Abstract
Resistance to bile is a prerequisite property of the gastrointestinal bacterial flora. Bile acids are powerful detergents, and resistance to sodium dodecyl sulfate (SDS) has therefore often been considered relevant to studies of bile resistance. We have studied the effects of bovine bile (BB) and SDS on Enterococcus faecalis V583 by traditional growth studies and microarrays. Transcriptional responses were studied by time course experiments. In the presence of BB (V583-BB) or SDS (V583-SDS), 308 and 209 genes were identified as differentially expressed at one or more time points, respectively. In V583 treated with both BB and SDS (V583-BB-SDS), 254 genes showed differential expression. Detergents exert their toxic effects primarily on the microbial membrane. The enrichment of differentially transcribed genes that encode proteins with membrane-associated functions and/or locations indicates a major impact of all three treatments on the integrity and functionality of the cell membrane. Two gene clusters involved in fatty acid biosynthesis were repressed in V583-BB and V583-BB-SDS and partly induced in V583-SDS. Furthermore, two EmrB/QacA family drug resistance transporters and a vacuolar-type ATPase were induced in V583-BB and V583-BB-SDS. None of the putative bile salt hydrolase homologs in V583 showed differential expression during the bile treatments. The transcriptional profile of V583-BB-SDS was qualitatively more similar to the response in V583-BB than to that in V583-SDS, suggesting that the presence of bile suppresses the effects of SDS in V583-BB-SDS. The overall results presented here indicate that different mechanisms are involved in detergent resistance in E. faecalis.
In order to survive in and colonize the gastrointestinal tract (GIT), a bacterium must overcome several biological barriers after consumption. Bacteria that survive the gastric acidity of the stomach transit to the intestine, where they encounter stress associated with variations in pH, low oxygen availability, elevated osmolarity, and relatively high concentrations of host-produced substances like bile. Bile is an aqueous solution whose major organic constituents are bile acids, cholesterol, and phospholipids (7). Bile acids are anionic-detergent-like biological compounds that are bactericidal because of their amphipathic structure (17). The primary bile acids are synthesized de novo from cholesterol in the liver and secreted as amino acid conjugates into the duodenum, where they aid in the digestion of fats and undergo enterohepatic circulation (17). During enterohepatic circulation, conjugated bile acids can undergo modifications by the intestinal microflora. Deconjugation of bile acids is catalyzed by bile salt hydrolases (BSHs), which hydrolyze the amide bond and liberate the glycine-taurine moiety from the steroid core (17).
Enterococcus faecalis is a natural member of the mammalian gastrointestinal bacterial flora. Previous work by Flahaut et al. (13, 14) demonstrated rapid killing of E. faecalis by a mixture of unconjugated bile salts (sodium cholate-sodium deoxycholate [1:1]) in vitro. However, E. faecalis is one of the most common lactic acid bacteria found in the GIT (12) and one of the predominant microbial species isolated from drainage devices used in biliary surgery (33). E. faecalis has also been isolated directly from the gallbladder (15). This indicates an inherent ability to tolerate high concentrations of bile under in vivo conditions. Several proteomic analyses of global changes in protein expression during bile salt exposure of gram-positive bacteria have been presented (13, 23, 31, 34). Bron et al. recently described a microarray-based approach for the identification of bile-responsive genes in Lactobacillus plantarum (9). Furthermore, random gene disruption strategies have been used to identify mutants that are more susceptible to bile salts than the wild-type strains in several gram-positive bacteria (6, 8, 20). In addition to emphasizing the effects of bile on membrane integrity and structure, these studies also suggested that bile imposes oxidative stress and DNA damage on the cell (summarized by Begley et al. [7]).
Sodium dodecyl sulfate (SDS) is an anionic detergent, and bacterial resistance to SDS has often been considered relevant to studies of bile resistance (13). Previous studies of bile salt and SDS stress responses in E. faecalis have shown significant cross-resistances between these two stress factors (13). This reported adaptation of E. faecalis to bile salts and SDS indicates that common resistance mechanisms are involved in the stress responses to these two detergents.
We have studied the susceptibility of E. faecalis V583 to bovine bile (BB) and SDS by using traditional growth studies and genome-wide microarrays. The aim of this work was to gain further insight into the effects of BB and SDS on growth and transcriptional events in V583. Since detergents exert their toxic effects primarily on the microbial membrane, it was expected that genes involved in cell membrane-associated functions would be affected by the detergent treatments. Moreover, in addition to genes related to DNA repair and oxidative response, proteins that take up or extrude bile and enzymes that modify and transform bile salts were also likely to play an important role in bile resistance.
MATERIALS AND METHODS
Bacterial strain and growth conditions.
E. faecalis V583 (32) was grown aerobically overnight (ON) in brain heart infusion (BHI) broth (Oxoid) with shaking (300 rpm) at 37°C. For determination of the number of CFU/ml, BHI agar plates were used. SDS and BB (B3883; Sigma) were solubilized in water to obtain 10% (wt/vol) solutions. Filter-sterilized stock solutions were added to autoclaved medium.
SDS and BB treatments.
