 sediment, Alphaproteobacteria sequences comprised ≥59% of 16S rRNA gene clone libraries, whereas Gamma- and Betaproteobacteria together comprised ≥76% of the RNA-based 16S rRNA clone libraries.
Different shifts in geochemistry and microbial community composition have been observed when contaminated sediments are amended with an electron donor in flowing systems for longer time periods. For example, lactate-amended, artificial groundwater was continuously circulated through U-contaminated FRC sediment for over 16 months (69). Effluent U concentrations decreased initially under iron-reducing conditions, which corresponded to an increase in Geobacteraceae- and Geothrix-related sequences in the column sediment. Effluent U concentrations subsequently increased under methanogenic conditions, and no decrease in Geobacteraceae- or Geothrix-related sequences was observed (8, 69). An in situ bioimmobilization study was conducted for a U- and sulfate-contaminated aquifer in Rifle, CO, by injecting acetate for ∼3 months (4). U concentrations initially decreased under iron-reducing conditions, which corresponded to increased Geobacteraceae-related sequences in groundwater. U concentrations subsequently increased under sulfate-reducing conditions, with a corresponding decrease in Geobacteraceae and an increase in sulfate-reducing-bacterium-related sequences in groundwater.
Laboratory and field studies have demonstrated the coupling between prevailing geochemistry and microbial community composition during bioimmobilization. However, spatial variability in microbial community composition and spatial correlations between community composition and geochemical conditions during long-term electron donor addition have not been described for FRC sediments. In a previous study, we continuously added ethanol to contaminated FRC site groundwater flowing through intermediate-scale, sediment-packed columns to model a potential field scale bioimmobilization strategy (42). Sediment and pore water analyses demonstrated that added ethanol effectively stimulated U and Tc removal for long time periods compared to a control with no donor added. The objective of this study was to characterize the sediment microbial community along flow paths within the ethanol-stimulated and control columns and to determine if microbial-community composition and geochemistry were spatially correlated.
MATERIALS AND METHODS
Materials and apparatus.
Above-ground, intermediate-scale columns were deployed and operated in area 2 of the FRC in Oak Ridge, TN, to serve as models of in situ permeable reactive barriers for removal of U and Tc from FRC groundwater (42). The columns were constructed from polyvinyl chloride pipe (6-in inside diameter by 8-ft length) and were packed with uncontaminated FRC site sediments. Contaminated FRC site groundwater (from well GW835) containing ∼0.8 mM nitrate, 1 mM sulfate, 4 μM U, and 580 pM Tc was continuously pumped through both columns to simulate groundwater flow. Ethanol was injected daily into the inlet and four locations along the length of one column (stimulated column); an identical column received no added ethanol (control). Pore water samples were routinely collected from eight sampling ports located along the length of each column, and changes in flow rates were monitored. Quantities of analytes removed during the experiment were quantified by integrating flow rates and differences in inlet and outlet concentrations. Sediment samples were collected for microbial community characterization from the sampling ports of the stimulated column after 13.5 months of operation and from the control after 9.5 months of operation. Detailed experimental procedures and geochemical results were summarized previously (42).
Lipid analyses.
Total lipids were extracted from the sediment samples using a modified Blyer and Dyer method (6, 72). Silicic acid chromatography was used to separate the total lipids into polar, neutral, and glycolipid fractions (22). The polar lipid fraction was subsequently transesterified using mild alkaline methanolysis to form fatty acid methyl esters (FAMEs) and convert plasmalogen ethers to dimethylacetals (DMAs) (22), with modifications (38). The neutral lipid fraction was analyzed for respiratory ubiquinone and menaquinone isoprenologues by high-performance liquid chromatography/atmospheric pressure photoionization tandem mass spectrometry (35). The FAMEs and DMAs were analyzed using a gas chromatogram (Agilent 6890) with a 55-m nonpolar column (0.25-mm inside diameter, 0.25-μm film) interfaced with a mass spectrometer (Agilent 5973). The conversion factor 2.5 × 104 cells per pmol phospholipid fatty acid (PLFA) was used to convert total PLFA extracted to cells per gram sediment (5). Individual PLFA analysis was limited to those with abundance greater than 0.5% in all stimulated and control samples.
Q-PCR analysis.
DNA was extracted from sediment samples (∼0.5 g each) using the FastDNA spin kit for soil (BIO101) and eluted in 100 μl 1/10 Tris-EDTA buffer. All quantitative PCR (Q-PCR) was performed by Microbial Insights Inc. (Rockford, TN). Each 30-μl TaqMan-based PCR assay mixture contained DNA template, 1× TaqMan universal PCR master mix (Applied Biosystems), TaqMan probe (100 to 500 nM), and forward and reverse primers (300 to 1,500 nM). TaqMan assays were performed on an ABI Prism 7300 sequence detection system (Applied Biosystems) with the following temperature program: 2 min at 50°C and 10 min at 95°C, followed by 50 cycles of 15 seconds at 95°C and 1 min at 58°C. The following groups of bacteria were targeted with the indicated TaqMan probe and forward/reverse primers, respectively: Eubacteria, TM1389, BACT1369/PROK1492R (57); Deltaproteobacteria, GBC2, 361F/685R (56); and Geobacteraceae, GBC2, 561F/825R (56). Each 30 μl SYBR green PCR assay mixture contained DNA template, 1× clone Pfu buffer (Stratagene), 0.4 mM MgCl2, 0.2 mM of each deoxynucleoside triphosphate (Roche Applied Science), SYBR green (1:30,000 dilution; Molecular Probes), 1 U PfuTurbo HotStart DNA polymerase (Stratagene), dimethyl sulfoxide (0 to 0.5 μl), and forward and reverse primers (500 to 2,500 nM). SYBR green assays were performed using an ABI Prism 7000 sequence detection system (Applied Biosystems) with temperature cycles varied based on primer set. SYBR green PCR was used to detect the following targets using the indicated forward/reverse primers: methanogens, ME1F/ME2R (24); type I and II methylotrophs, 9αF/519R and 10γF/519R, respectively (66); nirS gene, 1260F/1363R (21); and nirK gene, nirK876F/nirK1040R (27). Calibrations were obtained using a serial dilution of positive control DNA. The Sequence Detector program subtracted the background signal for each sample during cycles 3 through 15. The fluorescence threshold was computed as 10 times the standard deviation of the background signal, and the original concentration of DNA in each sample was determined by comparing the threshold cycle sample values with the calibration data. Gene copy numbers were calculated assuming 9.13 × 1014 bp/μg DNA.
Statistical analysis.
Redundancy analysis (RDA) is a linear, direct gradient ordination method by which response variables are constrained to be linear combinations of explanatory variables (62). In RDA, an eigenanalysis is performed to extract canonical factors from a product matrix containing response and predictor variable correlation coefficients. Factors are constrained to maximize the redundancy index, which is defined as the product of the variance in the predictor variable explained by the predictor factor and the variance in the response variable explained by the predictor factor (12, 55, 67). The sum of canonical eigenvalues in RDA equals the amount of variance in the response variable explained by the predictor variable. Our data set was well suited for RDA, with geochemical variables as predictor variables and community data as response variables, because the geochemical and community data varied over short distances in the columns and were reasonably represented by linear relationships (40). The response variable matrix contained the following community data: Q-PCR copy numbers, PLFA groups, PLFA ratios, DMAs, respiratory quinone ratios, and Shannon-Weiner diversity indices, which were calculated using concentrations of individual PLFAs for all stimulated and control sediment sample locations. Q-PCR values, originally in units of copy numbers per gram of sediment, were log transformed prior to analysis. The predictor variable matrix contained the following geochemical data: average U, Tc, sulfate, and nitrate concentrations for all stimulated- and control column locations prior to sediment collection for microbial community characterization. All column data were normalized to unit variance and zero mean prior to analysis to eliminate differences in magnitude yet preserve data trends. RDA was performed using the software Canoco version 4.53 (62). Monte Carlo permutations were performed (n = 3,000) to obtain a P value for the RDA results. Individual PLFAs were also analyzed via two-way cluster analysis using the software PC-ORD (41).
PCR-denaturing gradient gel electrophoresis (DGGE) analysis.
