Abstract
We investigated the fine-scale population structure of the “Candidatus Accumulibacter” lineage in enhanced biological phosphorus removal (EBPR) systems using the polyphosphate kinase 1 gene (ppk1) as a genetic marker. We retrieved fragments of “Candidatus Accumulibacter” 16S rRNA and ppk1 genes from one laboratory-scale and several full-scale EBPR systems. Phylogenies reconstructed using 16S rRNA genes and ppk1 were largely congruent, with ppk1 granting higher phylogenetic resolution and clearer tree topology and thus serving as a better genetic marker than 16S rRNA for revealing population structure within the “Candidatus Accumulibacter” lineage. Sequences from at least five clades of “Candidatus Accumulibacter” were recovered by ppk1-targeted PCR, and subsequently, specific primer sets were designed to target the ppk1 gene for each clade. Quantitative real-time PCR (qPCR) assays using “Candidatus Accumulibacter”-specific 16S rRNA and “Candidatus Accumulibacter” clade-specific ppk1 primers were developed and conducted on three laboratory-scale and nine full-scale EBPR samples and two full-scale non-EBPR samples to determine the abundance of the total “Candidatus Accumulibacter” lineage and the relative distributions and abundances of the five “Candidatus Accumulibacter” clades. The qPCR-based estimation of the total “Candidatus Accumulibacter” fraction as a proportion of the bacterial community as measured using 16S rRNA genes was not significantly different from the estimation measured using ppk1, demonstrating the power of ppk1 as a genetic marker for detection of all currently defined “Candidatus Accumulibacter” clades. The relative distributions of “Candidatus Accumulibacter” clades varied among different EBPR systems and also temporally within a system. Our results suggest that the “Candidatus Accumulibacter” lineage is more diverse than previously realized and that different clades within the lineage are ecologically distinct.
Phosphorus (P) is the critical factor leading to the eutrophication of many surface waters. Enhanced biological phosphorus removal (EBPR) has been applied for more than 3 decades to achieve low P levels in treated wastewater effluents. EBPR employs bacteria called polyphosphate [poly(P)]-accumulating organisms (PAOs), which can take up large amounts of phosphate and store it intracellularly in the form of poly(P) under cyclic anaerobic and aerobic conditions (14). Although EBPR is an economic and environmentally friendly means for achieving low effluent-P concentrations, this process is not always stable and troubleshooting is often not effective, due to an incomplete understanding of EBPR microbiology (15). Extensive research has been conducted to discover the identities of PAOs in an effort to understand the biochemical mechanisms involved and to further promote conditions favoring P removal. To date, no confirmed PAO exhibiting all of the characteristics observed during EBPR has been cultivated in isolation. Culture-independent approaches have identified bacteria affiliated with the Rhodocyclus group within the Betaproteobacteria as the primary PAOs in laboratory-scale reactors (3, 7), and members have been named “Candidatus Accumulibacter phosphatis.” Surveys of full-scale EBPR wastewater treatment plants (WWTPs) have confirmed the presence of bacteria related to “Candidatus Accumulibacter phosphatis,” but not all of them accumulate poly(P) at all times (10, 29). This observation, along with the disagreement regarding the ability of “Candidatus Accumulibacter” to reduce nitrate (4, 10), suggests finer phenotypic and ecological differences among members of this lineage.
