Abstract
A Vibrio parahaemolyticus strain isolated from the rhizosphere of the ecosystem dominant estuarine grass, Spartina alterniflora, was characterized and shown to carry nifH, the gene encoding the nitrogenase iron protein, and to fix N2. Nitrogen fixation may contribute substantially to the adaptability, niche breadth, and ecological significance of V. parahaemolyticus.
The family Vibrionaceae includes opportunistic pathogens of humans and animals (e.g., see references 10 and 28), as well as many less-pernicious species that are free-living chemoheterotrophs and/or commensals of marine fauna (e.g., see references 14 and 15). Outbreaks of Vibrio food poisonings, particularly those produced by Vibrio parahaemolyticus, are common in coastal areas during warmer months and are generally correlated with ingestion of raw or undercooked seafood. Maintenance of vibrios in estuarine environments during the winter months is often attributed to association with sediment or zooplankton (e.g., see references 17 to 19). However, the potential for other ecological niches, resulting in new reservoirs for V. parahaemolyticus, certainly exists.
Estuaries are frequently nitrogen limited, making the ability to reduce (“fix”) atmospheric N2 to ammonia highly advantageous. Only three Vibrio species, V. diazotrophicus, V. natriegens, and the opportunistic human pathogen Vibrio cincinnatiensis (5), have been shown to fix atmospheric N2 (30). Although V. parahaemolyticus has been recovered from the roots of salt marsh plants (1, 2), environments supporting high rates of N2 fixation (31), this organism was not previously shown to fix N2 or to have any other activity that might promote a stable interaction with plant hosts. Here we present evidence of N2 fixation by a strain of V. parahaemolyticus isolated from the rhizosphere of Spartina alterniflora, the dominant plant in salt marshes along the Atlantic and northern Gulf coasts of temperate North America. These results expand the niche breadth of this important potential human pathogen and contribute to understanding its maintenance in estuarine systems.
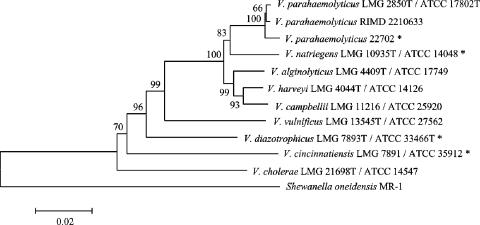
V. parahaemolyticus 22702 was isolated from S. alterniflora rhizosphere sediment collected from Sapelo Island, GA (8). Identification of this strain was accomplished using thiosulfate citrate bile salt sucrose agar (20) (BD Diagnostic Systems, Sparks, MD) and β-galactosidase activity (12) and confirmed by recovery and analysis of phylogenetically informative genes. DNA was extracted from overnight cultures using Prepman Ultra (ABI, Foster City, CA). Primers for PCR amplification of recA and rpoA were designed using the V. parahaemolyticus RIMD 2210633 sequences and Primer3 (25), recA 33F (5′-TGCGCTAGGTCAAATTGAAA-3′), recA 1008R (5′-AGCAGGTGCTTCTGGTTGAG-3′), rpoA 58F (5′-AGCTCGACTCACGCAAAAGT-3′), and rpoA 958R (5′-CTAGGCGCATACCCAGAGAC-3′). An internal recA primer, 622R (5′-TCGTTTCAGGGTTACCGAAC-3′), was also used in sequencing. The PCR program was 98°C for 30 s, followed by 35 cycles of 98°C for 10 s, 58°C for 30 s, and 72°C for 40 s and a final extension at 72°C for 10 min. Sequencing of amplicons was performed using an ABI Prism 3730 DNA analyzer, and sequences were edited and assembled using BioEdit version 7.0.5.2 (16). The 16S rRNA gene was amplified using primers 27F and 1492R (22). The following primers were employed for sequencing reactions: 519F (5′-CAGCAGCCGCGGTAA-3′), 529R (5′-CGCGGCTGCTGGCAC-3′), 907R (5′-CCCCGTCAATTCCTTTGAGTTT-3′), 1099F (5′-GCAACGAGCGCAACCC-3′), and 1240R (5′-CCATTGTAGCACGTGT-3′). The sequencing program was 34 cycles of 96°C for 10 s, 50°C for 5 s, and 60°C for 4 min, and sequences were determined using an ABI 3100 DNA analyzer. The 16S rRNA gene, recA, and rpoA sequences produced in this study or obtained from GenBank were joined (27) and then aligned using ClustalW (29). The concatenated sequences represented 87% of the full-length 16S rRNA gene and 71% and 78% of the coding regions for recA and rpoA, respectively. A neighbor-joining tree was constructed using the Jukes-Cantor correction (MEGA v. 3.1 [21]) (Fig. 1). This analysis placed 22702 with other V. parahaemolyticus strains and clearly differentiated it from species closely related to V. parahaemolyticus, such as Vibrio campbellii.
