Abstract
YjgB is one of five peptidoglycan hydrolases previously identified in Lactococcus lactis. Analysis of its amino acid sequence revealed that YjgB contains an NlpC/P60 domain, whereas no specific cell wall binding domain or motif could be identified. The NlpC/P60 family is characterized by three conserved residues, a cysteine, a histidine, and a polar residue. In agreement with the presence of a Cys residue in the catalytic site of YjgB, its enzymatic activity was enhanced in the presence of dithiothreitol. Peptidoglycan-hydrolyzing activity of YjgB was detected in growing cells of an L. lactis strain overexpressing YjgB, as revealed by the presence of disaccharide (DS)-dipeptide in the muropeptide composition of the overexpressing strain. YjgB hydrolyzes the peptide chains of L. lactis muropeptides between γ-d-Gln and l-Lys residues. Its hydrolytic activity was detected on DSs with tetra- and pentapeptide chains, whereas hydrolytic activity was very low on DS-tripeptides. Thus, we demonstrated that YjgB is an endopeptidase which cleaves γ-d-Gln-l-Lys bonds in peptide chains of L. lactis peptidoglycan.
The cell integrity of gram-positive bacteria is ensured by a cell wall which consists mainly of peptidoglycan, an amino-sugar polymer cross-linked with peptide bridges. Peptidoglycan metabolism is a necessary process in cell wall remodeling during growth and cell division, during which peptidoglycan hydrolysis is balanced with the synthesis of new polymer blocks (9, 31). While cell integrity is in this case maintained, it is compromised during cellular autolysis. Both processes require the action of enzymes called peptidoglycan hydrolases (PGHs), which catalyze cleavage of peptidoglycan sugar or peptide chains. In lactic acid bacteria such as Lactococcus lactis, which is used as a dairy starter, PGH activity during cheese ripening leads to cellular autolysis and release of the cellular content to the cheese curd. This provides the pool of enzymes acting in the development of organoleptic properties of cheese (22). Therefore, control of PGH activity in L. lactis bears the potential for control of cheese ripening in industrial conditions.
The PGH complement of L. lactis was analyzed previously and comprises five PGHs (16). It includes AcmA, AcmB, and AcmC, all of which exhibit N-acetylglucosaminidase hydrolytic specificity (16, 17, 32). AcmA is the major autolysin of L. lactis; it is involved in cell separation after cell division and in cellular autolysis when cells reach the stationary phase (6). The N-terminal part of AcmA contains the catalytic domain, while the C-terminal part comprises three LysM domains involved in cell wall binding (33). AcmB, the second PGH of L. lactis, is built of three domains: an S/T/P/N-rich domain at its N terminus followed by the catalytic domain and a C-terminal part of unknown function. AcmB contributes to cellular autolysis to a lesser extent than AcmA and does not participate in cell separation (17). The third L. lactis PGH, AcmC, does not contain any specific cell wall binding domain and contains only a catalytic domain, homologous to the corresponding domains of AcmA and AcmB. It demonstrates N-acetylglucosaminidase specificity, as tested with Bacillus subtilis peptidoglycan (16). The specificity of the fourth PGH of L. lactis, AcmD, has not yet been determined, but sequence similarity data suggest that it is an N-acetylglucosaminidase. Its peptidoglycan-hydrolyzing activity was confirmed in zymogram assays (16). The fifth PGH of L. lactis, YjgB, shows sequence similarity with the active site domain of peptidoglycan-specific endopeptidases, such as LytE and LytF from B. subtilis (18, 23, 24) or the γ-d-Glu-(l)-meso-diaminopimelic acid-endopeptidase from Bacillus sphaericus (15). Its peptidoglycan-hydrolyzing activity was previously detected by zymogram assay under native conditions, in a gel without sodium dodecyl sulfate (SDS), but its hydrolytic specificity was not determined (16). Also, its role in cellular autolysis was not evaluated previously.
In the present report, we show that YjgB hydrolyzes the peptide bond between γ-d-Gln and l-Lys in peptide chains of L. lactis peptidoglycan. On the basis of sequence similarity, it belongs to the NlpC/P60 superfamily of enzymes (1), which contains three conserved residues, a cysteine, a histidine, and a polar residue, that are involved in catalysis.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
Bacterial strains and plasmids used in this study are listed in Table 1. Escherichia coli strains were grown at 37°C in Luria-Bertani medium with shaking. L. lactis strains were grown in M17 medium (Difco Laboratories) supplemented with 0.5% (wt/vol) glucose at 30°C. Growth was monitored by optical density (OD) measurement at 600 nm (OD600) with a spectrophotometer (Uvicon 931; Biotek Kontron). The following antibiotics were added as selective agents when appropriate: erythromycin (5 μg ml−1 for L. lactis, 150 μg ml−1 for E. coli), chloramphenicol (5 μg ml−1 for L. lactis), ampicillin (100 μg ml−1 for E. coli), and kanamycin (25 μg ml−1 for E. coli).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant genotype and/or phenotype | Source or reference |
|---|---|---|
| Strains | ||
| Escherichia coli | ||
| TG1 | supE hsdΔ5 thi Δ(lacproAB) F′ [traD36proAB+lacIqlacZΔM15] | |
| TG1repA+ | TG1 derivative with repA gene integrated into the chromosome, allowing replication of L. lactis plasmids | P Renault, INRA, Jouy-en-Josas, France |
| M15(pREP4) | Derivative of E. coli K-12 containing pREP4 plasmid, ensuring the production of high levels of lac repressor protein; Kanr | QIAGEN |
| Lactococcus lactis | ||
| MG1363 | Plasmid free | 12 |
| MG1363acmAΔ1 | MG1363 derivative with a 701-bp deletion in acmA | 6 |
| MG1363acmB | MG1363 derivative with a 414-bp deletion in acmB and a Cmr cassette inserted; Cmr | 17 |
| MG1363acmAΔ1acmB | MG1363 derivative with deletions in both acmA and acmB; Cmr | 17 |
| MG1363yjgB | MG1363 derivative with a 333-bp deletion in yjgB | This study |
| TIL758 | MG1363 derivative, triple mutant acmA acmB yjgB; Cmr | This study |
| NZ9000 | MG1363, pepN::nisRK | 20 |
| htrA-NZ9000 | NZ9000 with htrA disrupted by single crossing-over recombination; Emr | 7 |
| TIL903 | htrA-NZ9000 containing pNZ8048 without insert | This study |
| TIL904 | htrA-NZ9000 containing pNZ8048::yjgB | This study |
| Plasmids | ||
| pQE60 | Expression vector for C-terminal six-His-tag fusion; Apr | QIAGEN |
| pG+host9 | Thermosensitive plasmid for gene replacement by double crossing-over integration; Eryr | 5 |
| pTIL752 | pG+host9 carrying a 1,602-bp fragment with a 333-bp deletion inside yjgB; Eryr | This study |
| pNZ8048 | Carries nisin-inducible nisA promoter for translational fusion; Cmr | 10 |
General recombinant DNA techniques.