Sediment-extracted DNA (stimulated column only) was PCR amplified using the 16S rRNA primer set 341F/519R with a 40-bp GC clamp on the forward primer (45). PCR product (20 μl product plus 5 μl loading dye) was added to the polyacrylamide denaturant gel (30 to 65%, formamide-urea) using the D-Code 16/16-cm gel system (Bio-Rad, Hercules, CA). The gel was run at 55 mV for 16 h in 0.5× Tris-acetate-EDTA buffer. Bands were subsequently excised and purified using the UltraClean PCR clean-up kit (MO BIO Laboratories Inc., Carlsbad, CA). Sequence analysis was performed as previously described (10).
16S rRNA gene clone libraries.
Clone libraries were constructed using sediment-extracted DNA from stimulated column ports 2 and 3 (near the column inlet) and port 8 (near the outlet) only. Sediment-extracted DNA was PCR amplified using primers uni8F/EUB805R, the product was purified using the Geneclean Turbo kit (BIO 101), and the purified product was cloned using the TOPO TA cloning kit for sequencing (Invitrogen, Carlsbad, CA). Plasmids from random clones were extracted, purified using the QIAprep spin miniprep kit (QIAGEN), and PCR amplified using plasmid-specific primers M13F (−20)/M13R. PCR products were analyzed by restriction fragment length polymorphism using the MspI or AluI restriction enzyme (New England BioLabs). Digests were run on an agarose gel, and unique patterns representing different operational taxonomic units (OTUs) were selected for sequencing at the Oklahoma Medical Research Foundation. The resulting sequences were compared with known sequences using the Basic Local Alignment Search Tool (BLAST) and Ribosomal Database Project II. The criterion for classification of sequences into OTUs was ≥97% similarity. Chimeric sequences were identified manually and by using the Bellerophon server (http://foo.maths.uq.edu.au/∼huber/bellerophon.pl). Nonchimeric sequences were aligned using ClustalX (64), and consensus phylogenetic trees were constructed using PAUP* (59), with the neighbor-joining tree algorithm, Jukes-Cantor correction, and 1,000 replicates for bootstrap values. Positions of the DGGE band sequences in the consensus phylogenetic trees were determined a priori in a separate alignment and analysis using all sequences.
Nucleotide sequence accession numbers.
Nonchimeric sequences have been submitted to GenBank and assigned accession numbers EF422252 to EF422266 and EF507963 to EF508031 for DGGE gel band sequences and clone sequences, respectively.
RESULTS
Prevailing geochemical and pore water conditions.
Differences between the stimulated and control column pore water chemistry are summarized in Fig. 1, which shows averaged pore water solute concentrations along the flow paths of both columns during the experiment until sediment samples were collected for microbial characterization. Nitrate, sulfate, U, and Tc concentrations initially decreased along the flow path in both columns during the first 2 weeks, and this trend continued in the stimulated column for the remainder of the experiment. Concentrations decreased to a lesser extent in the control, and within 3 months U and sulfate concentrations had stabilized along the control column flow path. During the 408 days of the experiment prior to sediment collection from the stimulated column, 1,450 liters of groundwater was passed through the column and 18 mol of ethanol, 0.78 μmol Tc, 6.0 mmol U, 0.94 mol nitrate, and 1.2 mol sulfate were removed. During the 290 days prior to sediment collection from the control column, 694 liters of groundwater was passed through the column and 0.16 μmol Tc, 0.83 mmol U, and 0.45 mol nitrate were removed. No sulfate was removed in the control column. Based on the routinely measured analytes shown in Fig. 1, no sulfate reduction occurred and nitrate was only partially removed in the control, whereas sulfate reduction occurred and nitrate was completely removed in the stimulated column. While sulfate reduction was routinely observed in the stimulated column, other biogeochemical processes likely occurred as well. Although Fe(II) was not measured during the experiment, Mössbauer spectra collected from three stimulated sediment samples and the pristine sediment mixture used to pack the columns indicated that Fe(III) reduction occurred in the stimulated column. Methane (∼1 mM), acetate (∼2.8 mM), and propionate (∼1.6 mM) were also detected in the stimulated column pore water during a single sampling event, indicating that methanogenesis and fermentation also occurred.
FIG. 1.
Average concentration profiles for the ethanol-stimulated (solid symbols, 13.5 months, n = 82) and control columns (open symbols, 9.5 months, n = 10) for the entire experiment. Inlet concentrations correspond to time zero, and error bars represent 1 standard deviation. Due to differences in pumping rates, pore water velocities were smaller and computed travel times were larger in the stimulated column than in the control; travel times ranged from 0 (inlet) to 105 h in the stimulated column and from 0 (inlet) to 55 h in the control.
PLFA results.
PLFA biomass was 2 orders of magnitude greater in the stimulated column than in the control, confirming that ethanol additions promoted microbial growth; differences in percentages of total PLFA within PLFA groups were also observed (Table 1). The percentage of terminally branched saturates, which are general indicators of gram-positive or anaerobic gram-negative bacteria (30, 48), and branched monounsaturates, which are general indicators of gram-negative sulfate-reducing bacteria (7, 16), were greater on average in the stimulated column than in the control. Percentages of both terminally branched saturates and branched monounsaturates followed a linearly decreasing trend along the control flow path (r2 = 0.81 and 0.61, respectively). Percentages of midchain-branched saturates and polyunsaturates, indicators of sulfate-reducing bacteria (14, 15) and eukaryotes (18), respectively, were smaller on average in the stimulated column than in the control. Both midchain-branched saturates and polyunsaturates followed a linearly increasing trend along the stimulated column flow path (r2 = 0.76 and 0.92, respectively). The percentage of normal saturates, which are common to a wide range of microorganisms (51), was smaller on average in the stimulated column than in the control. The percentages of monounsaturates, indicators of facultative and anaerobic bacteria (49, 73), were not significantly different between columns (P = 0.07), although an increasing trend was observed along the stimulated column flow path (r2 = 0.80). Hydroxy fatty acids, which are common though not exclusive to gram-negative bacteria (3, 16), comprised a small percentage of total PLFA and were also not significantly changed in the stimulated column compared to the control (P = 0.4). The Shannon-Weiner diversity index for the stimulated column was not different on average from that for the control, although a linearly increasing trend was observed along the stimulated column flow path (r2 = 0.90). Two-way cluster analysis of individual PLFAs revealed three visually dominant differences between stimulated and control column samples, corresponding primarily to increased PLFAs within the terminally branched saturate and monounsaturate groups and decreased PLFAs within the midchain-branched saturate groups (see Fig. S1 in the supplemental material).
TABLE 1.