Previous studies of the occurrence and phylogeny of “Candidatus Accumulibacter” in EBPR systems primarily relied on the use of 16S rRNA-based methods. A bacterial species defined using the most commonly cited cutoff (≥97% 16S rRNA gene sequence identity) (21) may sometimes contain microorganisms with substantial phenotypic and genetic differences. Although Stackebrandt and Ebers recommended 98.7 to 99% as the new taxonomic parameter to replace the old value of 97% (20), PCR and sequencing errors make it difficult to identify whether two microorganisms are truly from two different species by using this new cutoff. Cohan has proposed the “ecotype” concept and suggested that a typical named species contains many ecotypes, with each ecotype comprising a set of strains occupying the same ecological niche (2). Compared with a traditional species concept or definition, ecotypes are expected to be more biologically meaningful for studying microbial ecology. In the case of “Candidatus Accumulibacter,” some 16S rRNA sequences previously recovered from geographically and temporally distinct EBPR systems were nearly identical (4). We asked the question “Do these sequences represent essentially identical ‘Candidatus Accumulibacter’ populations or do they represent different ecotypes that have genotypic and/or phenotypic differences among populations and/or across EBPR systems?” A more divergent genetic locus than the 16S rRNA gene was required to study such fine-scale differences among “Candidatus Accumulibacter” populations.
Poly(P) kinase 1 (PPK1) (1) and poly(P) kinase 2 (PPK2) (28) are two different enzymes that have been identified to catalyze poly(P) synthesis in model organisms such as Escherichia coli. PPK1 catalyzes the reversible reaction of poly(P) formation from ATP, with the forward reaction preferred for synthesis of poly(P). In contrast, PPK2, an enzyme smaller than PPK1, operates in reverse to generate GTP from poly(P) and GDP (28). Metagenomic analysis of “Candidatus Accumulibacter”-enriched laboratory-scale reactors revealed that “Candidatus Accumulibacter” possessed genes encoding PPK1 and PPK2, and both genes were present as a single copy (4). These two enzymes may be important in poly(P) transformation by “Candidatus Accumulibacter” during EBPR. Previously, McMahon and coworkers designed degenerate PCR primers to retrieve ppk1 fragments from a “Candidatus Accumulibacter”-enriched laboratory-scale EBPR reactor (12) and several full-scale WWTPs (13), and a ppk1 primer set specifically targeting “Candidatus Accumulibacter” was further designed and validated (13). Because ppk1 appears to be evolving faster than 16S rRNA genes in “Candidatus Accumulibacter” (V. Kunin, S. He, F. Warnecke, S. B. Peterson, H. G. Martin, M. Haynes, N. Ivanova, L. L. Blackall, M. Breitbart, F. Rohwer, K. D. McMahon, and P. Hugenholtz, submitted for publication), ppk1 may provide enough resolution to observe fine-scale differences within this lineage (13). Thus, one objective of this study was to demonstrate the use of ppk1 as a genetic marker for studying the more resolved population structure of “Candidatus Accumulibacter” in different EBPR systems. For this purpose, “Candidatus Accumulibacter” 16S rRNA and ppk1 gene fragments were retrieved from both laboratory-scale and full-scale EBPR systems to reconstruct “Candidatus Accumulibacter” phylogenies. Quantitative real-time PCR (qPCR) methods were then developed to measure the abundances and relative distributions of “Candidatus Accumulibacter” clades to uncover the fine-scale population structure and provide more information about the ecology of this biotechnologically important group of organisms.
MATERIALS AND METHODS
Samples.
Two laboratory-scale, acetate-fed sequencing batch reactors, the University of Wisconsin—Madison (UWM) and University of California—Berkeley (UCB) reactors, were originally inoculated with activated sludge from the Nine Springs (NS) WWTP, Madison, WI, and the Southeast Water Pollution Control Plant, San Francisco, CA, respectively. Both reactors, with 4-day solid retention times (SRTs) and chemical oxygen demand/P mass ratios of 14, were operated under conditions nearly identical, except for differences in inoculation source, reactor volume, and laboratory location (5, 12). For the UWM reactor, samples were collected after reactor operation for 10 months and 2 years and designated samples UWMH and UWMS, respectively.
Activated sludge samples were also collected from nine full-scale EBPR facilities (CC, OL, SJ, NAN, VIR, DUR, NS, LV1, and LV2) and two conventional activated sludge plants not configured to perform EBPR (EB and OSK). Some important characteristics of these treatment plants are summarized in Table 1. More-detailed treatment plant configuration and performance information is available elsewhere (6, 13).