Molecular cloning techniques were performed using standard procedures (30). Restriction enzymes (Eurogentec or Roche Molecular Biochemicals), T4 DNA ligase (Epicenter), Taq DNA polymerase (Appligene Oncor), and calf intestinal alkaline phosphatase (New England Biolabs) were used according to the suppliers' recommendations. The oligonucleotides were purchased from Invitrogen. L. lactis DNA was isolated as described previously (27). PCR amplifications were carried out using GeneAmp PCR system 2400 (Perkin Elmer). pGEM-T Easy vector (Promega Corporation) was used to clone PCR products in E. coli TG1. Electroporation of L. lactis was performed as described previously (14), and transformants were plated on M17 glucose agar plates containing the required antibiotic. DNA sequences were determined using an Applied Biosystems 370A automated DNA sequencer and ABI PRISM Dye Terminator cycle sequencing and Dye Primer cycle sequencing kits (Perkin Elmer). DNA and protein sequences were assembled and analyzed with the Genetic Computer Group sequence analysis package (GCG Inc., Madison, WI) and the tools available on the Expasy (Expert Protein Analysis System) Biology Server of the Swiss Institute of Bioinformatics (http://www.expasy.org).
Purification of His-tagged protein from E. coli.
Six-His-tagged YjgB devoid of signal sequence (YjgB-His) was previously expressed in E. coli M15 (pREP4) with pQE60 vector (16). YjgB-His was purified from a 250-ml culture after preparation of soluble cell extract by use of Ni2+ nitrilotriacetic acid spin columns under native conditions as described previously (16). Fractions containing YjgB-His were combined. They were concentrated and dialyzed against 25 mM sodium citrate-50 mM sodium phosphate (pH 4.0) by ultrafiltration with a Centricon YM-10 concentrator.
SDS-PAGE and renaturing SDS-PAGE.
SDS-polyacrylamide gel electrophoresis (SDS-PAGE) was performed with 15% (wt/vol) polyacrylamide separating gel, and protein bands were stained with Coomassie blue (Biosafe; Bio-Rad). Renaturing SDS-PAGE was performed as described previously (21) with 0.2% (wt/vol) autoclaved Micrococcus luteus ATCC 4698 (Sigma) included in the polyacrylamide gel as an enzyme substrate. Gels were washed for 1 h in deionized water at room temperature and then incubated overnight at 37°C with shaking in 25 mM sodium citrate-50 mM sodium phosphate buffer (pH 4.0) containing 1% (vol/vol) Triton X-100 and 1 mM dithiothreitol (DTT). Gels were subsequently washed for 1 h in deionized water, stained for 2 h at room temperature with 0.1% methylene blue in 0.01% (wt/vol) KOH, and destained in deionized water. Gel images were generated with a Duoscan T1200 scanner (Agfa-Gevaert) customized for proper gel handling.
Inactivation of yjgB by double crossing-over integration.
A 1,935-bp fragment encompassing the yjgB gene was amplified by PCR with the two primers AU71 (5′-GGCACTTAAGATGCACGG-3′) and AU72 (5′-ATCTGCCAAGGTTTTATAAGTT-3′) with L. lactis MG1363 DNA as the template. The PCR product was cloned into pGEM-T Easy vector, resulting in plasmid pTIL350. Deletion of a 333-bp internal fragment from the yjgB gene (nucleotides 8 to 340 of the yjgB gene) was obtained by reverse PCR using the pTIL350 plasmid with primers AU73 (5′-CGGGGTACCTCTATAGAGAAGTTTTAGGTA-3′; KpnI site underlined) and AU74 (5′-CGGGGTACCTCAACATTGTTAGCAAGTATA-3′; KpnI site underlined). The resulting PCR-amplified fragment was digested with KpnI enzyme and religated. The 1,602-bp insert with an internal deletion was recovered with PstI and SacII digestion and was then ligated in the corresponding sites of plasmid pG+host9 after dephosphorylation. The ligation mixture was used to transform competent cells of MG1363, the MG1363acmB mutant, and the MG1363acmAΔ1acmB double mutant by electroporation. Clones containing recombinant pG+host9 with the insert (pTIL752) were selected. Integration of plasmid pTIL752 into the chromosome and subsequent excision were performed according to a previously developed protocol (5). Mutant strains were screened by PCR for the size of the PCR-amplified fragment with primers AU71 and AU72. The presence of a unique correct insertion event was further verified by Southern blotting.
Expression of yjgB under the control of a nisin-inducible promoter.