For broth assays, ON cultures were inoculated (50× dilution) into BHI broth containing various concentrations of (i) BB (V583-BB), (ii) SDS (V583-SDS), or (iii) a combination of BB and SDS (V583-BB-SDS). Cell growth was measured spectrophotometrically and by viable cell counts. Growth experiments were performed with a Bioscreen instrument (Bioscreen C). For V583-SDS, each well contained 3 μl SDS to which 297 μl bacterial inoculum in BHI broth was added. The final concentrations of SDS were 0.01 to 0.1%. For V583-BB and V583-BB-SDS assays, 150 μl of the appropriate concentration of BB-containing BHI was added to each well. SDS was added as described above. Bacterial inoculum in BHI broth was then added to a total volume of 300 μl. The final bile concentrations were 1 to 7.5%. Wells containing BHI with equal amounts of sterile BB and SDS were used as negative controls. Cultures were incubated at 37°C (sample plates were shaken for 10 s prior to each measurement), and the optical density at 600 nm (OD600) was measured at 30-min intervals for 24 h. All experiments were performed independently in triplicate. To determine the number of CFU/ml, viable cell counts were performed as follows. ON cultures in BHI were grown to an OD600 of ∼0.2, split into four, and centrifuged, and the pellets were resuspended in (i) fresh BHI (V583-BHI), (ii) BHI containing 1% BB, (iii) BHI containing 0.06% SDS, or (iv) BHI containing 1% BB and 0.06% SDS. Samples were collected immediately after the addition of BB and/or SDS and after 2, 4, 6, and 8 h. The number of CFU/ml was estimated by averaging the colony counts in three replicates per treatment after 24 h of incubation at 37°C.
Sample collection.
ON cultures were diluted 50× and grown in BHI to an OD600 of ∼0.2, split into two, and centrifuged, and pellets were resuspended in 0.5 volume of fresh BHI. For each stress treatment, 0.5 volume of BHI (control) and 0.5 volume of BHI containing BB and/or SDS were added to the two cultures. The final concentrations of BB and SDS were 1% and 0.06%, respectively. The two cultures were then further incubated, and 10-ml samples were collected immediately after the addition of BB and/or SDS (t0) and then after 10 (t10), 20 (t20), 30 (t30), and 60 (t60) min. Because of the difference in growth rate, samples were also collected after 120 min (t120) for V583-BB and V583-BB-SDS and after 360 min (t360) for V583-SDS. Samples were centrifuged at 5,000 rpm for 3 min in an Eppendorf 5804R tabletop centrifuge at 4°C, and pellets were flash frozen in liquid N2 prior to RNA extraction.
Microarrays.
The microarrays used in this study have been described previously by Aakra et al. (2). All PCR products, representing the predicted V583 open reading frames, were printed in three copies on each slide.
RNA isolation.
Total RNA was isolated from the samples collected at the time points described above by means of the RNeasy Mini kit (QIAGEN) with on-column DNA digestions with RNase-free DNase I (QIAGEN) according to the manufacturer's recommendations or by FastPrep (Bio 101/Savant) as follows. Frozen cell pellets were thawed, and 350 μl lysis buffer RLT (RNeasy Mini Kit; QIAGEN) supplemented with 0.1% (vol/vol) β-mercaptoethanol (Sigma) was added to each cell suspension. The suspensions were transferred to 2-ml screw-cap FastPrep tubes (Qbiogene) containing 0.6 g acid-washed glass beads (<106 μm; Sigma) and 250 μl chloroform (Merck). Cells were lysed by shaking the tubes for 20 s at 6 m/s in an FP120 FastPrep bead beater (Bio 101/Savant). After a short spin, the water phases were transferred to Eppendorf tubes and the Eppendorf tubes were centrifuged for 2 min at 13,000 rpm (Biofuge Pico; Heraeus). The RNA-containing supernatants were then collected in new Eppendorf tubes. Pellets were resuspended in 350 μl β-mercaptoethanol-containing lysis buffer RLT, and the cell suspensions were transferred back to the FastPrep tubes with the glass beads for a second extraction to increase the overall RNA yield. A 250-μl volume of ethanol was then added to the supernatant fluids from the two extractions, and after mixing, each sample was applied to an RNeasy spin column (QIAGEN). The column was further washed and RNA eluted according to the RNeasy Mini kit protocol (QIAGEN), with DNA digestions as described above. The concentrations of the RNA samples were measured by using the NanoDrop (NanoDrop Technologies), and the quality was assessed by using the RNA 600 Nano LabChip kit and the Bioanalyzer 2100 (Agilent Technologies). Ten micrograms of each RNA sample was used for cDNA synthesis.
cDNA synthesis, fluorescent labeling, and hybridization.
RNA was labeled with the FairPlay Microarray labeling kit (Stratagene) in accordance with the manufacturer's protocol, with the following modifications. For each labeling reaction, 10 μg of random hexamer primers (Amersham) and 10 μg of total RNA were initially preheated at 70°C for 10 min. A reverse transcription-PCR mixture (5× SuperScript III reaction buffer, a 20× deoxynucleoside triphosphate mixture, 0.1 M dithiothreitol, 20 U RNase block, and 520 U SuperScript III) was added to the annealed primers and RNA, and the reaction mixture was further incubated for 3 h at 46°C. After labeling, unincorporated dyes were removed from the samples by using the QIAquick PCR purification kit (QIAGEN). Prehybridization, hybridization, washing, and drying of the arrays were performed as described by Aakra et al. (2). Because of the reduced growth rates associated with growth in SDS, V583-SDS reached stationary phase after approximately 360 min while V583-BHI, V583-BB, and V583-BB-SDS reached stationary phase after approximately 120 min. We therefore chose to hybridize the t360 sample from V583-SDS against a sample that was harvested from faster-growing V583-BHI at a time point corresponding to the stationary phase. All of the other samples in the three experiments were cohybridized with control samples collected at the corresponding time points (e.g., V583-BB collected at t10 was hybridized along with V583-BHI collected at t10). Four replicate hybridizations were performed with two separate batches of RNA. The two batches of RNA were obtained in two separate growth experiments. The Cy3 and Cy5 dyes (Amersham) used during cDNA synthesis were swapped in two of the four replicate hybridizations. Hybridized arrays were scanned at wavelengths of 532 nm (Cy3) and 635 nm (Cy5) with an Agilent G2505B scanner (Agilent Technologies). Fluorescence intensities and spot morphologies were analyzed with GenePix Pro 6.0 (Molecular Devices), and spots were excluded on the basis of slide or morphology abnormalities. Raw data from each array were preprocessed independently, and testing for differentially expressed genes was done by using a random-effect analysis-of-variance approach, as described by Aakra et al. (2). All genes were functionally classified according to The Institute for Genomic Research comprehensive microbial resource (http://cmr.tigr.org/tigr-scripts/CMR/CmrHomePage.cgi), and functional groups were tested for significant enrichment among the differentially expressed genes by using the Fisher exact test and a significance level of P = 0.05.