Data summary of Q-PCR and PLFA groups for stimulated and control column sediment samples collected from sample ports 1 to 8
| Parameter | Value for sediment samples from:
|
Value for sediment samples from: | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Stimulated column
|
Control column
|
|||||||||||||||||||
| EtOH 1 | EtOH 2 | EtOH 3 | EtOH 4 | EtOH 5 | EtOH 6 | EtOH 7 | EtOH 8 | Avg | SD | C1 | C2 | C3 | C4 | C5 | C6 | C7 | C8 | Avg | SD | |
| PLFA | ||||||||||||||||||||
| Viable biomass (cells/g sediment) | 4.28E+08 | 6.46E+08 | 4.80E+08 | 6.49E+08 | 6.99E+08 | 5.18E+08 | 8.16E+08 | 4.53E+08 | 5.86E+08 | 1.28E+08 | 4.36E+06 | 4.39E+06 | 8.98E+06 | 1.48E+07 | 6.59E+06 | 4.43E+06 | 1.29E+07 | 2.08E+06 | 7.32E+06 | 4.24E+06 |
| Community structure (% total PLFA) | ||||||||||||||||||||
| Terminally branched saturates | 14.6 | 16.0 | 20.0 | 22.2 | 17.3 | 21.4 | 20.4 | 20.8 | 19.1 | 2.57 | 17.6 | 16.4 | 14.5 | 14.0 | 15.2 | 14.3 | 10.3 | 10.4 | 14.1 | 2.43 |
| Branched monounsaturates | 3.16 | 3.83 | 3.94 | 5.30 | 3.76 | 3.86 | 4.29 | 3.88 | 4.00 | 0.57 | 3.83 | 2.28 | 3.28 | 2.92 | 2.52 | 2.76 | 1.83 | 1.60 | 2.63 | 0.69 |
| Polyunsaturates | 0.242 | 0.230 | 0.448 | 0.357 | 0.423 | 0.434 | 0.510 | 0.788 | 0.43 | 0.16 | 1.75 | 1.05 | 1.67 | 1.59 | 1.54 | 1.39 | 1.25 | 0.72 | 1.37 | 0.33 |
| Midchain-branched saturates | 3.15 | 5.15 | 4.69 | 4.82 | 6.80 | 7.56 | 8.92 | 8.95 | 6.26 | 1.99 | 9.52 | 8.65 | 9.15 | 9.11 | 11.7 | 12.3 | 10.5 | 9.49 | 10.0 | 1.23 |
| Normal saturates | 16.6 | 21.3 | 18.9 | 18.8 | 19.9 | 20.3 | 20.6 | 20.6 | 19.6 | 1.39 | 22.2 | 26.0 | 22.8 | 23.7 | 23.9 | 23.8 | 27.1 | 28.5 | 24.7 | 2.07 |
| Monounsaturates | 61.9 | 53.4 | 52.0 | 48.4 | 51.8 | 46.3 | 45.1 | 44.9 | 50.5 | 5.31 | 45.1 | 45.7 | 48.6 | 48.7 | 45.2 | 45.4 | 49.0 | 49.3 | 47.1 | 1.81 |
| Hydroxy | 0.248 | 0.062 | 0.054 | 0.116 | 0.111 | 0.130 | 0.129 | 0.150 | 0.13 | 0.06 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 |
| Shannon-Weiner diversity index | 2.59 | 2.82 | 2.88 | 2.88 | 2.93 | 3.04 | 3.04 | 3.14 | 2.91 | 0.16 | 3.09 | 2.88 | 3.03 | 2.97 | 3.04 | 3.09 | 2.79 | 2.65 | 2.94 | 0.15 |
| Metabolic status (ratio) | ||||||||||||||||||||
| cy17:0/16:1w7c | 0.084 | 0.051 | 0.060 | 0.058 | 0.053 | 0.074 | 0.047 | 0.083 | 0.064 | 0.014 | 0.377 | 0.329 | 0.549 | 0.394 | 0.488 | 0.411 | 0.496 | 0.676 | 0.465 | 0.104 |
| cy19:0/18:17c | 0.057 | 0.080 | 0.094 | 0.103 | 0.113 | 0.096 | 0.181 | 0.129 | 0.107 | 0.035 | 0.260 | 0.267 | 0.293 | 0.231 | 0.256 | 0.262 | 0.224 | 0.213 | 0.251 | 0.024 |
| Total cyc/monoa | 0.141 | 0.131 | 0.154 | 0.161 | 0.166 | 0.169 | 0.228 | 0.212 | 0.170 | 0.031 | 0.637 | 0.596 | 0.842 | 0.625 | 0.744 | 0.673 | 0.720 | 0.890 | 0.716 | 0.098 |
| 16:1w7t/16:1w7c | 0.070 | 0.070 | 0.094 | 0.114 | 0.099 | 0.122 | 0.102 | 0.155 | 0.103 | 0.026 | 0.063 | 0.054 | 0.080 | 0.065 | 0.062 | 0.069 | 0.054 | 0.063 | 0.064 | 0.008 |
| 18:1w7t/18:1w7c | 0.034 | 0.036 | 0.033 | 0.033 | 0.033 | 0.035 | 0.064 | 0.075 | 0.043 | 0.016 | 0.054 | 0.038 | 0.076 | 0.063 | 0.062 | 0.066 | 0.036 | 0.072 | 0.058 | 0.014 |
| Total trans/cisb | 0.104 | 0.106 | 0.127 | 0.146 | 0.131 | 0.157 | 0.165 | 0.230 | 0.146 | 0.038 | 0.117 | 0.092 | 0.156 | 0.127 | 0.124 | 0.135 | 0.090 | 0.134 | 0.122 | 0.021 |
| i15:0/a15:0 | 0.831 | 0.767 | 0.725 | 0.453 | 0.617 | 0.540 | 0.670 | 0.838 | 0.680 | 0.129 | 1.21 | 1.37 | 1.28 | 1.47 | 1.39 | 1.38 | 1.45 | 1.38 | 1.368 | 0.081 |
| i17:0/a17:0 | 0.245 | 0.198 | 0.193 | 0.181 | 0.172 | 0.229 | 0.249 | 0.295 | 0.220 | 0.039 | 0.695 | 0.859 | 0.648 | 0.736 | 0.702 | 0.713 | 0.720 | 0.945 | 0.752 | 0.092 |
| Total iso/anteisoc | 1.08 | 0.965 | 0.917 | 0.635 | 0.789 | 0.769 | 0.919 | 1.13 | 0.900 | 0.154 | 1.91 | 2.23 | 1.92 | 2.21 | 2.09 | 2.10 | 2.17 | 2.33 | 2.120 | 0.138 |
| DMAs (pmol/g sediment) | 206 | 343 | 397 | 326 | 479 | 531 | 1,012 | 612 | 488 | 231 | 16.5 | 1.79 | 28.2 | 18.3 | 15.9 | 8.65 | 14.9 | 8.19 | ||
| UQ/MQ ratio | 2.53 | 2.14 | 2.36 | 4.07 | 4.33 | 3.68 | 3.19 | 0.87 | 3.49 | 2.77 | 2.97 | 2.45 | 2.12 | 1.68 | 1.38 | 0.390 | 2.16 | 0.926 | ||
| Q-PCR (copies/g sediment)d | ||||||||||||||||||||
| Eubacterial 16S rRNA | 1.23E+09 | 3.94E+08 | 2.08E+09 | 1.88E+09 | 3.11E+09 | 2.00E+09 | 2.65E+09 | 2.07E+09 | 1.93E+09 | 7.76E+08 | 5.00E+07 | 5.83E+07 | 5.83E+07 | 6.91E+06 | 2.11E+07 | 1.59E+07 | 8.22E+06 | 3.68E+06 | 3.08E+07 | 2.71E+07 |
| nirS | 2.89E+08 | 3.08E+07 | 3.06E+08 | 3.55E+08 | 4.35E+08 | 4.53E+08 | 5.99E+08 | 1.17E+09 | 4.55E+08 | 3.10E+08 | 1.02E+07 | 8.20E+06 | 7.69E+06 | 1.17E+06 | 2.80E+06 | 2.44E+06 | 9.57E+06 | 1.65E+06 | 5.46E+06 | 3.55E+06 |
| nirK | 9.31E+07 | 5.56E+07 | 1.85E+08 | 2.07E+08 | 2.20E+08 | 2.46E+08 | 2.45E+08 | 3.97E+08 | 2.06E+08 | 9.72E+07 | 4.48E+06 | 9.99E+06 | 1.80E+07 | 5.12E+06 | 9.35E+06 | 4.43E+06 | 2.11E+04 | ND | 6.42E+06 | 5.55E+06 |
| Deltaproteobacteria | 3.88E+05 | ND | 4.67E+07 | 3.46E+08 | 1.78E+08 | 2.22E+08 | 1.49E+08 | 3.52E+08 | 1.62E+08 | 1.32E+08 | 3.08E+07 | 3.25E+06 | 4.05E+06 | 2.44E+04 | 5.42E+05 | 5.42E+05 | 2.44E+04 | 2.36E+03 | 4.91E+06 | 9.90E+06 |
| Geobacteraceae | 108* | ND | 205* | 642* | 4.64E+04 | 1.88E+04 | 4.82E+03 | 1.98E+04 | 1.13E+04 | 1.54E+04 | <100 | <100 | <100 | ND | ND | <100 | 112* | ND | ||
| Methanogens | ND | ND | ND | ND | ND | ND | ND | ND | 6.61E+05 | 5.56E+06 | 2.25E+05 | 4.18E+05 | 2.64E+05 | 3.27E+05 | 2.64E+05 | 3.22E+05 | 1.01E+06 | 1.73E+06 | ||
| Methylotrophs | 6.24E+06 | ND | 5.54E+05 | 1.60E+05 | 1.35E+05 | 1.31E+05 | 1.72E+05 | 4.34E+05 | 9.78E+05 | 2.00E+06 | 3.72E+05 | 2.15E+07 | 5.53E+05 | 2.82E+05 | 1.90E+05 | 5.14E+05 | 3.63E+05 | ND | 2.97E+06 | 7.00E+06 |
cyc/mono, ratio of cyclopropane to monoenoic precursor fatty acids.
trans/cis, ratio of monounsaturated trans to cis isomers.
iso/anteiso, ratio of iso- to anteiso-saturated fatty acids.