The yjgB gene was cloned under the control of a nisin-inducible promoter (19) in pNZ8048 vector (obtained from NIZO, The Netherlands) (10). yjgB was amplified by PCR using MG1363 template DNA and primers AU81 (5′-CATGCCATGGTGAAAAAAATAATTATTTCC; NcoI restriction site underlined) and AU82 (5′-AACTGCAGTTAGTATTCTAAGGCAAAATC; PstI site underlined). The PCR product was purified and cloned into pGEM-T Easy vector. The insert was excised with NcoI and PstI digestion and ligated with pNZ8048 restricted with the same enzymes. The ligation mixture was used to transform L. lactis strain htrA-NZ9000 (7) by electroporation. The resulting strain was named TIL904. A control strain (TIL903) was obtained by transformation of htrA-NZ9000 with empty pNZ8048 vector.
Western blotting of cellular fractions.
L. lactis TIL904 and TIL903 were grown in M17 medium at 30°C to an OD600 of 0.4, and nisin was added at a concentration of 2.5 ng ml−1. Growth was continued for 4 h, and cells were harvested by centrifugation. Culture supernatant was filtered through a 0.45-μm-pore-size filter and precipitated with 20% trichloroacetic acid prior to SDS-PAGE. Cell fractionation was performed as described previously (26). Briefly, the bacterial pellet was resuspended in Tris-EDTA buffer containing 25% sucrose, and 0.5 mg ml−1 lysozyme and 100 μg ml−1 mutanolysin (Sigma Chemicals) were added. The suspension was incubated for 1 h at 37°C. The protoplasts were pelleted by centrifugation at 14,000 rpm for 5 min at 4°C. The supernatant (cell wall extract) was subjected to 20% trichloroacetic acid precipitation prior to electrophoresis. The isolated cell fractions were then analyzed by SDS-PAGE and Western blotting.
Antibodies directed against YjgB were obtained by injection of purified YjgB-His recombinant protein into a rabbit, according to a standard protocol of immunization (Eurogentec). Samples of isolated cell fractions were separated by SDS-PAGE with a 15% polyacrylamide gel and transferred to a nitrocellulose membrane. The membrane was successively incubated with anti-YjgB antiserum and with horseradish peroxidase-conjugated protein G (Bio-Rad), and blots were revealed with an Opti-4CN kit (Bio-Rad) according to the manufacturer's instructions.
N-terminal sequencing.
N-terminal amino acid sequences were determined with an automatic protein sequencer (Applied Biosystems 494A Procise) after blotting of the protein onto a polyvinylidene difluoride membrane (Problott; Applied Biosystems).
Peptidoglycan structural analysis.
L. lactis strain TIL904 was grown in M17 glucose, and expression of yjgB was induced at an OD600 of 0.15 by the addition of 1 ng ml−1 nisin. Control strain TIL903 was treated the same way. Cells from both cultures were harvested at an OD600 of 1.5. Peptidoglycan was extracted as described previously (8) and digested with mutanolysin (2,500 U ml−1) for 16 h at 37°C with stirring. Muropeptides were reduced with sodium borohydride and separated by reverse-phase high-pressure liquid chromatography (RP-HPLC) using a Hypersyl ODS column (C18; ThermoHypersil-Keystone) (250 by 4.6 mm; 5 μm) at 50°C as described previously (8). Muropeptides were eluted for 5 min with 10 mM ammonium phosphate buffer (pH 5.6) (buffer A) and then with a 270-min methanol (0% to 20%) linear gradient in buffer A at a flow rate of 0.5 ml min−1. Sodium azide (180 μg liter−1 in buffer A) was added to avoid baseline drift at 202 nm. Muropeptide analysis by matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) mass spectrometry was performed with a Voyager DE STR mass spectrometer (Perseptive Biosystem, Framingham, MA) as described previously (8) using 1 μl of sample with an alpha-cyano-4-hydroxy-cinnamic acid matrix. Calibration was performed with an external mass-calibration standard containing bradykinin fragment (1-5), angiotensin I, neurotensin, and melittin.
Hydrolytic specificity of purified YjgB for L. lactis muropeptides.
Muropeptides from L. lactis MG1363 peptidoglycan digested with mutanolysin were separated by RP-HPLC after reduction as described above (8). Peptidoglycan monomers were collected, dried in a vacuum using a Speed-Vac concentrator, and resuspended in 100 μl of deionized water containing 2 mM DTT. A volume of 100 μl containing approximately 50 μg of purified and concentrated YjgB-His in sodium citrate-phosphate buffer (pH 4.0) was added to each muropeptide, and the mixture was incubated for 20 h at 37°C. After incubation, the samples were analyzed by RP-HPLC with a Hypersyl ODS column with the same gradient as described above. Disaccharide-dipeptide (DS-di) purified by RP-HPLC from an L. lactis TIL904 peptidoglycan mutanolysin digest and identified by MALDI-TOF mass spectrometry was used as a standard for identification of the YjgB digestion product. The percentage of digested muropeptide was calculated as the ratio of the DS-di peak area to the sum of the peak areas of nondigested muropeptide substrate and DS-di product.
RESULTS AND DISCUSSION
YjgB belongs to the NlpC/P60 superfamily of enzymes.
YjgB was initially identified in the L. lactis IL1403 genome by its sequence similarity to known peptidoglycan-specific endopeptidases (16). Further analysis of sequence conservation of the catalytic domain of YjgB revealed that it contains an NlpC/P60 domain (Pfam PF00877) (3). The NlpC/P60 enzyme superfamily contains several related but distinct catalytic activities such as peptidoglycan cleavage, acyl transfer, and amide hydrolysis (1). Proteins of this superfamily are characterized by two conserved motifs containing highly conserved cysteine and histidine amino acid residues which are essential for catalysis along with an intermediate motif containing a highly conserved Gly that is often followed by an Asp (1). Conserved Cys and His residues are present in the catalytic domain of L. lactis MG1363 YjgB at positions 107 and 155, respectively, together with a Gly-Asp pair at positions 145 and 146 (see the supplemental material). Several proteins with an NlpC/P60 domain are cysteine peptidases included in the MEROPS C40 peptidase family, such as dipeptidyl-peptidase VI from B. sphaericus which hydrolyzes γ-d-Glu-l-Lys bonds or LytE and LytF characterized as γ-d-glutamyl-meso-diaminopimelate muropeptidases.