Microarray data accession number.
The microarray data obtained in this study have been deposited in the ArrayExpress database (http://www.ebi.ac.uk/arrayexpress/) under accession number E-TABM-242.
RESULTS
Growth of E. faecalis V583 in the presence of SDS and BB.
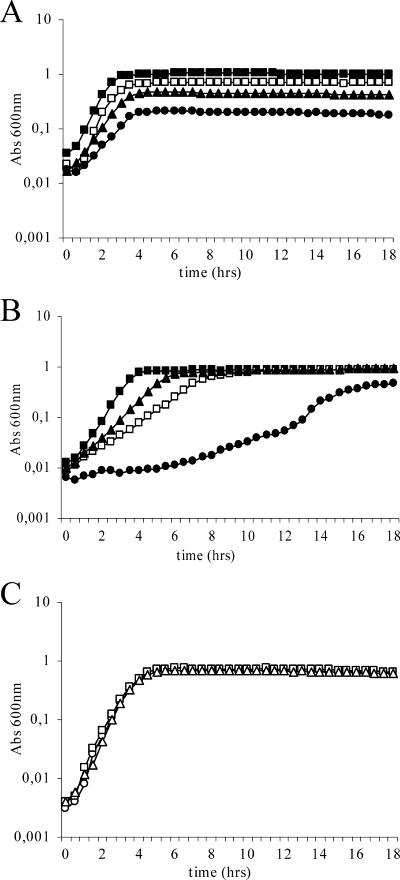
V583 was able to grow in BHI broth containing up to 7.5% BB and up to 0.1% SDS, with an extended lag phase, in addition to reduced growth rates and lower cell densities in the stationary phase of growth (Fig. 1A and B). In cultures treated with BB (V583-BB), the growth rates and final cell densities decreased with increasing concentrations of BB (Fig. 1A). The final cell density of V583 grown in medium containing 7.5% BB was approximately one-fifth of the final OD600 of untreated cultures (V583-BHI). However, V583-BHI and V583-BB with various concentrations of BB reached stationary phase after approximately the same time of growth. In V583 cultures treated with SDS (V583-SDS), growth rates decreased more drastically with increasing detergent concentrations than did those of V583-BB. On the other hand, the effect of SDS on cell density appeared to be more concentration dependent in V583-SDS. As opposed to V583-BB cultures, V583-SDS cultures containing less than 0.06% SDS were able to reach stationary-phase cell densities similar to those of V583-BHI cultures after prolonged incubation. In V583-SDS cultures containing higher concentrations of SDS, the stationary-phase cell densities decreased. The final cell density reached by V583 grown in the presence of 0.1% SDS was approximately half of the final cell density of V583-BHI (Fig. 1B). These growth experiments suggest that BB and SDS affect E. faecalis V583 differently.
FIG. 1.
Growth of E. faecalis V583 treated with BB (A; left untreated [▪] or treated with 1 [□], 5 [▴], or 7.5% [•] BB), SDS (B; left untreated [▪] or treated with 0.03 [▴], 0.06 [□], or 0.1% [•] SDS), or 1% BB plus SDS (C; 0 [○], 0.03 [□], or 0.06% [▵] SDS). Abs, absorbance.
With the results of the V583-BB and V583-SDS growth experiments in mind, it was of interest to test the effects of mixtures of the two compounds on V583. Experiments were carried out by a constant amount of BB with various concentrations of SDS added to the BHI prior to inoculation. On the basis of the reported cross-resistance between bile salt- and SDS-adapted cells (13), both additive and synergistic effects on V583 were expected. However, the effect of the detergent mixture turned out to be partly antagonistic (Fig. 1C). The presence of BB seemed to abolish the inhibitory effects of SDS on V583 growth. Viable cell counts of V583 treated with selected concentrations of each detergent (1% BB, 0.06% SDS, and a mixture of 1% BB and 0.06% SDS) at different time points during growth confirmed the results obtained by OD measurements (data not shown).
Transcriptional profiling of V583-SDS, V583-BB, and V583-BB-SDS.