*, estimated value; <100, below detection limit; ND, not detected (sample not processed).
Elevated ratios of both cyclopropyl fatty acids to their monoenoic precursors and ratios of monounsaturated trans to cis isomers have been linked with starvation, stationary-phase growth, and nutrient deprivation (22, 23, 31, 63). The ratio of cyclopropyl fatty acids to their monoenoic precursors was greater on average in the control than in the stimulated column, suggesting that microbial growth in the control was substrate limited. The ratio of cyclopropyl fatty acids to monoenoic precursors also increased linearly along the stimulated column flow path (r2 = 0.81). The ratio of monounsaturated trans to cis isomers was not significantly changed in the stimulated column compared to the control (P = 0.08) but was observed to increase linearly along the stimulated column flow path (r2 = 0.81). Elevated ratios of iso- to anteiso-saturated fatty acids have been linked with bacterial membrane fluidity changes in response to environmental stress, particularly temperature (30, 52, 70). Though both models were maintained at the same temperature during the experiment (∼21°C), the ratio of iso- to anteiso-saturated fatty acids was decreased in the stimulated column compared to the control, suggesting a change in bacterial community. DMAs, indicators of clostridia and some gram-negative bacteria (43, 44), were greater on average in the stimulated column and followed a linearly increasing trend along the stimulated column flow path (r2 = 0.63). Respiratory ubiquinones are associated with high-energy electron acceptors such as oxygen and nitrate, while menaquinones are associated with anaerobic respiration (26); thus, elevated ratios of ubiquinones to menaquinones (UQ/MQ) indicate aerobic respiration. The UQ/MQ ratio was not significantly changed in the stimulated model compared to the control, but a linearly decreasing trend along the control flow path was observed (r2 = 0.94). The UQ/MQ ratio was elevated near the control inlet (3.5), then decreased linearly to 0.4 near the control outlet, suggesting the presence of more aerobes and denitrifiers near the inlet. The UQ/MQ ratios for the stimulated column were lower in ports near the inlet (2.5) but increased in ports near the outlet (∼4). Nitrate reduction was an important process in the stimulated column, particularly in ports near the inlet, as nitrate was typically completely removed by port 2. Sulfate reduction was consistently observed in the stimulated column, and so increased UQ/MQ ratios along the stimulated column flow path were unexpected. It is interesting that Dehalococcoides sp. obligate anaerobes were recently found to have more ubiquinones, which they may use to manage oxidative stress (71).
Q-PCR results.
Eubacterial 16S rRNA gene copy numbers were greater in the stimulated column than in the control, further substantiating that ethanol additions promoted microbial growth (Table 1). Eubacterial 16S rRNA gene copy numbers were also observed to decrease linearly with distance along the control column flow path (r2 = 0.66). Copy numbers of the dissimilatory nitrite reductase genes, nirS and nirK, were greater in the stimulated column than in the control and increased linearly along the stimulated column flow path (r2 = 0.70 and 0.84, respectively). Shifts were also detected in several general groups of Bacteria and Archaea. For example, Deltaproteobacteria were more abundant in the stimulated column than in the control and also increased linearly along the stimulated column flow path (r2 = 0.54). Geobacteraceae were also more abundant on average in the stimulated column, and a marked increase was observed near the stimulated column outlet, where soluble electron acceptors were largely depleted. Although elevated methane concentrations were previously detected in stimulated column pore water (42), Methanogens were not detected using our methods in stimulated column sediments. Methanogens were increased in the control column compared to the stimulated column, though not significantly (P = 0.07). Methylotrophs also not differ significantly between the stimulated and control sediments.
RDA results.
RDA results were summarized in a joint plot containing geochemistry-derived sample scores (points), PLFA, Q-PCR, and geochemical variable scores (Fig. 2). A brief guide to joint plot interpretation follows (see references 60 and 61 for more detailed information). Arrow length represents the magnitude of the correlation coefficient with the geochemistry-derived canonical axes. Arrows pointing in the same direction indicate strong positive correlations, perpendicular arrows indicate no correlation, and arrows pointing in opposite directions indicate strong negative correlations. Distance between sample points is proportional to the magnitude of the difference in community composition within samples. Nearly 52% of community data variance was explained by the first geochemistry-derived canonical axis (P = 0.0003), reflected graphically in a clear separation of stimulated and control sample scores on the first axis. PLFA biomass, Q-PCR eubacterial biomass, and DMA scores were elevated in stimulated-column samples and negatively correlated with geochemical variables, whereas biomarker stress ratios (iso- to anteiso-saturated fatty acids and cyclopropane to monoenoic precursor fatty acids), normal saturates, polyunsaturates, midchain-branched saturates, and methanogen scores were elevated in control sediment samples and positively correlated with geochemical variables. Branched monounsaturates, terminally branched saturates, Geobacteraceae, Deltaproteobacteria, UQ/MQ ratios, and nirK scores were also elevated in stimulated-sediment samples and negatively correlated with geochemical variables. Both monounsaturates and pore water concentrations were greatest in ethanol-stimulated sediment from port 1 (EtOH 1) relative to those in subsequent ports, the combined effect being that monounsaturates were less negatively correlated with geochemical variables. Community composition shifts along the stimulated column flow path were confirmed by the separation of stimulated samples on the second canonical axis. Stimulated samples near the inlet (ports 1 to 3) showed positive loadings, while subsequent stimulated samples showed negative loadings, with the exception of those from port 7. PLFA biomass was elevated in port 7 sediment, which resulted in a slightly positive loading on the second axis. The Shannon-Weiner diversity indices and methylotroph scores for stimulated and control samples were similar and were uncorrelated with geochemical variables.
FIG. 2.
Ordination joint plot of RDA results. Points represent geochemistry-derived sample scores for ethanol-stimulated (EtOH 1 to 8) and control sediment samples (C1 to C8), small-tipped arrows represent geochemistry-derived community scores (PLFA groups, ratios, and Q-PCR targets), and large-tipped arrows represent geochemical variable scores. Axis labels indicate the percentage of community variance explained by the environmental variables.
Community composition of stimulated-column sediment.
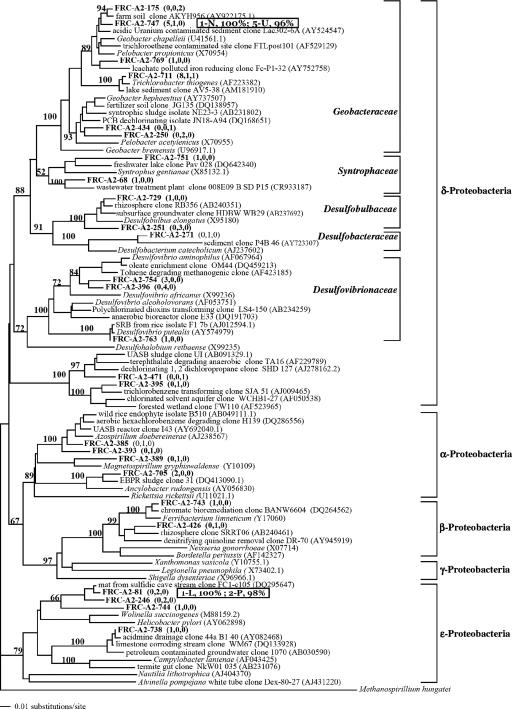
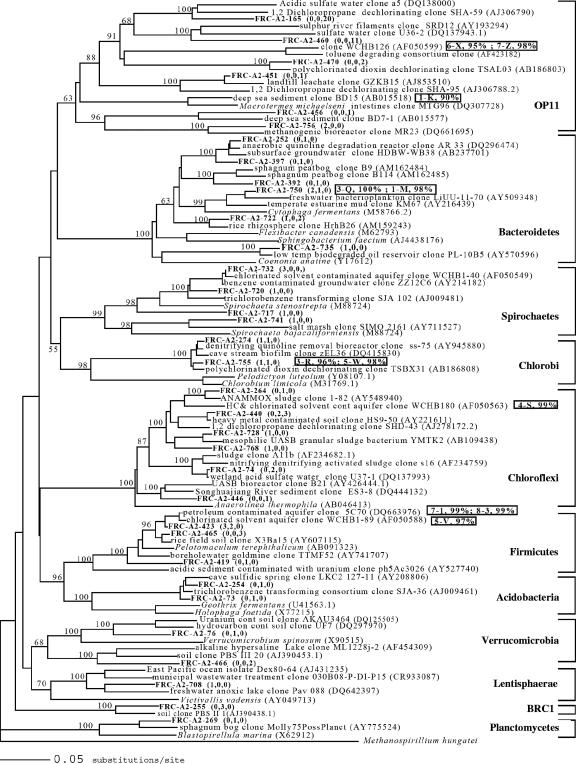
Port 1 had the highest number of visually dominant DGGE bands, while the adjacent port 2 contained only one visually dominant band (see Fig. S2 in the supplemental material). Selected band sequences from stimulated sediments were most similar to the candidate division OP11 (port 1-K), Epsilonproteobacteria (port 1-L, port 2-P), Bacteroidetes (port 1-M, port 3-Q), Geobacteraceae (port 1-N, port 5-U), Chlorobi (port 3-R, port 5-W), and Chloroflexi (port 4-S) (Fig. 3 and 4). Multiple sequences from ports near the column outlet were similar to those of Firmicutes (port 5-V, port 7-1, port 8-3) and candidate division OP11 (port 6-X, port 7-Z).