Peptidoglycan-hydrolyzing activity of YjgB in zymogram assays.
When the peptidoglycan-hydrolyzing activity of recombinant six-His-tagged YjgB was previously examined by a conventional zymogram assay, with autoclaved M. luteus cells as a substrate, it was revealed in native PAGE but not in SDS-PAGE (16). Taking into account the homology of YjgB to the proteins of the NlpC/P60 family, Cys107 should be essential for the catalytic activity of YjgB. The catalytic mechanism of known enzymes of the NlpC/P60 superfamily presumably involves nucleophilic attack with the conserved Cys residue as a nucleophile, which implies that it has to be in a reduced state for activity to occur (1). Therefore, we tested the recombinant YjgB-His activity in a zymogram assay with renaturing buffer containing 1 mM DTT. The presence of a reducing agent allowed detection of YjgB activity even after denaturing SDS-PAGE (Fig. 1). This finding suggests the requirement of reduced Cys for YjgB peptidoglycan-hydrolyzing activity.
FIG. 1.
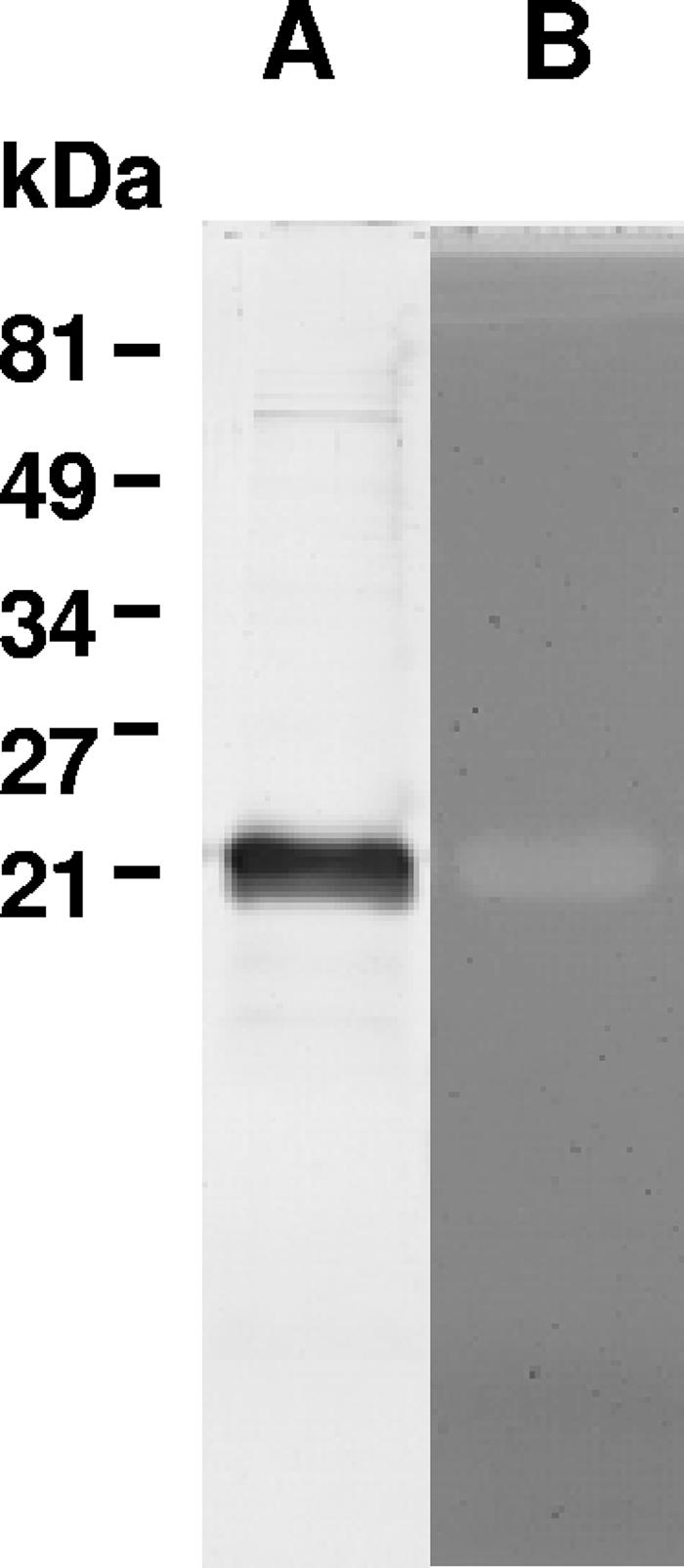
SDS-PAGE with protein bands stained with Coomassie blue (A) and zymogram assay (B) of purified recombinant YjgB-His. The zymogram assay was performed after SDS-PAGE in a 15% polyacrylamide gel containing M. luteus cells as a substrate. Renaturing buffer contained 1 mM DTT.
YjgB is dispensable for growth and is not involved in autolysis.