FIG. 1.
Neighbor-joining phylogeny of concatenated sequences of 16S rRNA, recA, and rpoA genes from species of Vibrio. A neighbor-joining tree was constructed using the Jukes-Cantor correction. Bootstrap values represent 1,000 replications, and values of <50% are not shown. Diazotrophic strains are marked with asterisks.
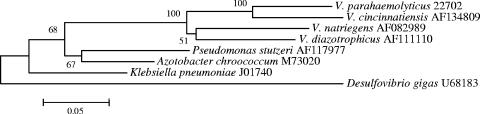
Strain 22702 maintained growth in the absence of combined nitrogen sources over several transfers and was subsequently tested for the presence of the nitrogenase iron protein gene (nifH), a widely used marker for diazotrophic bacteria. The nifH gene of strain 22702 was amplified using primers nifH F [5′-TACGG(P/K)AAKGG(P/G)GG(P/K)ATPGG-3′] and nifH R [5′-CGCCCGCCGCGCCCCGCGCCCGTCCCGCCGCCCCCGCCCG(G/C)ACGATGTAGATPTCCTG-3′] as described previously (23). Amplicons were cloned into pGEM-T (9) and sequenced using the Sp6 and T7 primers and the methods described above. Partial nifH nucleotide sequences were aligned, and a neighbor-joining phylogeny was constructed in MEGA using pairwise deletion of gaps and missing data and the Kimura two-parameter correction. Phylogenetic analysis showed a clear affiliation of the V. parahaemolyticus strain 22702 nifH sequence with sequences from other Vibrio species (Fig. 2) and also clearly differentiated the strain 22702 nifH sequence from those of other N2-fixing vibrios.
FIG. 2.
Neighbor-joining phylogeny of Vibrio and reference nifH sequences. A neighbor-joining tree was constructed using the Kimura two-parameter correction. Bootstrap values represent 1,000 replications, and values of <50% are not shown.
V. parahaemolyticus strain 22702 was tested for its ability to fix N2 using the acetylene reduction assay. Serum vials containing 10 ml of mineral salts medium (2) and a N2 headspace were inoculated with strain 22702, acetylene injected (10% of headspace volume), and the vials incubated at 30°C for 15 h. Four randomly selected replicate vials were sacrificed at each of 11 time intervals over the course of this incubation. Acetylene reduction activity was assayed by gas chromatography (1). Cell protein was quantified using the Rose Bengal dye binding assay (11). Negative controls were uninoculated medium, and rates were adjusted to account for “background” ethylene. Protein and ethylene quantities were determined from standard curves, and specific activities were reported as means ± standard deviations (n = 4). While the acetylene reduction rate of strain 22702 (1.31 ± 0.40 nmol C2H2·min−1·mg−1 protein; this study) is lower than those of previously described diazotrophic vibrios (38, 51, and 17 nmol C2H2·min−1·mg−1 protein for V. diazotrophicus ATCC 33466, V. natriegens ATCC 14048, and V. cincinnatiensis ATCC 35912, respectively [30]), it nonetheless provides definitive evidence of N2 fixation by V. parahaemolyticus. This capability enables this organism to maintain viability and growth in culture under strongly nitrogen-limited conditions and is expected to contribute to its capacity to colonize and grow on roots of salt marsh grasses under similarly nitrogen-limited conditions in salt marsh ecosystems.
Vibrionaceae, including V. parahaemolyticus, may play a substantial role in N2 fixation in the salt marsh ecosystem. The N2-fixing bacterial assemblage associated with the rhizosphere of salt marsh plants is a significant contributor of utilizable nitrogen to the system (reviewed in reference 24). Salt marsh vibrios can participate in N2 fixation, as indicated by recovery from S. alterniflora rhizosphere samples of a nifH cDNA sequence allied with nifH sequences from Vibrio species (6). Previous studies (2, 3, 23) recovered numerous diazotrophs from the roots of Spartina alterniflora, Juncus roemerianus, and Salicornia virginica. Vibrios comprised more than half of this culture collection, and about 14% of the Vibrio strains were identified as V. parahaemolyticus. These findings imply that this potential human pathogen could be a substantial rhizosphere resident and a contributing member of the salt marsh diazotroph community. Vibrio species are abundant on the roots of salt marsh plants, and cultivation-independent quantification of these organisms is ongoing (J. D. Criminger and C. R. Lovell, unpublished data).