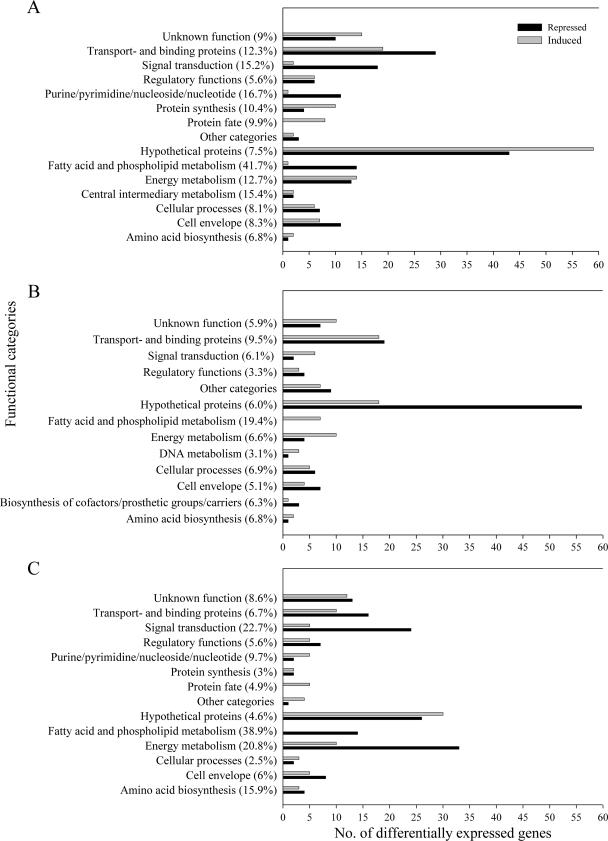
A genome-wide amplicon-based microarray containing 3,160 genes (94.7% of the genes in the E. faecalis V583 genome) (2) was used for profiling of the transcriptional responses in V583-BB, V583-SDS, and V583-BB-SDS. Transcriptional events were studied by time course experiments. The time course experiments permitted us to investigate immediate responses, as well as adaptations to the more permanent presence of BB or/and SDS. We chose a stringent significance level of P = 0.01 for the determination of differential transcription. We also used a conservative Bonferroni correction to control the family-wise error rate under multiple testing to ensure a low false discovery rate. By these criteria, 308 genes were identified as differentially expressed at one or more of the six time points during the course of growth in V583-BB. In V583-SDS, 209 genes were differently expressed, while 254 genes showed differential expression when both compounds were present (V583-BB-SDS). Figure 2 contains the details. All functional groups were tested for significant enrichment among the differentially transcribed genes by using the Fisher exact test and a significance level of P = 0.05 (data not shown).
FIG. 2.
Genes differentially expressed in V583-BB (A), V583-SDS (B), and V583-BB-SDS (C) grouped by functional classification according to The Institute for Genomic Research comprehensive microbial resource (http://cmr.tigr.org/tigr-scripts/CMR/CmrHomePage.cgi). Black bars refer to repressed genes, and gray bars refer to induced genes. Values in parenthesis are percentages of the total number of genes within each functional class in the genome. Functional classes with fewer than two regulated genes are gathered under “Other categories.” Genes that were both up- and downregulated during the time course were counted twice.
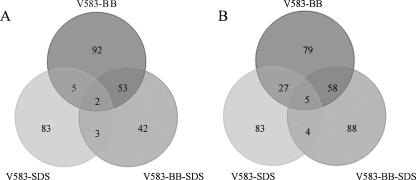
By comparison of the three different transcriptional profiles, 24 genes were identified as differentially expressed in V583-SDS, V583-BB, and V583-BB-SDS. Only two genes, EF0108 (C4-dicarboxylate transporter, putative) and EF0288 (hypothetical protein), were induced during all three treatments, while three genes (EF1027, which codes for a putative membrane protein; EF1207, which codes for a citrate carrier protein; and EF2983, which codes for a putative glutamyl-tRNA amidotransferase) were repressed. In addition, two genes that encode ABC transporters (EF2049 and -50) were both induced and repressed in V583-BB but only repressed in V583-SDS and V583-BB-SDS. Apparently, the expression profile of V583-BB-SDS was more similar to the expression profile of V583-BB than to that of V583-SDS (Fig. 3; see Tables S4, S6, and S7 in the supplemental material). Of the 24 commonly differentially expressed genes, 22 showed qualitatively similar responses to both V583-BB and V583-BB-SDS (see Table S4 in the supplemental material). Comparison of the transcriptional events in V583-SDS and V583-BB revealed 68 genes that were differentially expressed during both treatments, 38 of which showed similar expression patterns (either up- or downregulated) in both experiments (see Table S5 in the supplemental material).
FIG. 3.
Distribution of genes upregulated (A) and downregulated (B) in V583-BB, V583-SDS, and V583-BB-SDS. Genes that were both up- and downregulated during the time course were counted twice.
Interestingly, the two gene clusters EF0282 to -84 and EF2875 to -86, which encode proteins involved in type II fatty acid synthesis (FAS) in V583, were significantly repressed in V583-BB and V583-BB-SDS, while EF2875 to -80 and EF2883 showed increased transcriptional activity at t10 to t20 in V583-SDS (Fig. 4). This suggests that the presence of BB eradicated the stress imposed on FAS in V583-SDS. Other genes that were found to be induced in V583-BB and V583-BB-SDS but not induced in V583-SDS were genes that encode two EmrB/QacA family drug resistance transporters (EF0420 and EF1814) and a vacuolar-type (V-type) ATPase (EF1492 to -1500).
FIG. 4.
Changes in mRNA levels of selected genes in V583-SDS, V583-BB, and V583-BB-SDS during the course of growth, represented in gray (induced), black (repressed), and white (no significant regulation). CoA, coenzyme A.
For the log2 ratios of genes differentially expressed in V583-BB, V583-SDS, and V583-BB-SDS, see Tables S1 to S3 in the supplemental material.
DISCUSSION
In order to survive, the bacterial flora of the GIT must tolerate relatively high concentrations of host-produced inhibitory compounds like bile. Previous studies have shown strong cross-resistance between cells adapted to bile salts and the anionic detergent SDS (13), indicating that the physiological responses to bile salts and SDS are closely related in E. faecalis. The strain used in this study, V583, however, showed different responses to these compounds with respect to both growth and transcription.