TABLE 1.
Treatment plant characteristics at time of samplinga
| Facility(ies) | Location | Configuration | Size (MGD) | SRT (no. of days) |
|---|---|---|---|---|
| CC | Martinez, CA | High-rate anaerobic selector (AN/AE) | 45 | 1.5 |
| OL | San Leandro, CA | High-rate anaerobic selector (AN/AE) | 15 | 2.5 |
| SJ | Alviso, CA | Four-stage AX/AE with step feed | 118 | 7.5 |
| NAN | Suffolk, VA | VIP (AN/AX/AE) | 20 | 8.5 |
| VIR | Norfolk, VA | VIP (AN/AX/AE) | 30 | 9.5 |
| DUR | Durham, OR | A2O (AN/AX/AE) | 22 | 11 |
| LV1, LV2b | Las Vegas, NV | Modified UCT (AN/AX/AE) | 30 | 8.4 |
| NS | Madison, WI | Combined UCT (AN/AX/AE) and A/O (AN/AE) | 42 | 9 |
| EB | Oakland, CA | Conventional activated sludge | 80 | 1.4 |
| OSK | Oshkosh, WI | Conventional activated sludge | 12 | ∼6 to 10 |
More-detailed operational conditions and performance information for treatment plants DUR, NS, VIR, NAN, LV1, and LV2 (designated facilities A, B, C, D, E1, and E2, respectively) were described elsewhere (6). More-detailed information about treatment plants CC, OL, SL, and EB were described elsewhere (13). Samples in those studies were taken at the same time as the corresponding samples in the current study. AN, anaerobic; AX, anoxic (with nitrate as the electron acceptor); AE, aerobic; VIP, Virginia Initiative Plant; A2O, anaerobic-anoxic-oxic; A/O, anaerobic-oxic; MGD, millions of gallons per day.
LV1 and LV2 were parallel trains in the same WWTP with different SRT histories. Samples were taken 5 days after the SRTs in both trains were switched to the same value (8.4 days).
Among the 11 EBPR systems (2 laboratory scale and 9 full scale), samples UCB, CC, OL, and SJ were previously included by McMahon et al. (12, 13) while studying ppk1 gene phylogeny. Therefore, in the current study, the retrieval of “Candidatus Accumulibacter” 16S rRNA and ppk1 genes was performed only on the other EBPR systems, but qPCR analysis was performed on all samples.
Genomic DNA extraction.
Genomic DNA was extracted by a modified enzyme digestion method followed by phenol-chloroform purification and isopropanol precipitation (18). DNA was further purified by precipitation with sodium acetate and ethanol to remove RNA contamination and other PCR-inhibitory materials. DNA integrity was checked by agarose gel electrophoresis, and purity was evaluated by spectrometry at 260 nm and 280 nm. DNA concentrations were measured by a PicoGreen double-stranded-DNA assay kit (Invitrogen, Carlsbad, CA) on a Gemini SpectraMax spectrofluorometer (Bucher Biotech, Basel, Switzerland).
16S rRNA gene retrieval and phylogenetic analysis.
Retrieval of 16S rRNA genes from “Candidatus Accumulibacter” was described elsewhere (5). Briefly, the 16S and internally transcribed spacer region of the rRNA operon was amplified from community genomic DNA from samples UWMH, NAN, VIR, DUR, LV1, and LV2 by using bacterium-specific 8f and 23Sr primers (16). Ninety-six clones from each library were picked randomly. For sample UWMH, unique clones were determined by restriction fragment length polymorphism (RFLP). For the other samples, “Candidatus Accumulibacter”-like clones were identified by screening with real-time PCR (24), using a mixture of forward primers (RHC 439f, PAO 651f, and PAO 846bf) (3, 7, 29) and 1492r (16). The sequences were grouped into operational taxonomic units (OTUs) based on 99% DNA sequence identity within each library. The sequences were aligned with the GCG version 11.1 software package (Accelrys, CA), and a mask was generated using a 33% majority rule. MrBayes version 3.1.2 was used for Bayesian analyses (8) with a general-time-reversible model with gamma-distributed rate variation. A total of 3,000,000 generations were run, with sampling every 100 generations, to create 30,000 trees. The consensus tree was generated by setting the burn-in value to discard trees generated before the chain reached stabilization.
ppk1 clone library construction and phylogenetic analysis.