The significance of V. parahaemolyticus and other vibrios in salt marsh ecology requires further study, but the implications of N2 fixation by potential human pathogens are of immediate interest. Vibrio cholerae, V. parahaemolyticus, and Vibrio vulnificus are potent human pathogens (4, 7, 13, 32, 33), and prior to this study, none of these species were known to fix N2. The capability of some V. parahaemolyticus strains to fix N2 may support their maintenance in nitrogen-limited coastal marine environments, contributing to a broader distribution of pathogenic strains. This key function may also define a new niche for V. parahaemolyticus as a rhizosphere-associated heterotrophic diazotroph. Finally, association with plant roots may provide a particularly important advantage to this organism as a refugium from cooler, growth-limiting temperatures and from predation (26). Disturbance of the rhizosphere by infaunal burrowing, severe weather events, and human activities may then facilitate transfer of this organism to the overlying waters, mobilizing pathogenic strains and potentially contributing to infectious outbreaks. Ultimately, maintenance and propagation of this organism is of significant human health concern, and identifying natural habitats and functions that may broaden its distribution is essential to predicting epidemic outbreaks of V. parahaemolyticus.
Nucleotide sequence accession numbers.
GenBank accession numbers for sequences of V. parahaemolyticus strain 22702 determined in this work are as follows: for recA and rpoA, EU018456 and EU018455, respectively; for the 16S rRNA gene, EF203421; and for nifH, EF203422.
Acknowledgments
This research was supported by NSF award MCB-0237854 (C.R.L.) and by a grant in aid of research from the Slocum-Lunz Foundation (J.D.C.).
We acknowledge Mike Friez for assistance with DNA sequencing.
Footnotes
Published ahead of print on 3 August 2007.
REFERENCES
- 1.Bagwell, C. E., and C. R. Lovell. 2000. Persistence of selected Spartina alterniflora rhizoplane diazotrophs exposed to natural and manipulated environmental variability. Appl. Environ. Microbiol. 66:4625-4633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bagwell, C. E., Y. M. Piceno, A. Ashburne-Lucas, and C. R. Lovell. 1998. Physiological diversity of the rhizosphere diazotroph assemblages of selected salt marsh grasses. Appl. Environ. Microbiol. 64:4276-4282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bergholz, P. W., C. E. Bagwell, and C. R. Lovell. 2001. Physiological diversity of rhizoplane diazotrophs of the saltmeadow cordgrass, Spartina patens: implications for host specific ecotypes. Microb. Ecol. 42:466-473. [DOI] [PubMed] [Google Scholar]
- 4.Blake, P. A. 1994. Historical perspectives on pandemic cholera, p. 293-295. In I. K. Wachsmuth, P. A. Blake, and O. Olsvik (ed.), Vibrio cholerae and cholera: molecular to global perspectives. ASM Press, Washington, DC.
- 5.Brayton, P. R., R. B. Bode, R. R. Colwell, M. T. MacDonell, H. L. Hall, D. J. Grimes, P. A. West, and T. N. Bryant. 1986. Vibrio cincinnatiensis sp. nov., a new human pathogen. J. Clin. Microbiol. 23:104-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brown, M. M., M. J. Friez, and C. R. Lovell. 2003. Expression of nifH genes by diazotrophic bacteria in the rhizosphere of short form Spartina alterniflora. FEMS Microbiol. Ecol. 43:411-417. [DOI] [PubMed] [Google Scholar]
- 7.Centers for Disease Control and Prevention. 1996. Vibrio vulnificus infections associated with eating raw oysters—Los Angeles, 1996. Morb. Mortal. Wkly. Rep. 45:621-624. [PubMed] [Google Scholar]
- 8.Cook, M. A., A. M. Osborn, J. Bettandorff, and P. A. Sobecky. 2001. Endogenous isolation of replicon probes for assessing plasmid ecology of marine sediment microbial communities. Microbiology 147:2089-2101. [DOI] [PubMed] [Google Scholar]
- 9.Dang, H., and C. R. Lovell. 2000. Bacterial primary colonization and early succession on surfaces in marine waters as determined by amplified rRNA gene restriction analysis and sequence analysis of 16S rRNA genes. Appl. Environ. Microbiol. 66:467-475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Daniels, N. A., L. MacKinnon, R. Bishop, S. Altekruse, B. Ray, R. M. Hammond, S. Thompson, S. Wilson, N. H. Bean, P. M. Griffin, and L. Slutsker. 2000. Vibrio parahaemolyticus infections in the United States, 1973-1998. J. Infect. Dis. 181:1661-1666. [DOI] [PubMed] [Google Scholar]