FIG. 3.
Phylogenetic relationships of cloned 16S rRNA genes and selected sequences (Proteobacteria only). Nodal values represent bootstrap probabilities based on 1,000 replicates. Clones are designated FRC-A2-clone number, with frequencies detected in port 2, port 3, and port 8 shown in parentheses. DGGE band sequences are designated by the port number and band letter and are positioned adjacent to the most similar sequence determined in a separate alignment and analysis.
FIG. 4.
Phylogenetic relationships of cloned 16S rRNA genes and selected sequences (excluding Proteobacteria). Nodal values represent bootstrap probabilities based on 1,000 replicates. Clones are designated FRC-A2-clone number, with frequencies detected in port 2, port 3, and port 8 shown in parentheses. DGGE band sequences are designated by the port number and band letter and are positioned adjacent to the most similar sequence determined in a separate alignment and analysis.
Most clones detected in port 2 and 3 sediment 16S rRNA gene libraries (46 to 52%) belonged to the phylum Proteobacteria (Table 2). In port 2 sediments, 41% of sequences detected belonged to Gammaproteobacteria; other sequences detected belonged to Spirochaetes (12%), Bacteroidetes (10%), Firmicutes (8%), Chloroflexi (4%), Acidobacteria (2%), other Proteobacteria (10%), and the candidate division OP11 (4%). Sequences belonging to these phyla were also detected the port 3 library, with the exception of candidate division OP11. In contrast to ports 2 and 3, where Proteobacteria sequences dominated, 67% of clones in port 8 belonged to candidate division OP11; other sequences detected belonged to Delta- and Epsilonproteobacteria (11%), Chloroflexi (7%), Firmicutes (5%), Spirochaetes (4%), and Bacteroidetes (2%).
TABLE 2.
Distribution of clones within the ethanol-stimulated sediment clone libraries from ports 2, 3, and 8
| Phylum, class, or candidate division | % of clones in clone library fora:
|
||
|---|---|---|---|
| EtOH 2 (52, 32, 3.217) | EtOH 3 (45, 32, 3.352) | EtOH 8 (55, 17, 2.193) | |
| Deltaproteobacteria | 41 | 29 | 9 |
| Spirochaetes | 12 | 4 | 4 |
| Bacteroidetes | 10 | 9 | 2 |
| OP11 | 4 | 67 | |
| Chloroflexi | 4 | 11 | 7 |
| Firmicutes | 8 | 7 | 5 |
| Epsilonproteobacteria | 4 | 9 | 2 |
| Alphaproteobacteria | 4 | 7 | |
| Betaproteobacteria | 2 | 2 | |
| Acidobacteria | 2 | 7 | |
| Others | 8 | 16 | 4 |
Values in parentheses are as follows: number of taxa, number of OTUs, Shannon-Weiner diversity index.
Deltaproteobacteria clone sequences grouped into five families, including Geobacteraceae, Syntrophaceae, Desulfobulbaceae, Desulfobacteraceae, and Desulfovibrionaceae (Fig. 3). Geobacteraceae is a well known family of iron- and U-reducing bacteria and may be important to successful implementation of in situ bioimmobilization (4). DGGE band sequences (port 1-N, port 5-U) and clone sequences from this study shared close homology with a Geobacteraceae clone detected in electron donor-stimulated FRC sediment (50). Although Desulfobacterium autotrophicum was shown not to enzymatically reduce U(VI) (34), Desulfobacteraceae sequences are commonly detected in electron donor-stimulated, uranium-contaminated sediments (4, 9). A single clone (271) shared close homology with a Desulfobacteraceae clone detected in other U-reducing sediment (58). Within the Epsilonproteobacteria, DGGE band sequences (port 1-L, port 2-P) and clone sequences from this study shared close homology with clones detected in other sulfidic environments (36). Betaproteobacteria clone sequences were similar to those of clones detected in other denitrifying (33) and metal-contaminated environments as well (clone BANW6604, unpublished). The majority of port 8 clone sequences shared close homology with clones detected in sulfur-rich environments within the candidate division OP11 (Fig. 4). Also within the candidate division OP11, DGGE band sequences (port 6-X, port 7-Z) shared close homology with clones detected in hydrocarbon-degrading environments (13, 19) and DGGE band sequence from port 1-K was most similar (90%) to a deep-sea sediment clone (32). Within the phylum Firmicutes, DGGE band sequences (port 7-1, port 8-3, port 5-V) and clone sequences were similar to clones detected in hydrocarbon- and chlorinated-solvent-contaminated environments (13) and also grouped with a low-G+C, gram-positive clone detected in electron donor-stimulated FRC sediment (50). Within the phylum Verrucomicrobia, clone sequences shared close homology with a clone detected in unstimulated FRC sediment (8). Bacteroidetes clone sequences grouped with clones detected in anaerobic environments (e.g., peat bogs and rice paddies), while DGGE band sequences (port 3-Q, port 1-M) shared close homology with a freshwater bacterial consortium clone (17). Within the phylum Chlorobi, DGGE band sequences (port 3-R, port 5-W) and clone sequences from this study were similar to clones detected in denitrifying (33) and dechlorinating (74) environments. Within the phylum Chloroflexi, DGGE band sequence port 4-S shared close homology with a clone detected in a hydrocarbon- and chlorinated-solvent-contaminated environment (13).
DISCUSSION
While in situ field studies are critically important in understanding bioimmobilization processes, extensive sampling during such studies is often difficult due to site inaccessibility or labor- and cost-intensive sediment core collection. In this study, we used flowing site groundwater and site sediments to simulate the operation of an in situ biobarrier above ground for over 13 months. The experimental design allowed for continuous monitoring of pore water geochemistry and collection of sediment samples at relatively small spatial scales (∼25 cm) along geochemical gradients and column flow paths. RDA provided a direct, quantitative test of our hypothesis that microbial community composition was correlated with pore water geochemistry. The model results showed that geochemical variables were good predictors of microbial community composition, as measured by PLFA and Q-PCR analyses, with 68% of the community variance explained on the first two canonical axes. The strong negative correlation of stimulated and control column sample scores on the first axis and the significance of the model results (P = 0.0003) clearly indicate that added ethanol effectively stimulated a distinct microbial community that promoted and sustained removal of U and Tc from site groundwater.
Levels of nitrate and metal contamination are important determining factors in microbial community composition both before (2, 20) and following biostimulation of FRC sediments (46, 53, 54a). In other FRC studies, microbial communities in contaminated site sediments were characterized following in situ biostimulation with pH-neutralized site groundwater containing extreme nitrate and ethanol concentrations (∼125 mM nitrate and 350 mM ethanol) (47, 54a). Spain et al. observed that under denitrifying conditions Betaproteobacteria sequences were dominant (50 to 79%) in clone libraries, and members of the genus Castellaniella were identified as important denitrifiers (54a). In this study, nitrate levels were much lower (0.8 mM), and although denitrification occurred in port 2 and 3 sediments in this study, only 2% of sequences detected belonged to Betaproteobacteria and no Castellaniella sequences were detected. In contrast, several nitrate-reducing clones within the Alpha- and Betaproteobacteria detected in this study were related to iron-reducing bacteria capable of using nitrate as an electron acceptor for growth: Magnetospirillum gryphiswaldense (37, 54) and Ferribacterium limneticum (11). In a separate FRC study, North et al. observed that under iron-reducing conditions Geobacteraceae and Anaeromyxobacter sequences were equally dominant and together comprised 37% of sequences detected in clone libraries (46). In this study, Geobacteraceae sequences were detected in significant proportions in port 2 and 3 sediments but no Anaeromyxobacter sequences were detected.