To investigate the in vivo function of YjgB, a series of yjgB mutants was created. The chromosomal copy of yjgB was replaced by yjgB internally disrupted by deletion of a 333-bp fragment as described in Material and Methods, using thermosensitive pG+host9 vector. yjgB gene was inactivated in the wild-type L. lactis MG1363 strain as well as in the acmB and acmAacmB mutants characterized previously (17). Phenotypes of the resulting MG1363yjgB, MG1363acmByjgB, and MG1363acmAacmByjgB mutants (Table 1) were analyzed by a number of custom assays used for characterization of peptidoglycan hydrolases (17). These included analysis of cellular growth rate, measurement of cell autolysis in the stationary phase both by decrease in optical density and by activity of concomitantly released intracellular peptidase PepX, and analysis of membrane permeabilization in the stationary phase with the fluorescent DNA marker propidium iodide (25). The assays demonstrated no visible phenotypic change associated with yjgB inactivation in either of the created mutants compared to the wild type (data not shown). Finally, the peptidoglycan structure of the triple mutant was investigated by RP-HPLC analysis of its muropeptide composition but no difference was found between the triple mutant and wild-type MG1363 muropeptide profiles (data not shown).
All these results suggest that either YjgB plays a secondary role in the peptidoglycan hydrolysis of L. lactis or YjgB activity is regulated so that it plays a more significant role in peptidoglycan hydrolysis in response to environmental signals. It is also conceivable that an undetected redundant peptidoglycan-specific peptidase is present in L. lactis. It is worth noting that another putative protein (llmg_0506) with an NlpC/P60 domain is present in the MG1363 complete genome sequence published recently (34), which could constitute a sixth putative PGH in L. lactis. In addition, the NlpC/P60 domain (Pfam PF00877) is related to the CHAP (cysteine, histidine-dependent amidohydrolases/peptidases) domain (Pfam PF05257). The NlpC/P60 enzyme family defined by Anantharaman and Aravind (1) was initially considered to be identical to the CHAP superfamily (4, 29), with similar conserved residues, Cys and His. The CHAP family also includes peptidoglycan hydrolases. In L. lactis MG1363, four (putative) proteins contain a CHAP domain: autolysin AcmB at its C terminus and major secreted protein Usp45 of unknown function as well as two other putative proteins, llmg_0904 and llmg_1890. We cannot exclude the possibility that the CHAP domain present in one of these proteins has peptidoglycan hydrolase activity.
YjgB is secreted into culture supernatant.
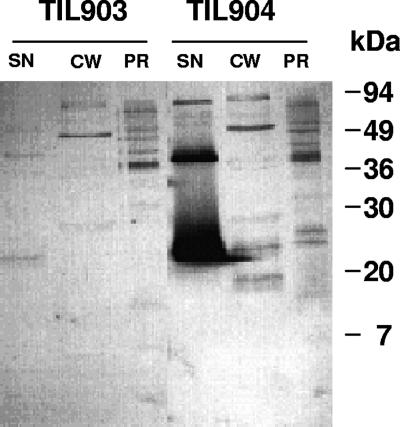
In order to evaluate the impact of increased expression of yjgB in L. lactis, the gene was expressed under the control of nisin-inducible promoter (10). Strain TIL904 bearing plasmid pNZ8048 with a yjgB insertion was analyzed for YjgB production following nisin induction and was compared to the control TIL903 strain bearing pNZ8048 without an insert. Isolated cell wall and protoplast fractions were prepared according to a previously described procedure (26) and analyzed along with the culture supernatant by Western blotting using specific anti-YjgB antibodies. This analysis revealed that, in strain TIL904, YjgB is exported to the extracellular environment, as the bulk of the protein was found in the culture supernatant whereas only minor amounts of YjgB could be detected in cell wall and protoplast fractions (Fig. 2). In the control TIL903 strain bearing plasmid pNZ8048 without an insert, a faint protein band was recognized by anti-YjgB antibodies in culture supernatant but no bands of the expected molecular mass could be detected in cell wall or protoplast fractions (Fig. 2).
FIG. 2.
Western blotting of L. lactis cellular fractions revealed with anti-YjgB antibodies. TIL903, control strain; TIL904, strain overexpressing yjgB under the control of a nisin-inducible promoter. Nisin was added at a concentration of 2.5 ng ml−1 and an OD600 of 0.4 and growth was continued for 4 h before cells were harvested for fractionation. SN, culture supernatant; CW, cell wall fraction; PR, protoplast fraction.
YjgB export to the extracellular environment is predicted by the presence of a putative N-terminal 26-amino-acid signal peptide in the MG1363 YjgB sequence (EMBL/GenBank accession number AJ414527) (16). The calculated molecular mass of the exported protein devoid of signal peptide is 17.6 kDa, while the apparent molecular mass of the immunodetected protein was 21 kDa, which corresponds to the molecular mass of an intact YjgB polypeptide. Therefore, to find out whether the predicted signal peptide was removed from YjgB upon secretion, we performed N-terminal protein sequencing of the YjgB present in the culture supernatant. The determined N-terminal sequence (DSVFEDN) corresponded to the YjgB protein devoid of signal peptide; thus, the apparent molecular mass of 21 kDa was most probably due to a protein migration artifact. Alternatively, the possibility of posttranslational protein modification cannot be excluded.
We compared growth rate and autolysis in the stationary phase of strain TIL904 overexpressing YjgB and of control strain TIL903 after nisin induction. No significant differences were observed between the two strains (data not shown).
Effect of YjgB overexpression in L. lactis on peptidoglycan structure.