- 11.Elliot, J. F., and S. M. Brewer. 1978. The inactivation of yeast enolase by 2,3-butanedione. Arch. Biochem. Biophys. 190:351-357. [DOI] [PubMed] [Google Scholar]
- 12.Farmer, J. J., and J. M. Janda. 2005. Family I. Vibrionaceae Véron 1965, 5245,AL p. 491-555. In D. J. Brenner, N. R. Krieg, J. T. Staley, and G. M. Garrity (ed.), Bergey's manual of systematic bacteriology, 2nd ed., vol. 2. Springer, New York, NY. [Google Scholar]
- 13.Finkelstein, R., S. Edelstein, and G. Mahamid. 2002. Fulminant wound infections due to Vibrio vulnificus. Isr. Med. Assoc. J. 4:654-655. [PubMed] [Google Scholar]
- 14.Grimes, D. J., P. Brayton, R. R. Colwell, and S. H. Gruber. 1985. Vibrios as autochthonous flora of neritic sharks. Syst. Appl. Microbiol. 6:221-226. [Google Scholar]
- 15.Guerinot, M. L., and D. G. Patriquin. 1981. N2-fixing vibrios isolated from the gastrointestinal tract of sea urchins. Can. J. Microbiol. 27:311-317. [DOI] [PubMed] [Google Scholar]
- 16.Hall, T. A. 1999. Bio-Edit: a user-friendly biological sequence alignment editor for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41:95-98. [Google Scholar]
- 17.Jonas, R. B., E. N. Buckley, and F. K. Pfaender. 1978. A note on the isolation of the bacterium Vibrio parahaemolyticus from estuarine North Carolina. Estuaries 1:264-266. [Google Scholar]
- 18.Kaneko, T., and R. R. Colwell. 1973. Ecology of Vibrio parahaemolyticus in Chesapeake Bay. J. Bacteriol. 113:24-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kaneko, T., and R. R. Colwell. 1974. Distribution of Vibrio parahaemolyticus and related organisms in the Atlantic Ocean off South Carolina and Georgia. Appl. Environ. Microbiol. 28:1009-1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kobayashi, T., S. Enomoto, R. Sakazaki, and S. Kuwahara. 1963. A new selective isolation medium for the Vibrio group; on a modified Nakanishi's medium (thiosulfate citrate bile salt sucrose agar medium). Nippon Saikingaku Zasshi 18:387-392. [DOI] [PubMed] [Google Scholar]
- 21.Kumar, S., K. Tamura, and M. Nei. 2004. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief. Bioinform. 5:150-163. [DOI] [PubMed] [Google Scholar]
- 22.Lane, D. J. 1991. 16S/23S rRNA sequencing, p. 115-175. In E. Stackebrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. John Wiley and Sons, Chichester, United Kingdom.
- 23.LaRocque, J. R., P. W. Bergholz, C. E. Bagwell, and C. R. Lovell. 2004. Influence of host plant-derived and abiotic environmental parameters on the composition of the diazotroph assemblage associated with roots of Juncus roemerianus. Antonie Leeuwenhoek 86:249-261. [DOI] [PubMed] [Google Scholar]
- 24.Lovell, C. R. 2002. Plant-microbe interactions in the marine environment, p. 2539-3554. In G. Bitton (ed.), Encyclopedia of environmental microbiology, vol. 5. John Wiley and Sons, New York, NY. [Google Scholar]
- 25.Rozen, S., and H. J. Skaletsky. 2000. Primer3 on the WWW for general users and for biologist programmers, p. 365-386. In S. Krawetz and S. Misener (ed.), Bioinformatics methods and protocols: methods in molecular biology. Humana Press, Totowa, NJ. [DOI] [PubMed]
- 26.Sherr, E. B., and B. F. Sherr. 2002. Significance of predation by protists in aquatic microbial food webs. Antonie Leewenhoek 81:293-308. [DOI] [PubMed] [Google Scholar]
- 27.Thompson, F. L., D. Gevers, C. C. Thompson, P. Dawyndt, S. Naser, B. Hoste, C. B. Munn, and J. Swings. 2005. Phylogeny and molecular identification of vibrios on the basis of multilocus sequence analysis. Appl. Environ. Microbiol. 71:5107-5115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thompson, F. L., T. Iida, and J. Swings. 2004. Biodiversity of vibrios. Microbiol. Mol. Biol. Rev. 68:403-431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Urdaci, M. C., L. J. Stal, and M. Marchand. 1988. Occurrence of nitrogen fixation among Vibrio spp. Arch. Microbiol. 150:224-229. [Google Scholar]
- 31.Whiting, G. J., E. L. Gandy, and D. C. Yoch. 1986. Tight coupling of root-associated nitrogen fixation and plant photosynthesis in the salt marsh grass Spartina alterniflora and carbon dioxide enhancement of nitrogenase activity. Appl. Environ. Microbiol. 52:108-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.World Health Organization. 2001. Cholera, 2000. Wkly. Epidemiol. 76:233-240. [PubMed] [Google Scholar]
- 33.World Health Organization. 2002. Cholera, 2001. Wkly. Epidemiol. 77:257-268. [PubMed] [Google Scholar]