An additional and notable difference between this study and other FRC studies is the apparent importance of the candidate division OP11, which comprised 67% of sequences in the port 8 sediment clone library. DGGE band sequences (port 6-X, port 7-Z) in ports 6 and 7 were also most similar to candidate division OP11, and a similar band was present in port 8 but absent in all other ports, confirming the significance of OP11 in sediment near the column outlet. Although little is known about the physiology of this group, its members are often detected in anaerobic environments linked with sulfur cycling, hydrocarbon contamination, and methanogenesis (25, 28). Many OP11 sequences detected in this study were similar to other OP11 sequences detected in dechlorinating and sulfate-rich environments, suggesting that OP11 may play an important role in sulfur or other anaerobic cycles in reducing FRC sediments. A recent environmental genome sequencing study of unstimulated FRC area 2 sediment also showed a relatively high number of sequences related to the candidate division OP11 (1).
Other long-term bioimmobilization studies of flowing systems have shown that microbial communities and geochemistry may shift in such a way as to not favor bioimmobilization. Microbial community changes were observed in groundwater in a single acetate-stimulated monitoring well during an in situ U bioimmobilization study (4). Clone libraries of 16S rRNA genes showed that under iron- and U-reducing conditions Geobacteraceae sequences were dominant, but after 80 days and under sulfate-reducing conditions, U was remobilized and sulfate-reducing bacterium sequences were dominant. Eight months later, acetate injection resumed, and microbial community changes were monitored in both groundwater and sediment after another 40 days (68). As before, Geobacteraceae sequences were dominant in groundwater where the greatest U and iron reduction occurred. In a separate laboratory study, artificial lactate-amended groundwater was continuously pumped through U-contaminated FRC sediments (69). U concentrations initially decreased under iron-reducing conditions but subsequently increased under methanogenic conditions. Q-PCR showed that Geothrix and Geobacteraceae sequences increased during U reduction and did not decrease during U remobilization, which occurred under methanogenic conditions (8, 69).
In this study we characterized the microbial community after 13 months of U and Tc removal, although the system continued to sustain contaminant removal for a total of 20 months (42). Geobacteraceae sequences were detected in significant proportions near the inlet where U and Tc reduction occurred, as were sequences within the sulfate-reducing families Desulfovibrionaceae and Desulfobulbaceae. Two important differences between our study and the two aforementioned studies are (i) uncontaminated sediment containing no sorbed U(VI) was used in this study and (ii) we observed no increase in U concentrations under sulfate-reducing conditions, although methanogenesis, fermentation, and iron reduction were also confirmed in single samples (42). Observed U concentration changes in flowing, electron donor-stimulated systems result from a combination of abiotic (i.e., desorption, abiotic reduction, or oxidation) and microbially catalyzed reactions, which occur at different rates. Under iron-reducing conditions, the rate of microbially catalyzed U(VI) reduction may exceed that of U(VI) desorption from contaminated sediments, but under sulfate-reducing or methanogenic conditions, the rate of U(VI) desorption from contaminated sediments may exceed the combined rate of abiotic and microbially catalyzed U(VI) reduction (47). We speculate, therefore, that U remobilization would likely have been observed if sediment containing significant sorbed U(VI) had been used in this study. Rates of U(VI) desorption should be considered and accounted for when designing in situ bioimmobilization treatments in contaminated formations without the use of constructed, uncontaminated fill material.
Supplementary Material
Acknowledgments
This research was supported by grants FG03-02ER63443, DE-FC02-96ER62278, and FG02-00ER62986 (subcontract FSU F48792) from the Office of Biological and Environmental Research of the Office of Science, U.S. Department of Energy, Natural and Accelerated Bioremediation Research Program. Additional support was provided by an Integrative Graduate Education and Research Traineeship grant from the National Science Foundation.
This work would not have been possible without the tireless efforts of Mary Anna Bogle, who collected weekly samples and maintained the experimental systems. Special thanks also to Dave Watson, who provided extensive support at the Field Research Center.
Footnotes
Published ahead of print on 13 July 2007.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Abulencia, C. B., D. L. Wyborski, J. A. Garcia, M. Podar, W. Q. Chen, S. H. Chang, H. W. Chang, D. Watson, E. L. Brodie, T. C. Hazen, and M. Keller. 2006. Environmental whole-genome amplification to access microbial populations in contaminated sediments. Appl. Environ. Microbiol. 72:3291-3301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Akob, D. M., H. J. Mills, and J. E. Kostka. 2007. Metabolically active microbial communities in uranium-contaminated subsurface sediments. FEMS Microbiol. Ecol. 59:95-107. [DOI] [PubMed] [Google Scholar]
- 3.Alugupalli, S. P., and F. L. Larrson. 1994. Systematic study of the 3-hydroxy fatty acid composition of mycobacteria. J. Bacteriol. 176:2962-2969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anderson, R. T., H. A. Vrionis, I. Ortiz-Bernad, C. T. Resch, P. E. Long, R. Dayvault, K. Karp, S. Marutzky, D. R. Metzler, A. Peacock, D. C. White, M. Lowe, and D. R. Lovely. 2003. Stimulating the in situ activity of Geobacter species to remove uranium from the groundwater of a uranium-contaminated aquifer. Appl. Environ. Microbiol. 69:5884-5891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Balkwill, D. L., F. R. Leach, J. T. Wilson, J. F. McNabb, and D. C. White. 1988. Equivalence of microbial biomass measures based on membrane lipid and cell wall components, adenosine triphosphate, and direct counts in subsurface aquifer sediments. Microb. Ecol. 16:73-84. [DOI] [PubMed] [Google Scholar]
- 6.Bligh, E. G., and W. J. Dyer. 1959. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37:911-917. [DOI] [PubMed] [Google Scholar]
- 7.Boon, J. J., J. W. de Leeuw, G. J. Hoek, and J. H. Vosjan. 1977. Significance and taxonomic value of iso and anteiso monoenoic fatty acids and branched β-hydroxy acids in Desulfovibrio desulfuricans. J. Bacteriol. 129:1183-1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brodie, E. L., T. Z. DeSantis, D. C. Joyner, S. M. Baek, J. T. Larsen, G. L. Andersen, T. C. Hazen, P. M. Richardson, D. J. Herman, T. K. Tokunaga, J. M. Wan, and M. K. Firestone. 2006. Application of a high-density oligonucleotide microarray approach to study bacterial population dynamics during uranium reduction and reoxidation. Appl. Environ. Microbiol. 72:6288-6298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chang, Y., P. E. Long, R. Geyer, A. D. Peacock, C. T. Resch, K. Sublette, S. Pfiffner, A. Smithgall, R. T. Anderson, H. A. Vrionis, J. R. Stephen, R. Dayvault, I. Ortiz-Bernad, D. R. Lovely, and D. C. White. 2005. Microbial incorporation of 13C-labeled acetate at the field scale: detection of microbes responsible for reduction of U(VI). Environ. Sci. Technol. 39:9039-9048. [DOI] [PubMed] [Google Scholar]
- 10.Chang, Y. J., J. R. Stephen, A. P. Richter, A. D. Venosa, J. Bruggemann, S. J. Macnaughton, G. A. Kowalchuk, J. R. Haines, E. Kline, and D. C. White. 2000. Phylogenetic analysis of aerobic freshwater and marine enrichment cultures efficient in hydrocarbon degradation: effect of profiling method. J. Microbiol. Methods 40:19-31. [DOI] [PubMed] [Google Scholar]
- 11.Cummings, D. E., F. Caccavo, S. Spring, and R. F. Rosenzweig. 1999. Ferribacterium limneticum, gen. nov., sp. nov., an Fe(III)-reducing microorganism isolated from mining-impacted freshwater lake sediments. Arch. Microbiol. 171:183-188. [Google Scholar]
- 12.DeSarbo, W. S., and K. Jedd. 1986. Redundancy analysis, p. 622-666. In S. Kotz and N. L. Johnson (ed.), Encyclopedia of statistical sciences, vol. 7. John Wiley & Sons, Toronto, Canada. [Google Scholar]
- 13.Dojka, M. A., P. Hugenholtz, S. K. Haack, and N. R. Pace. 1998. Microbial diversity in a hydrocarbon- and chlorinated-solvent-contaminated aquifer undergoing intrinsic bioremediation. Appl. Environ. Microbiol. 64:3869-3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dowling, N. J., F. Widdel, and D. C. White. 1986. Phospholipid ester-linked fatty acid biomarkers of acetate-oxidizing sulfate reducers and other sulfide forming bacteria. J. Gen. Microbiol. 132:1815-1825. [Google Scholar]
- 15.Dowling, N. J. E., P. D. Nichols, and D. C. White. 1988. Phospholipid fatty acid and infra-red spectroscopic analysis of a sulphate-reducing consortium. FEMS Microbiol. Lett. 53:325-333. [Google Scholar]
- 16.Edlund, A. N., P. D. Nichols, R. Roffey, and D. C. White. 1985. Extractable and lipopolysaccharide fatty acid and hydroxy acid profiles from Desulfovibrio species. J. Lipid Res. 26:982-988. [PubMed] [Google Scholar]
- 17.Eiler, A., and S. Bertilsson. 2004. Composition of freshwater bacterial communities associated with cyanobacterial blooms in four Swedish lakes. Environ. Microbiol. 6:1228-1243. [DOI] [PubMed] [Google Scholar]
- 18.Erwin, J. A. 1973. Comparative biochemistry of fatty acids in eukaryotic microorganisms, p. 41-143. In J. A. Erwin (ed.), Lipids and biomembranes of eukaryotic microorganisms. Academic Press, New York, NY.