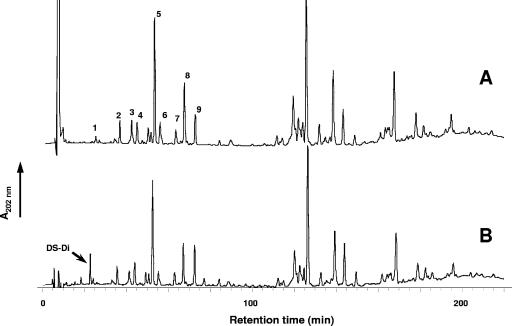
We then examined whether the increased amount of YjgB synthesized in TIL904 cells has an impact on peptidoglycan structure by analyzing the constituent muropeptides of TIL904 after nisin induction. The assay was performed using the HPLC-based method developed for L. lactis peptidoglycan structural analysis (8). Overexpression of YjgB in TIL904 led to the appearance of a new product in the muropeptide profile, eluted from the column with the retention time of 23 min, compared to the muropeptide profile of the control strain with empty plasmid (TIL903) (Fig. 3). Mass spectrometry analysis of this new product revealed that it corresponds to DS-di, with amidation at a γ-d-Glu residue ([M + Na]+ of 720.29). Accumulation of DS-di concomitant with YjgB overexpression strongly suggests that YjgB hydrolyzes peptide bonds between γ-d-Gln and l-Lys in peptidoglycan peptide chains (Fig. 4). These results demonstrate the presence of YjgB activity during bacterial growth and confirm its putative specificity as predicted on the basis of sequence similarity. The absence of DS-di muropeptide in the wild-type peptidoglycan was most probably a result of YjgB activity that was too low in the wild-type strain.
FIG. 3.
RP-HPLC separation profile of muropeptides from L. lactis TIL903 (control strain) (A) and TIL904 (strain overexpressing yjgB) (B) induced with nisin. Peak numbers refer to Table 2. DS-di, disaccharide dipeptide (GlcNAc-MurNAc-l-Ala-γ-d-Gln).
FIG. 4.
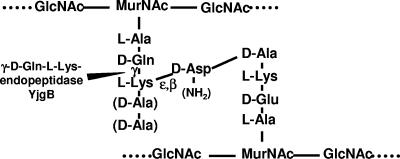
Schematic structure of L. lactis peptidoglycan and site of cleavage of YjgB. GlcNAc, N-acetylglucosamine; MurNAc, N-acetylmuramic acid. d-Ala residues at positions 4 and 5 of the acceptor peptide chains may be present or not, and d-Asp may be amidated or not, according to previous L. lactis peptidoglycan structural analysis (8).
Contrary to the results seen with numerous characterized peptidoglycan hydrolases, YjgB lacks a specific cell wall binding domain. No LysM domain (2) or other conserved motifs found in cell wall-associated proteins (11) are present in YjgB. In agreement with this, we detected YjgB in culture supernatant. Nevertheless, when YjgB is overexpressed, it cleaves bonds inside the cell wall peptidoglycan of growing bacteria. Taken together, our results suggest either that YjgB binds transiently to the cell wall from the outside of the cell or that a small amount of enzyme is trapped inside the cell wall upon secretion.
Determination of YjgB hydrolytic specificity with respect to L. lactis muropeptides.
To validate the finding that YjgB specifically cleaves the γ-d-Gln-l-Lys peptide bond in L. lactis peptidoglycan and to further investigate the enzyme specificity, we assayed YjgB activity on purified muropeptides as substrates. Nine individual muropeptides were isolated from L. lactis peptidoglycan digested with mutanolysin, which is a muramidase. They are numbered from 1 to 9 in the muropeptide HPLC separation profile presented in Fig. 3. These muropeptides were identified previously as disaccharide tri-, tetra-, and pentapeptides with variations in their peptide moieties (8) (Table 2). They contain reduced MurNAc, since reduction was performed before HPLC separation to avoid the presence of several anomeric forms for each muropeptide (13). Purified recombinant YjgB was incubated with each muropeptide, and the reaction products were analyzed after separation by RP-HPLC. The relative amounts of DS-di produced from the various muropeptide substrates demonstrate that YjgB-His exhibits hydrolytic activity with respect to disaccharide tetra- and pentapeptides, whereas its activity with respect to disaccharide-tripeptides is very low (Table 2). YjgB activity was not compromised by the presence of side chain d-Asp or d-Asn bound to l-Lys. Thus, we have demonstrated experimentally that YjgB is an endopeptidase which specifically hydrolyzes the γ-d-Gln-l-Lys peptide bond of peptidoglycan peptide chains. All natural muropeptides used as substrates in this study contained α-amidated γ-d-Glu residue (8), and we did not assay YjgB activity with respect to muropeptides containing a γ-d-Glu without amidation at the same position. Therefore, we term YjgB a γ-d-glutaminyl-l-lysyl-endopeptidase instead of the γ-d-glutamyl-l-lysyl-endopeptidase usually found in the literature.
TABLE 2.
Substrate specificity of purified YjgB-His with respect to purified reduced muropeptides
| Peaka | Muropeptideb | % Digested muropeptidec |
|---|---|---|
| 1 | DS-Tri | 5.4 |
| 2 | DS-Tetra | 70.5 |
| 3 | DS-Tri-D | 2.1 |
| 4 | DS-Tri*-N | 1.7 |
| 5 | DS-Tri-N | 3.3 |
| 6 | DS-Tetra-D | 84.2 |
| 7 | DS-Penta-D | 98.3 |
| 8 | DS-Tetra-N | 92.2 |
| 9 | DS-Penta-N | 92.5 |
Peak numbers refer to chromatogram shown in Fig. 3.
DS-Tri, disaccharide tripeptide (l-Ala-γ-D-Gln-l-Lys); DS-Tri*, disaccharide tripeptide (l-Ala-γ-d-Glu-l-Lys); DS-Tetra, disaccharide tetrapeptide (l-Ala-γ-d-Gln-l-Lys-d-Ala); DS-Penta, disaccharide pentapeptide disaccharide (l-Ala-γ-d-Gln-l-Lys-d-Ala-d-Ala); disaccharide, GlcNAc-MurNAc (GlcNAc, N-acetylglucosamine, MurNAc, N-acetylmuramic acid); D, d-Asp; N, d-Asn.
The percentage of digested muropeptide was calculated as the ratio of the DS-di peak area to the sum of the peak areas of the nondigested muropeptide substrate and the DS-di product.
Concluding remarks.