V583 was able to grow in medium containing up to 7.5% BB and 0.1% SDS (Fig. 1A and B). Growth in 7.5% bile exceeds the physiological levels in the intestine (0.2 to 2%) (16). Previous susceptibility studies of E. faecalis have reported lower resistance to both bile salts and SDS (13, 14, 18), and the present results indicate large variations in resistance to these detergents within this species. The apparent divergent susceptibility to bile among E. faecalis strains may be a consequence of true strain-specific variation in bile resistance. A recently published microarray-based survey of genomic diversity in E. faecalis showed that regions corresponding to putative mobile genetic elements in V583 are missing in several E. faecalis strains (1), and one may suggest that genes within these mobile genetic elements could be required for the detergent resistance seen in V583. Divergent susceptibility may, however, also reflect particular traits of the mixture of bile acids used in the experiments reported (6, 7). Increased lag times associated with growth in the presence of SDS have previously been attributed to the time necessary to accumulate sufficient intracellular K+ for logarithmic growth of gram-negative bacteria (4).
The addition of BB to cultures containing different concentrations of SDS apparently suppressed the inhibitory effects of SDS on growth in V583 (Fig. 1C). This observation may be explained by intercalation of bile acids with membrane lipids (22). As a consequence, the membrane may become more resistant to detergents because of enhanced lipid membrane stability. However, the enhanced resistance of V583 to SDS in the presence of BB might also be a result of bile acids and SDS forming micelles, preventing SDS from adhering to the bacterial cells (19). Such a rationale may, in turn, also explain the qualitatively similar responses in V583-BB and V583-BB-SDS.
To gain deeper insight into the BB and SDS resistance of V583, global transcriptional profiling was performed by use of DNA microarrays. The stress conditions applied caused significantly decreased growth compared to that of unstressed cultures. We decided to cohybridize the test samples with reference samples collected at the same time points and to correct for the differences in growth phase at only one time point, where the most obvious deviation in growth phase occurred (see Materials and Methods). This design allowed a direct comparison between stressed and nonstressed bacteria at a given time, as opposed to the use of a common reference, e.g., an untreated t0 control sample, where differential expression may be an outcome of the stress conditions applied to the cells but also an effect of time. We therefore considered cohybridization of the treated samples with reference samples collected at the same time points as more precise than hybridization against a common reference. For the identification of significantly differentially expressed genes, the conservative Bonferroni correction was used as a strategy to reduce the occurrence of type I errors (false positives), at the expense of increasing type II errors (false negatives). To address the possible loss of information due to type II errors, an independent verification of the results for selected genes could be considered.
Testing for significant enrichment among the differentially expressed genes by using the Fisher exact test and a significance level of P = 0.05 (data not shown) showed that genes engaged in fatty acid and phospholipid metabolism were significantly enriched in all three treatments. Genes that encode the components of type II FAS are located in two separate clusters in the V583 genome (EF0282 to -84 and EF2875 to -86). Our data suggest that the type II FAS genes are a key target for both BB and SDS in V583. However, it was surprising to observe that the two compounds affect these genes so differently. Random gene disruption in E. faecalis has previously revealed bile-sensitive mutants that produce homologues of proteins involved in membrane composition and cell wall synthesis (20). Moreover, bile-induced changes in membrane composition have been reported in other gram-positive bacteria. In Lactobacillus reuteri (36), a fivefold reduction in the phospholipid fraction of the total membrane lipid content was seen in response to bile salts. Such a bile-induced decline in phospholipids is in line with the observed repression of genes involved in fatty acid and phospholipid metabolism in V583-BB and V583-BB-SDS. In Streptococcus pneumoniae, FabF and FabK are believed to be involved in the regulation of membrane fatty acid composition (26). These observations may be applicable to the induction of EF2880 (FabF2) and EF2883 (FabK) in V583-SDS. The high number of genes involved in membrane-associated functions with altered expression indicates a major impact of all three detergent treatments on the integrity and functionality of the cytoplasmic membrane.
Genes involved in energy metabolism were enriched among the genes differentially expressed in V583-BB-SDS (Fisher exact test; data not shown). The bkd operon (EF1658 to -63), which encodes the branched-chain α-keto acid dehydrogenase complex in V583, was downregulated at t120 in V583-BB-SDS. The branched-chain α-keto acid dehydrogenase complex catalyzes the oxidative decarboxylation of branched-chained α-keto acids with the concomitant reduction of NAD+, supposedly generating the corresponding acyl coenzyme A and ATP via substrate level phosphorylation (39, 40). To our knowledge, there is no evidence linking the enterococcal bkd gene cluster to the biosynthesis of branched-chain fatty acids, as found in Bacillus subtilis (39). The repression of this operon in V583-BB-SDS should therefore not be correlated to the observed repression of genes involved in type II FAS.
In V583-BB and V583-BB-SDS, two gene clusters assigned to functions in energy metabolism were induced, EF0104 to -08 and EF1492 to -1500 (Fig. 4). EF0104 to -06 encode the three enzymatic steps of the arginine deiminase pathway, generating ATP through fermentative arginine catabolism (5). EF1492 to -1500 encode a V-type ATPase. V-type ATPases are membrane-bound proteins that function in active ion pumping at the expense of ATP hydrolysis. The induction of the gene cluster that encodes the V-type ATPase in V583-BB and V583-BB-SDS suggested that the generation of a proton gradient is important for survival of the cell during exposure to bile. Bile-mediated stress responses have previously been linked to maintenance of the proton motive force (PMF) in L. plantarum and Bifidobacterium sp. (9, 34, 35). Sánchez et al. recently proposed that increased ATPase activity in Bifidobacterium animalis is an attempt to compensate for the PMF-dissipating and internal pH-decreasing effects of bile (35).