- 19.Ficker, M., K. Krastel, S. Orlicky, and E. Edwards. 1999. Molecular characterization of a toluene-degrading methanogenic consortium. Appl. Environ. Microbiol. 65:5576-5585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fields, M. W., T. Yan, S.-K. Rhee, S. L. Carroll, P. M. Jardine, D. B. Watson, C. S. Criddle, and J. Zhou. 2005. Impacts on microbial communities and cultivable isolates from groundwater contaminated with high levels of nitric acid-uranium waste. FEMS Microbiol. Ecol. 53:417-428. [DOI] [PubMed] [Google Scholar]
- 21.Gruntzig, V., S. C. Nold, J. Zhou, and J. M. Tiedje. 2001. Pseudomonas stutzeri nitrite reductase gene abundance in environmental samples measured by real-time PCR. Appl. Environ. Microbiol. 67:760-768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guckert, J. B., C. P. Antworth, P. D. Nichols, and D. C. White. 1985. Phospholipid, ester-linked fatty acid profiles as reproducible assays for changes in prokaryotic community structure of estuarine sediments. FEMS Microbiol. Ecol. 31:147-158. [Google Scholar]
- 23.Guckert, J. B., M. A. Hood, and D. C. White. 1986. Phospholipid ester-linked fatty acid profile changes during nutrient deprivation of Vibrio cholerae: increases in the trans/cis ratio and proportions of cyclopropyl fatty acids. Appl. Environ. Microbiol. 52:794-801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hales, B. A., C. Edwards, D. A. Ritchie, G. Hall, R. W. Pickup, and J. R. Saunders. 1996. Isolation and identification of methanogen-specific DNA from blanket bog peat by PCR amplification and sequence analysis. Appl. Environ. Microbiol. 62:668-675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Harris, J. K., S. T. Kelley, and N. R. Pace. 2004. New perspective on uncultured bacterial phylogenetic division OP11. Appl. Environ. Microbiol. 70:845-849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hedrick, D. B., and D. C. White. 1986. Microbial respiratory quinones in the environment. I. A sensitive liquid chromatographic method. J. Microbiol. Methods 5:243-254. [Google Scholar]
- 27.Henry, S., E. Baudoin, J. C. Lopez-Gutierrez, F. Martin-Laurent, A. Brauman, and L. Philippot. 2004. Quantification of denitrifying bacteria in soils by nirK gene targeted real-time PCR. J. Microbiol. Methods 59:327-335. [DOI] [PubMed] [Google Scholar]
- 28.Hugenholtz, P., C. Pitulle, K. L. Hershberger, and N. R. Pace. 1998. Novel division level bacterial diversity in a Yellowstone hot spring. J. Bacteriol. 180:366-376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Istok, J. D., J. M. Senko, L. R. Krumholz, D. Watson, M. A. Bogle, A. Peacock, Y. J. Chang, and D. C. White. 2004. In situ bioreduction of technetium and uranium in a nitrate-contaminated aquifer. Environ. Sci. Technol. 38:468-475. [DOI] [PubMed] [Google Scholar]
- 30.Kaneda, T. 1991. Iso- and anteiso-fatty acids in bacteria: biosynthesis, function, and taxonomic significance. Microbiol. Rev. 55:288-302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kieft, T. L. R., D. B. Ringelberg, and D. C. White. 1994. Changes in ester-linked phospholipid fatty acid profiles of subsurface bacteria during starvation and desiccation in a porous medium. Appl. Environ. Microbiol. 60:3292-3299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li, L., C. Kato, and K. Horikoshi. 1999. Bacterial diversity in deep-sea sediments from different depths. Biodivers. Conserv. 8:659-677. [Google Scholar]
- 33.Liu, B., F. Zhang, X. Feng, Y. Liu, X. Yan, X. Zhang, L. Wang, and L. Zhao. 2006. Thauera and Azoarcus as functionally important genera in a denitryifying quinoline-removal bioreactor as revealed by microbial community structure comparison. FEMS Microbiol. Ecol. 55:274-286. [DOI] [PubMed] [Google Scholar]
- 34.Lovley, D. R., E. E. Roden, E. J. P. Phillips, and J. C. Woodward. 1993. Enzymatic iron and uranium reduction by sulfate-reducing bacteria. Mar. Geol. 113:41-53. [Google Scholar]
- 35.Lytle, C. A. G., Y. D. Gan, K. Salone, and D. C. White. 2001. Sensitive charachterization of microbial ubiquinones from biofilms by electrospray/mass spectrometry. Environ. Microbiol. 3:265-272. [DOI] [PubMed] [Google Scholar]
- 36.Macalady, J. L., E. H. Lyon, B. Koffman, L. K. Albertson, K. Meyer, S. Galdenzi, and S. Mariani. 2006. Dominant microbial populations in limestone-corroding stream biofilms, Frasassi cave system, Italy. Appl. Environ. Microbiol. 72:5596-5609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Matsunaga, T., and N. Tsujimura. 1993. Respiratory inhibitors of a magnetic bacterium Magnetospirillum sp. AMB-1 capable of growing aerobically. Appl. Microbiol. Biotechnol. 39:368-371. [Google Scholar]
- 38.Mayberry, W. R., and J. R. Lane. 1993. Sequential alkaline saponification/acid hydrolysis/esterification: a one-tube method with enhanced recovery of both cyclopropane and hydroxylated fatty acids. J. Microbiol. Methods 18:21-32. [Google Scholar]
- 39.McCullough, J., T. C. Hazen, and S. M. Benson. 1999. Bioremediation of metals and radionuclides. What it is and how it works. Lawrence Berkeley National Laboratory, Berkeley, CA.
- 40.McCune, B., and J. B. Grace. 2002. Analysis of ecological communities. John Wiley & Sons, Toronto, Canada.
- 41.McCune, B., and M. Medford. 1999. PC-ORD multivariate analysis of ecological data, version 5.19β. MjM Software Design, Gleneden Beach, OR.
- 42.Michalsen, M. M., B. A. Goodman, S. D. Kelly, K. M. Kemner, J. P. McKinley, J. W. Stucki, and J. D. Istok. 2006. Uranium and technetium bio-immobilization in intermediate-scale physical models of an in situ bio-barrier. Environ. Sci. Technol. 40:7048-7053. [DOI] [PubMed] [Google Scholar]
- 43.Mikell, A. T., T. J. Phelps, and D. C. White. 1986. Phospholipids to monitor microbial ecology in anaerobic digesters, p. 413-444. In W. H. S. and J. R. Frank (ed.), Methane from biomass, a systems approach. Elsevier Publishing, New York, NY.