In this study, we investigated the hydrolytic specificity of L. lactis YjgB peptidoglycan hydrolase and we showed that YjgB is a γ-d-glutaminyl-l-lysyl endopeptidase. YjgB activity was demonstrated both in vivo for the cell wall peptidoglycan of the growing L. lactis strain overexpressing YjgB and in vitro for purified muropeptides. Disaccharide-tetra- and pentapeptides were hydrolyzed with efficiency that was significantly higher than that seen with disaccharide-tripeptides, which were barely hydrolyzed. Besides YjgB, the L. lactis PGH complement comprises three other characterized enzymes, AcmA, AcmB, and AcmC, which exhibit N-acetyl-glucosaminidase specificity, and a fourth putative glucosaminidase, AcmD (16). This makes YjgB the only identified peptidoglycan hydrolase of L. lactis capable of hydrolyzing the peptide moiety of peptidoglycan in L. lactis.
Based on the confirmed specificity of YjgB, we propose to rename the protein PgpA (for peptidoglycan peptidase A). This enzyme belongs to the C40 family of cysteine peptidases of the MEROPS peptidase database (http://merops.sanger.ac.uk) (28) and has been assigned MEROPS identification number C40.006.
The absence of a phenotype in yjgB mutants could result from undetected redundant peptidase activity specific for peptidoglycan. Indeed, several other putative proteins containing NlpC/P60 domains (Pfam PF0087) or a CHAP domain (Pfam PF05257) are encoded in the complete MG1363 genome sequence. Since both families include enzymes with peptidoglycan hydrolase activity, the PGH complement of L. lactis could comprise more than five members. Further evaluation of the peptidoglycan hydrolase activity of these putative candidates is required.
Supplementary Material
Acknowledgments
We are very grateful to G. Buist for the gift of MG1363acmA mutant, to J.-C. Huet for N-terminal protein sequencing, and to F. Wessner for helpful technical advice.
Y.R. was the recipient of an INRA fellowship. The work was carried out with the financial support of the Commission of the European Communities (FAIR contract CT98-4396, LAB-lysis).
Footnotes
Published ahead of print on 20 July 2007.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Anantharaman, V., and L. Aravind. 2003. Evolutionary history, structural features and biochemical diversity of the NlpC/P60 superfamily of enzymes. Genome Biol. 4:R11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bateman, A., and M. Bycroft. 2000. The structure of a LysM domain from E. coli membrane-bound lytic murein transglycosylase D (MltD). J. Mol. Biol. 299:1113-1119. [DOI] [PubMed] [Google Scholar]
- 3.Bateman, A., L. Coin, R. Durbin, R. D. Finn, V. Hollich, S. Griffiths-Jones, A. Khanna, M. Marshall, S. Moxon, E. L. Sonnhammer, D. J. Studholme, C. Yeats, and S. R. Eddy. 2004. The Pfam protein families database. Nucleic Acids Res. 32:D138-D141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bateman, A., and N. D. Rawlings. 2003. The CHAP domain: a large family of amidases including GSP amidase and peptidoglycan hydrolases. Trends Biochem. Sci. 28:234-237. [DOI] [PubMed] [Google Scholar]
- 5.Biswas, I., A. Gruss, S. D. Ehrlich, and E. Maguin. 1993. High-efficiency gene inactivation and replacement system for gram-positive bacteria. J. Bacteriol. 175:3628-3635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buist, G., J. Kok, K. J. Leenhouts, M. Dabrowska, G. Venema, and A. J. Haandrikman. 1995. Molecular cloning and nucleotide sequence of the gene encoding the major peptidoglycan hydrolase of Lactococcus lactis, a muramidase needed for cell separation. J. Bacteriol. 177:1554-1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cortes-Perez, N. G., I. Poquet, M. Oliveira, J. J. Gratadoux, S. M. Madsen, A. Miyoshi, G. Corthier, V. Azevedo, P. Langella, and L. G. Bermudez-Humaran. 2006. Construction and characterization of a Lactococcus lactis strain deficient in intracellular ClpP and extracellular HtrA proteases. Microbiology 152:2611-2618. [DOI] [PubMed] [Google Scholar]
- 8.Courtin, P., G. Miranda, A. Guillot, F. Wessner, C. Mezange, E. Domakova, S. Kulakauskas, and M. P. Chapot-Chartier. 2006. Peptidoglycan structure analysis of Lactococcus lactis reveals the presence of an l,d-carboxypeptidase involved in peptidoglycan maturation. J. Bacteriol. 188:5293-5298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Delcour, J., T. Ferain, M. Deghorain, E. Palumbo, and P. Hols. 1999. The biosynthesis and functionality of the cell-wall of lactic acid bacteria. Antonie Leeuwenhoek 76:159-184. [PubMed] [Google Scholar]
- 10.de Ruyter, P. G., O. P. Kuipers, and W. M. de Vos. 1996. Controlled gene expression systems for Lactococcus lactis with the food-grade inducer nisin. Appl. Environ. Microbiol. 62:3662-3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Desvaux, M., E. Dumas, I. Chafsey, and M. Hebraud. 2006. Protein cell surface display in Gram-positive bacteria: from single protein to macromolecular protein structure. FEMS Microbiol. Lett. 256:1-15. [DOI] [PubMed] [Google Scholar]
- 12.Gasson, M. J. 1983. Plasmid complements of Streptococcus lactis NCDO 712 and other lactic streptococci after protoplast-induced curing. J. Bacteriol. 154:1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Glauner, B. 1988. Separation and quantification of muropeptides with high-performance liquid chromatography. Anal. Biochem. 172:451-464. [DOI] [PubMed] [Google Scholar]