Amplification of “Candidatus Accumulibacter” ppk1 fragments was carried out on genomic DNA extracted from samples UWMH, UWMS, NAN, VIR, DUR, NS, LV1, and LV2. The 25-μl reaction mixtures contained 1× PCR buffer II, 3.0 mM MgCl2, 200 μM of each deoxynucleoside triphosphate, 400 nM of each forward and reverse primer (Acc-ppk1-254f and Acc-ppk1-1376r) (13), 5% of dimethyl sulfoxide, and 0.05 U/μl of AmpliTaq Gold DNA polymerase (Applied Biosystems, Foster City, CA). The PCR was conducted on an iCycler (Bio-Rad, Hercules, CA), with the program consisting of an initial 10-min denaturation step at 95°C, followed by 25 cycles of 95°C for 45 s, 68°C for 1 min, and 72°C for 2 min, and then a final extension at 72°C for 5 min. To minimize the artifacts from PCR, a reconditioning step was conducted by adding 2.5 μl of the PCR product resulting from the amplification described above to 22.5 μl of a fresh reaction mixture with the same composition and five cycles were conducted using the parameters specified above (22).
The amplified ppk1 fragments were then purified from agarose gels and cloned using a TOPO TA cloning kit (Invitrogen, CA) according to the manufacturer's instructions. From each library (except UWMH and UWMS, from which 96 clones were picked), 30 to 45 clones were randomly picked and subsequently screened by RFLP analysis with MspI (Promega, Madison, WI) to identify unique sequence types. Representatives from each unique genotype defined by RFLP patterns were sequenced, and these sequences were further grouped into OTUs based on 99% identity of DNA sequences within each library. Sequence alignment and phylogenetic tree reconstruction were conducted as described above.
Primer design.
Primer sets were designed to exclusively target the ppk1 gene of each “Candidatus Accumulibacter” clade (Table 2). Specificity was checked against sequences available in GenBank. For accurate quantification using qPCR, primers were designed to avoid long amplicon length and degenerate bases. Specificity was ensured for both forward and reverse primers for all clades except IIC. OTU NS D3 in clade IIC was only distantly related to other sequences in this clade and had sequence regions similar to those in other clades, making it difficult to design a primer set targeting the whole clade without using degenerate bases. Primer Acc-ppk1-460r combined with general “Candidatus Accumulibacter” ppk1 primer Acc-ppk1-254f (13) can target the whole clade, but it has only one mismatch with sequences in clade I. Therefore, primer Acc-ppk1-1123f was also designed to combine with general “Candidatus Accumulibacter” ppk1 primer Acc-ppk1-1376r (13) to target the clade to the exclusion of OTU NS D3. Qualitative PCR was performed on all samples, using all the ppk1 primer sets designed in this study. Single bands on an agarose gel at the expected amplicon sizes were obtained for all the positive results (data not shown), indicating that the amplification was from ppk1 genes.
TABLE 2.