Another possible rationale for the cell to generate such a proton gradient in response to bile stress is the importance of PMF in the active extrusion of bile acids. The two drug resistance transporter genes EF0420 and EF1814 (drug resistance transporters, EmrB/QacA family proteins) (29) showed enhanced transcriptional activity in V583-BB and V583-BB-SDS (Fig. 4). Previous studies have shown that inactivation of efflux pumps has a deleterious effect on the ability of the bacterium to tolerate bile (24, 37), and bile resistance has been linked to ATP-dependent active efflux in gram-positive bacteria (41). However, EF0420 and EF1814 in E. faecalis are related to the known PMF-dependent efflux systems EmrB in Escherichia coli (25) and QacA in Staphylococcus aureus (10). EmrB has previously been reported to be involved in the active efflux of bile salts in E. coli (37), but to our knowledge, this is the first report of a PMF-dependent transport system linked to bile resistance in gram-positive bacteria.
A two-component system (EF3289 and -90; croRS) was induced in V583-BB and V583-BB-SDS at t20. CroRS shows significant homology (60.4% similarity) to a bile-inducible two-component regulatory system in Lactobacillus acidophilus (30). The expression of croRS has recently been shown to be repressed when E. faecalis enters the stationary phase, implying that expression toward the end of the time course was not to be expected (21). Moreover, CroRS is believed to be involved in the regulation of the stress-inducible secreted protein SalB (EF0394) in E. faecalis (28). salB has previously been referred to as sagA and is identical to the disrupted locus in one of the bile-sensitive E. faecalis mutants identified by Le Breton et al. (20). salB was predicted to encode a putative surface-exposed, virulence-related protein in V583 by Paulsen et al. (29). It has previously been shown that growth of E. faecalis strains in ox bile altered the physicochemical surface properties of the bacterium and resulted in increased adhesion to bile drainage materials (38). EF3245, which encodes a cell envelope-associated acid phosphatase, was induced at four and five of the six time points in V583-BB and V583-BB-SDS, respectively. Several other genes listed as putative virulence-related genes by Paulsen et al. (29) also showed significantly increased expression at one or more time points in V583-BB (see Table S1 in the supplemental material).
The gene cluster EF2977 to -83 was differentially transcribed at the same time points as EF3245. EF2977 to -81 encode a PTS system and a σ54-dependent DNA-binding response regulator with significant homology to the mpo operon and its σ54-dependent activator in Listeria monocytogenes (3). Inactivation of mpoA resulted in intermediate bacteriocin resistance in L. monocytogenes. The downregulation of EF2977 to -81 in V583-BB and V583-BB-SDS led us to the suggestion that exposure to bile enhances the tolerance to certain class IIa bacteriocins (11) in E. faecalis. Preliminary bacteriocin plate assays (27) performed with Pediococcus acidilactici (PacI) and Lactobacillus sakei (SakP) did show that while V583 was sensitive to PacI and SakP in the absence of bile, the presence of 2% bile rendered V583 significantly more resistant to these class IIa bacteriocins (results not shown).
In view of previous studies, our data imply that at least certain aspects of the response to bile are conserved among gram-positive bacteria. However, contrary to previous reports, this study does not reflect any impact of bile on DNA repair and oxidative response in E. faecalis V583. Intrinsic resistance to bile has also previously been associated with the presence of BSH activity in gram-positive bacteria (6, 8), but none of the putative bsh homologs in V583 (EF0521 and EF3005) showed significant differential expression in our experiments.
Comparison of the three different transcriptional profiles (V583-BB, V583-SDS, and V583-BB-SDS) revealed only 24 genes that were differentially transcribed in all three experiments. In V583-SDS and V583-BB, 68 genes were differentially expressed during both treatments, 38 of which showed similar expression patterns (either up- or downregulated) in both experiments (see Table S5 in the supplemental material). Cross-protection is generally regarded as a result of different stress conditions causing similar effects on cellular physiology. However, in a previous report by Flahaut et al. (13), the responses of E. faecalis to treatments with bile salts and SDS appeared to be less related on the protein level than suggested by the observed cross-resistance. The low number of commonly differentially expressed genes in the two transcriptional profiles presented here suggests that the responses to bile and SDS in E. faecalis involve different changes, also on the transcriptional level. So, although the toxicity pattern of bile acids resembles that of detergents such as SDS, a mechanism of resistance common to both remains to be established.
Supplementary Material
Acknowledgments
This work was financially supported by grants from the Research Council of Norway to I.F.N.
We thank Bjørn E. Kristiansen, The Norwegian Microarray Consortium, Oslo, for printing and scanning of the microarray slides.