- 14.Holo, H., and I. F. Nes. 1995. Transformation of Lactococcus by electroporation. Methods Mol. Biol. 47:195-199. [DOI] [PubMed] [Google Scholar]
- 15.Hourdou, M. L., C. Duez, B. Joris, M. J. Vacheron, M. Guinand, G. Michel, and J. M. Ghuysen. 1992. Cloning and nucleotide sequence of the gene encoding the gamma-d-glutamyl-l-diamino acid endopeptidase II of Bacillus sphaericus. FEMS Microbiol. Lett. 70:165-170. [DOI] [PubMed] [Google Scholar]
- 16.Huard, C., G. Miranda, Y. Redko, F. Wessner, S. J. Foster, and M. P. Chapot-Chartier. 2004. Analysis of the peptidoglycan hydrolase complement of Lactococcus lactis: identification of a third N-acetylglucosaminidase, AcmC. Appl. Environ. Microbiol. 70:3493-3499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huard, C., G. Miranda, F. Wessner, A. Bolotin, J. Hansen, S. J. Foster, and M. P. Chapot-Chartier. 2003. Characterization of AcmB, an N-acetylglucosaminidase autolysin from Lactococcus lactis. Microbiology 149:695-705. [DOI] [PubMed] [Google Scholar]
- 18.Ishikawa, S., Y. Hara, R. Ohnishi, and J. Sekiguchi. 1998. Regulation of a new cell wall hydrolase gene, cwlF, which affects cell separation in Bacillus subtilis. J. Bacteriol. 180:2549-2555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kleerebezem, M., M. M. Beerthuyzen, E. E. Vaughan, W. M. de Vos, and O. P. Kuipers. 1997. Controlled gene expression systems for lactic acid bacteria: transferable nisin-inducible expression cassettes for Lactococcus, Leuconostoc, and Lactobacillus spp. Appl. Environ. Microbiol. 63:4581-4584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kuipers, O. P., P. G. de Ruyter, M. Kleerebezem, and W. M. de Vos. 1998. Quorum sensing-controlled gene expression in lactic acid bacteria. J. Biotechnol. 64:15-21. [Google Scholar]
- 21.Lepeuple, A.-S., E. Van Gemert, and M.-P. Chapot-Chartier. 1998. Analysis of the bacteriolytic enzymes of the autolytic Lactococcus lactis subsp. cremoris strain AM2 by renaturing polyacrylamide gel electrophoresis: identification of a prophage-encoded enzyme. Appl. Environ. Microbiol. 64:4142-4148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lortal, S., and M. P. Chapot-Chartier. 2005. Role, mechanisms and control of lactic acid bacteria lysis in cheese. Int. Dairy J. 15:857-871. [Google Scholar]
- 23.Margot, P., M. Pagni, and D. Karamata. 1999. Bacillus subtilis 168 gene lytF encodes a gamma-d-glutamate-meso-diaminopimelate muropeptidase expressed by the alternative vegetative sigma factor, sigmaD. Microbiology 145:57-65. [DOI] [PubMed] [Google Scholar]
- 24.Margot, P., M. Wahlen, A. Gholamhoseinian, P. Piggot, D. Karamata, and A. Gholamhuseinian. 1998. The lytE gene of Bacillus subtilis 168 encodes a cell wall hydrolase. J. Bacteriol. 180:749-752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Niven, G. W., and F. Mulholland. 1998. Cell membrane integrity and lysis in Lactococcus lactis: the detection of a population of permeable cells in post-logarithmic phase cultures. J. Appl. Microbiol. 84:90-96. [DOI] [PubMed] [Google Scholar]
- 26.Piard, J. C., I. Hautefort, V. A. Fischetti, S. D. Ehrlich, M. Fons, and A. Gruss. 1997. Cell wall anchoring of the Streptococcus pyogenes M6 protein in various lactic acid bacteria. J. Bacteriol. 179:3068-3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pospiech, A., and B. Neumann. 1995. A versatile quick-prep of genomic DNA from gram-positive bacteria. Trends Genet. 11:217-218. [DOI] [PubMed] [Google Scholar]
- 28.Rawlings, N. D., F. R. Morton, and A. J. Barrett. 2006. MEROPS: the peptidase database. Nucleic Acids Res. 34:D270-D272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rigden, D. J., M. J. Jedrzejas, and M. Y. Galperin. 2003. Amidase domains from bacterial and phage autolysins define a family of gamma-d,l-glutamate-specific amidohydrolases. Trends Biochem. Sci. 28:230-234. [DOI] [PubMed] [Google Scholar]
- 30.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 31.Shockman, G. D., and J. V. Höltje. 1994. Microbial peptidoglycan (murein) hydrolases, p. 131-167. In J.-M. Ghuysen and R. Hakenbeck (ed.), New comprehensive biochemistry. Bacterial cell wall. Elsevier Science, Amsterdam, The Netherlands.
- 32.Steen, A., G. Buist, G. J. Horsburgh, G. Venema, O. P. Kuipers, S. J. Foster, and J. Kok. 2005. AcmA of Lactococcus lactis is an N-acetylglucosaminidase with an optimal number of LysM domains for proper functioning. FEBS J. 272:2854-2868. [DOI] [PubMed] [Google Scholar]
- 33.Steen, A., G. Buist, K. J. Leenhouts, M. El Khattabi, F. Grijpstra, A. L. Zomer, G. Venema, O. P. Kuipers, and J. Kok. 2003. Cell wall attachment of a widely distributed peptidoglycan binding domain is hindered by cell wall constituents. J. Biol. Chem. 278:23874-23881. [DOI] [PubMed] [Google Scholar]
- 34.Wegmann, U., M. O'Connell-Motherway, A. Zomer, G. Buist, C. Shearman, C. Canchaya, M. Ventura, A. Goesmann, M. J. Gasson, O. P. Kuipers, D. van Sinderen, and J. Kok. 2007. Complete genome sequence of the prototype lactic acid bacterium Lactococcus lactis subsp. cremoris MG1363. J. Bacteriol. 189:3256-3270. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.