- 44.Moore, L. V. H. B., D. M. Bourne, and W. E. C. Moore. 1994. Comparative distribution and taxonomic value of cellular fatty acids in thirty-three genera of anaerobic gram-negative bacteria. Int. J. Syst. Bacteriol. 44:338-347. [DOI] [PubMed] [Google Scholar]
- 45.Muyzer, G., E. C. de Waal, and A. G. Uitterlinden. 1993. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 59:695-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.North, N. N., S. L. Dollhopf, L. Petrie, J. D. Istok, D. L. Balkwill, and J. E. Kostka. 2004. Change in bacterial community structure during in situ biostimulation of subsurface sediment cocontaminated with uranium and nitrate. Appl. Environ. Microbiol. 70:4911-4920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nyman, J. L., T. L. Marsh, M. A. Ginder-Vogel, M. Gentile, S. Fendorf, and C. Criddle. 2006. Heterogeneous response to biostimulation for U(VI) reduction in replicated sediment microcosms. Biodegradation 17:303-316. [DOI] [PubMed] [Google Scholar]
- 48.O'Leary, W. M., and S. G. Wilkinson. 1988. Gram-positive bacteria, p. 117-185. In C. Ratledge and S. G. Wilkinson (ed.), Microbial lipids, vol. 1. Academic Press, San Diego, CA. [Google Scholar]
- 49.Parkes, R. J., and J. Taylor. 1983. The relationship between fatty acid distribution and bacterial respiratory types in contemporary marine sediments. Estuar. Coast. Shelf Sci. 16:173-189. [Google Scholar]
- 50.Petrie, L., N. N. North, S. L. Dollhopf, D. L. Balkwill, and J. E. Kostka. 2003. Enumeration and characterization of iron(III)-reducing microbial communities from acidic subsurface sediments contaminated with uranium(VI). Appl. Environ. Microbiol. 69:7467-7479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pinkart, H. C., D. B. Ringelberg, Y. M. Piceno, S. J. MacNaughton, and D. C. White. 2000. Biochemical approaches to biomass measurements and community structure analysis, p. 101-113. In C. J. Hurst, R. L. Crawford, G. R. Knudsen, M. J. McInerny, and L. D. Stetzenbach (ed.), Manual of environmental microbiology, 2nd ed. ASM Press, Washington, DC.
- 52.Rilfors, L., A. Wieslander, and S. Stahl. 1978. Lipid and protein composition of membranes of Bacillus megaterium variants in the temperature range 5 to 70 degrees C. J. Bacteriol. 135:1043-1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shelobolina, E. S., K. O'Neill, K. T. Finneran, L. A. Hayes, and D. R. Lovley. 2003. Potential for in situ bioremediation of a low-pH, high-nitrate uranium-contaminated groundwater. Soil Sediment Contam. 12:865-884. [Google Scholar]
- 54.Short, K. A., and R. P. Blakemore. 1986. Iron respiration-driven proton translocation in aerobic bacteria. J. Bacteriol. 167:729-731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54a.Spain, A. M., A. D. Peacock, J. D. Istok, M. S. Elshahed, F. Z. Najar, B. A. Roe, D. C. White, and L. R. Krumholz. 2007. Identification and isolation of a Castellaniella species important during biostimulation of an acidic nitrate- and uranium-contaminated aquifer. Appl. Environ. Microbiol. 73:4892-4904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stewart, D. K., and W. A. Love. 1968. A general canonical correlation analysis index. Psychol. Bull. 70:160-163. [DOI] [PubMed] [Google Scholar]
- 56.Stults, J. R., O. Snoeyenbos-West, B. Methe, D. R. Lovely, and D. P. Chandler. 2001. Application of the 5′ fluorogenic exonuclease assay (TaqMan) for quantitative ribosomal DNA and rRNA analysis in sediments. Appl. Environ. Microbiol. 67:2781-2789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Suzuki, M. T., L. T. Taylor, and E. F. DeLong. 2000. Quantitative analysis of small-subunit rRNA genes in mixed microbial populations via 5′-nuclease assays. Appl. Environ. Microbiol. 66:4605-4614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Suzuki, Y., S. D. Kelly, K. M. Kemner, and J. F. Banfield. 2005. Direct microbial reduction and subsequent preservation of uranium in natural near-surface sediment. Appl. Environ. Microbiol. 71:1790-1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Swofford, D. L. 2000. PAUP*: phylogenetic analysis using parsimony (*and other methods), 4.0b10 ed. Sinauer Associates, Sunderland, MA.
- 60.ter Braak, C. J. F. 1996. Unimodal models to relate species to environment. Agricultural Mathematics Group, Wageningen, The Netherlands.
- 61.ter Braak, C. J. F., and P. Simlauer. 2002. CANOCO reference manual and CanoDraw for Windows user's guide. Microcomputer Power, Ithaca, NY.
- 62.ter Braak, C. J. F., and P. Simlauer. 2004. Canoco for Windows version 4.53. Plant Research International, Wageningen, The Netherlands.
- 63.Thomas, T. D., and R. D. Batt. 1969. Degradation of cell constituents by starved Streptococcus lactis in relation to survival. J. Gen. Microbiol. 58:347-362. [DOI] [PubMed] [Google Scholar]
- 64.Thompson, J. D., T. J. Gibson, F. Pleioniak, F. Jeanmougin, and D. G. Higgings. 1997. The ClustalX interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tokunaga, T. K., J. M. Wan, M. K. Firestone, T. C. Hazen, K. R. Olson, D. J. Herman, S. R. Sutton, and A. Lanzirotti. 2003. In situ reduction of chromium(VI) in heavily contaminated soils through organic carbon amendment. J. Environ Qual. 32:1641-1649. [DOI] [PubMed] [Google Scholar]
- 66.Tsien, H. C., B. J. Bratina, K. Tsuji, and R. S. Hanson. 1990. Use of oligodeoxynucleotide signature probes for identification of physiological groups of methylotrophic bacteria Appl. Environ. Microbiol. 56:2858-2865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.van den Wollenberg, A. L. 1977. Redundancy analysis an alternative for canonical correlation analysis. Psychometrika 42:207-219. [Google Scholar]
- 68.Vrionis, H. A., R. T. Anderson, I. Ortiz-Bernad, K. R. O'Neill, C. T. Resch, A. D. Peacock, R. Dayvault, D. C. White, P. E. Long, and D. R. Lovley. 2005. Microbiological and geochemical heterogeneity in an in situ uranium bioremediation field site. Appl. Environ. Microbiol. 71:6308-6318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wan, J. M., T. K. Tokunaga, E. Brodie, Z. M. Wang, Z. P. Zheng, D. Herman, T. C. Hazen, M. K. Firestone, and S. R. Sutton. 2005. Reoxidation of bioreduced uranium under reducing conditions. Environ. Sci. Technol. 39:6162-6169. [DOI] [PubMed] [Google Scholar]
- 70.Weerkamp, A., and W. Heinen. 1972. Effect of temperature on the fatty acid composition of the extreme thermophiles Bacillus caldolyticus and Bacillus caldotenax. J. Bacteriol. 109:443-446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.White, D. C., R. Geyer, A. D. Peacock, D. B. Hendrick, S. S. Koenigsberg, Y. Sung, J. He, and F. E. Loffler. 2005. Phospholipid furan fatty acids and ubiquinone-8: lipid biomarkers that may protect Dehalococcoides strains from free radicals. Appl. Environ. Microbiol. 71:8426-8433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.White, D. C., and D. B. Ringelberg. 1998. Signature lipid biomarker analysis, p. 255-272. In R. A. R. S. Burlage, D. Stahl, G. Geesey, and G. Sayler (ed.), Techniques in microbial ecology. Oxford University Press, New York, NY.
- 73.Wilkinson, S. G. 1988. Gram-negative bacteria, p. 299-488. In C. Ratledge and S. G. Wilkinson (ed.), Microbial lipids, vol. 1. Academic Press, San Diego, CA. [Google Scholar]
- 74.Yoshida, N., N. Takahashi, and A. Hiraishi. 2005. Phylogenetic characterization of a polychlorinated-dioxin-dechlorinating microbial community by use of microcosm studies. Appl. Environ. Microbiol. 71:4325-4334. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.