Footnotes
Published ahead of print on 27 July 2007.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Aakra, A., O. L. Nyquist, L. Snipen, T. S. Reiersen, and I. F. Nes. 2007. Survey of genomic diversity among Enterococcus faecalis strains by microarray-based comparative genomic hybridization. Appl. Environ. Microbiol. 73:2207-2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aakra, A., H. Vebo, L. Snipen, H. Hirt, A. Aastveit, V. Kapur, G. Dunny, B. E. Murray, and I. F. Nes. 2005. Transcriptional response of Enterococcus faecalis V583 to erythromycin. Antimicrob. Agents Chemother. 49:2246-2259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arous, S., K. Dalet, and Y. Hechard. 2004. Involvement of the mpo operon in resistance to class IIa bacteriocins in Listeria monocytogenes. FEMS Microbiol. Lett. 238:37-41. [DOI] [PubMed] [Google Scholar]
- 4.Aspedon, A., and K. W. Nickerson. 1993. A two-part energy burden imposed by growth of Enterobacter cloacae and Escherichia coli in sodium dodecyl sulfate. Can. J. Microbiol. 39:555-561. [DOI] [PubMed] [Google Scholar]
- 5.Barcelona-Andrés, B., A. Marina, and V. Rubio. 2002. Gene structure, organization, expression, and potential regulatory mechanisms of arginine catabolism in Enterococcus faecalis. J. Bacteriol. 184:6289-6300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Begley, M., C. G. Gahan, and C. Hill. 2002. Bile stress response in Listeria monocytogenes LO28: adaptation, cross-protection, and identification of genetic loci involved in bile resistance. Appl. Environ. Microbiol. 68:6005-6012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Begley, M., C. G. Gahan, and C. Hill. 2005. The interaction between bacteria and bile. FEMS Microbiol. Rev. 29:625-651. [DOI] [PubMed] [Google Scholar]
- 8.Bron, P. A., M. Marco, S. M. Hoffer, E. Van Mullekom, W. M. de Vos, and M. Kleerebezem. 2004. Genetic characterization of the bile salt response in Lactobacillus plantarum and analysis of responsive promoters in vitro and in situ in the gastrointestinal tract. J. Bacteriol. 186:7829-7835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bron, P. A., D. Molenaar, W. M. De Vos, and M. Kleerebezem. 2006. DNA micro-array-based identification of bile-responsive genes in Lactobacillus plantarum. J. Appl. Microbiol. 100:728-738. [DOI] [PubMed] [Google Scholar]
- 10.Brown, M. H., and R. A. Skurray. 2001. Staphylococcal multidrug efflux protein QacA. J. Mol. Microbiol. Biotechnol. 3:163-170. [PubMed] [Google Scholar]
- 11.Ennahar, S., T. Sashihara, K. Sonomoto, and A. Ishizaki. 2000. Class IIa bacteriocins: biosynthesis, structure and activity. FEMS Microbiol. Rev. 24:85-106. [DOI] [PubMed] [Google Scholar]
- 12.Facklam, R. R., M. Carvalho, and L. M. Teixeira. 2002. History, taxonomy, biochemical characteristics and antibiotic susceptibility testing of enterococci, p. 1-54. In M. S. Gilmore (ed.), The enterococci: pathogenesis, molecular biology and antibiotic resistance. ASM Press, Washington, DC.
- 13.Flahaut, S., J. Frere, P. Boutibonnes, and Y. Auffray. 1996. Comparison of the bile salts and sodium dodecyl sulfate stress responses in Enterococcus faecalis. Appl. Environ. Microbiol. 62:2416-2420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Flahaut, S., A. Hartke, J. C. Giard, A. Benachour, P. Boutibonnes, and Y. Auffray. 1996. Relationship between stress response toward bile salts, acid and heat treatment in Enterococcus faecalis. FEMS Microbiol. Lett. 138:49-54. [DOI] [PubMed] [Google Scholar]
- 15.Flores, C., I. Maguilnik, E. Hadlich, and L. Z. Goldani. 2003. Microbiology of choledochal bile in patients with choledocholithiasis admitted to a tertiary hospital. J. Gastroenterol. Hepatol. 18:333-336. [DOI] [PubMed] [Google Scholar]
- 16.Hofmann, A. 1998. Bile secretion and the enterohepatic circulation of bile acids, p. 937-948. In F. Feldman and M. Sleisenger (ed.), Sleisenger and Fordtran's gastrointestinal and liver disease, 6th ed. W. B. Saunders Co., Philadelphia, PA.
- 17.Hofmann, A. F. 1994. Bile acids, p. 677-718. In I. M. Arias (ed.), The liver: biology and pathobiology, 3rd. ed. Raven Press Ltd., New York, NY.
- 18.Keyhani, J., E. Keyhani, F. Attar, and A. Haddadi. 2006. Sensitivity to detergents and plasmid curing in Enterococcus faecalis. J. Ind. Microbiol. Biotechnol. 33:238-242. [DOI] [PubMed] [Google Scholar]
- 19.Kimoto, H., S. Ohmomo, and T. Okamoto. 2002. Enhancement of bile tolerance in lactococci by Tween 80. J. Appl. Microbiol. 92:41-46. [DOI] [PubMed] [Google Scholar]
- 20.Le Breton, Y., A. Maze, A. Hartke, S. Lemarinier, Y. Auffray, and A. Rince. 2002. Isolation and characterization of bile salts-sensitive mutants of Enterococcus faecalis. Curr. Microbiol. 45:434-439. [DOI] [PubMed] [Google Scholar]
- 21.Le Breton, Y., C. Muller, Y. Auffray, and A. Rincé. 2007. New insights into the Enterococcus faecalis CroRS two-component system obtained using a differential-display random arbitrarily primed PCR approach. Appl. Environ. Microbiol. 73:3738-3741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Le Maire, M., P. Champeil, and J. V. Moller. 2000. Interaction of membrane proteins and lipids with solubilizing detergents. Biochim. Biophys. Acta 1508:86-111. [DOI] [PubMed] [Google Scholar]
- 23.Leverrier, P., D. Dimova, V. Pichereau, Y. Auffray, P. Boyaval, and G. Jan. 2003. Susceptibility and adaptive response to bile salts in Propionibacterium freudenreichii: physiological and proteomic analysis. Appl. Environ. Microbiol. 69:3809-3818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lin, J., O. Sahin, L. O. Michel, and Q. Zhang. 2003. Critical role of multidrug efflux pump CmeABC in bile resistance and in vivo colonization of Campylobacter jejuni. Infect. Immun. 71:4250-4259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lomovskaya, O., and K. Lewis. 1992. Emr, an Escherichia coli locus for multidrug resistance. Proc. Natl. Acad. Sci. USA 89:8938-8942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lu, Y. J., and C. O. Rock. 2006. Transcriptional regulation of fatty acid biosynthesis in Streptococcus pneumoniae. Mol. Microbiol. 59:551-566. [DOI] [PubMed] [Google Scholar]
- 27.Mortvedt, C. I., and I. F. Nes. 1990. Plasmid-associated bacteriocin production by a Lactobacillus-sake strain. J. Gen. Microbiol. 136:1601-1607. [Google Scholar]
- 28.Muller, C., Y. Le Breton, T. Morin, A. Benachour, Y. Auffray, and A. Rince. 2006. The response regulator CroR modulates expression of the secreted stress-induced SalB protein in Enterococcus faecalis. J. Bacteriol. 188:2636-2645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Paulsen, I. T., L. Banerjei, G. S. Myers, K. E. Nelson, R. Seshadri, T. D. Read, D. E. Fouts, J. A. Eisen, S. R. Gill, J. F. Heidelberg, H. Tettelin, R. J. Dodson, L. Umayam, L. Brinkac, M. Beanan, S. Daugherty, R. T. DeBoy, S. Durkin, J. Kolonay, R. Madupu, W. Nelson, J. Vamathevan, B. Tran, J. Upton, T. Hansen, J. Shetty, H. Khouri, T. Utterback, D. Radune, K. A. Ketchum, B. A. Dougherty, and C. M. Fraser. 2003. Role of mobile DNA in the evolution of vancomycin-resistant Enterococcus faecalis. Science 299:2071-2074. [DOI] [PubMed] [Google Scholar]
- 30.Pfeiler, E. A., M. A. Azcarate-Peril, and T. R. Klaenhammer. 2007. Characterization of a novel bile-inducible operon encoding a two-component regulatory system in Lactobacillus acidophilus. J. Bacteriol. 189:4624-4634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Phan-Thanh, L., and T. Gormon. 1997. Stress proteins in Listeria monocytogenes. Electrophoresis 18:1464-1471. [DOI] [PubMed] [Google Scholar]
- 32.Sahm, D. F., J. Kissinger, M. S. Gilmore, P. R. Murray, R. Mulder, J. Solliday, and B. Clarke. 1989. In vitro susceptibility studies of vancomycin-resistant Enterococcus faecalis. Antimicrob. Agents Chemother. 33:1588-1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Saito, T., K. Senda, S. Takakura, N. Fujihara, T. Kudo, Y. Iinuma, T. Kiuchi, M. Tanimoto, and S. Ichiyama. 2003. Biliary bacteria in living related liver transplant recipients: microbiology and rapid detection system using flow cytometry. Clin. Chem. Lab. Med. 41:159-163. [DOI] [PubMed] [Google Scholar]
- 34.Sánchez, B., M. C. Champomier-Verges, P. Anglade, F. Baraige, C. G. de Los Reyes-Gavilan, A. Margolles, and M. Zagorec. 2005. Proteomic analysis of global changes in protein expression during bile salt exposure of Bifidobacterium longum NCIMB 8809. J. Bacteriol. 187:5799-5808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sánchez, B., C. G. de los Reyes-Gavilan, and A. Margolles. 2006. The F1F0-ATPase of Bifidobacterium animalis is involved in bile tolerance. Environ. Microbiol. 8:1825-1833. [DOI] [PubMed] [Google Scholar]
- 36.Taranto, M. P., M. L. Fernandez Murga, G. Lorca, and G. F. de Valdez. 2003. Bile salts and cholesterol induce changes in the lipid cell membrane of Lactobacillus reuteri. J. Appl. Microbiol. 95:86-91. [DOI] [PubMed] [Google Scholar]
- 37.Thanassi, D. G., L. W. Cheng, and H. Nikaido. 1997. Active efflux of bile salts by Escherichia coli. J. Bacteriol. 179:2512-2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Waar, K., H. C. van der Mei, H. J. Harmsen, J. E. Degener, and H. J. Busscher. 2002. Adhesion to bile drain materials and physicochemical surface properties of Enterococcus faecalis strains grown in the presence of bile. Appl. Environ. Microbiol. 68:3855-3858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ward, D. E., R. P. Ross, C. C. van der Weijden, J. L. Snoep, and A. Claiborne. 1999. Catabolism of branched-chain α-keto acids in Enterococcus faecalis: the bkd gene cluster, enzymes, and metabolic route. J. Bacteriol. 181:5433-5442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ward, D. E., C. C. van Der Weijden, M. J. van Der Merwe, H. V. Westerhoff, A. Claiborne, and J. L. Snoep. 2000. Branched-chain α-keto acid catabolism via the gene products of the bkd operon in Enterococcus faecalis: a new, secreted metabolite serving as a temporary redox sink. J. Bacteriol. 182:3239-3246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yokota, A., M. Veenstra, P. Kurdi, H. W. van Veen, and W. N. Konings. 2000. Cholate resistance in Lactococcus lactis is mediated by an ATP-dependent multispecific organic anion transporter. J. Bacteriol. 182:5196-5201. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.