Primer information, qPCR conditions, and performancea
| Primer | Sequence (5′-3′) | Target | Amplicon size (bp) | No. of mismatches with NCb | qPCR condition
|
qPCR performance
|
||||
|---|---|---|---|---|---|---|---|---|---|---|
| Primer concn (nM) | Betaine concn (M) | Ta (oC) | Standard curve correlation coefficient | PCR efficiency (%)c | Specific fractioning limit (%)d | |||||
| Acc-ppk1-763f | GACGAAGAAGCGGTCAAG | Acc-I ppk1 | 408 | 2 | 500 | 0 | 61 | 0.999 ± 0.001 | 94.2 ± 1.4 | 0.001 |
| Acc-ppk1-1170r | AACGGTCATCTTGATGGC | 2 | ||||||||
| Acc-ppk1-893f | AGTTCAATCTCACCGAGAGC | Acc-IIA ppk1 | 105 | 2 | 550 | 0.7 | 61 | 1.000 ± 0.001 | 90.7 ± 1.0 | 1 |
| Acc-ppk1-997r | GGAACTTCAGGTCGTTGC | 1 | ||||||||
| Acc-ppk1-870f | GATGACCCAGTTCCTGCTCG | Acc-IIB ppk1 | 133 | 2 | 400 | 0 | 61 | 1.000 ± 0.000 | 95.7 ± 0.4 | 0.001 |
| Acc-ppk1-1002r | CGGCACGAACTTCAGATCG | 2 | ||||||||
| Acc-ppk1-254fe | TCACCACCGACGGCAAGAC | Acc-IIC ppk1 | 207 | 0 | 400 | 1.0 | 66 | 0.999 ± 0.001 | 88.3 ± 4.1 | 10 |
| Acc-ppk1-460r | CCGGCATGACTTCGCGGAAG | 1 | ||||||||
| Acc-ppk1-1123f | GAACAGTCCGCCAACGACC | Acc-IIC ppk1 excluding OTU NS D3 | 254 | 2 | 500 | 0.7 | 63 | 1.000 ± 0.001 | 91.2 ± 2.1 | 0.1 |
| Acc-ppk1-1376re | ACGATCATCAGCATCTTGGC | 0 | ||||||||
| Acc-ppk1-375f | GGGTATCCGTTTCCTCAAGCG | Acc-IID ppk1 | 148 | 2 | 400 | 0.7 | 63 | 0.999 ± 0.001 | 91.3 ± 1.3 | 1 |
| Acc-ppk1-522r | GAGGCTCTTGTTGAGTACACGC | 1 | ||||||||
| 518f | CCAGCAGCCGCGGTAAT | Acc 16S rRNA genes | 351 | 0 | 400 | 0 | 65 | 0.999 ± 0.001 | 92.6 ± 2.8 | NA |
| PAO-846r | GTTAGCTACGGCACTAAAAGG | 2 | ||||||||
| 341f | CCTACGGGAGGCAGCAG | Bacterial 16S rRNA genes | 194 | NA | 500 | 0 | 60 | 0.999 ± 0.001 | 90.7 ± 0.6 | NA |
| 534r | ATTACCGCGGCTGCTGG | NA | ||||||||
Acc, “Candidatus Accumulibacter”; NA, not applicable; Ta, annealing temperature.
NC indicates the negative control, which is a plasmid containing a nontarget “Candidatus Accumulibacter” ppk1 fragment that has the fewest mismatches with the ppk1 primer set or, in the case of primer PAO-846r, the plasmid containing non-“Candidatus Accumulibacter” 16S rRNA with the fewest mismatches.
PCR efficiency was estimated from the slope of the standard curve by the formula 10−1/slope − 1.
The specific fractioning limit is the specific detection limit for the fraction of an “Candidatus Accumulibacter” clade in the total “Candidatus Accumulibacter” lineage (see text).
Acc-ppk1-254f and Acc-ppk1-1376r are the primers that target total “Candidatus Accumulibacter” ppk1 genes (13).
qPCR was also used to quantify 16S rRNA genes from total bacteria and “Candidatus Accumulibacter” in order to estimate the relative abundance of the total “Candidatus Accumulibacter” lineage in the bacterial community. Primer set 341f/534r was used to quantify total bacterial 16S rRNA genes (26). Previously, a probe mixture (PAO 462, PAO 651, and PAO 846) was developed to detect “Candidatus Accumulibacter”-related PAOs by fluorescent in situ hybridization (FISH) (3). To quantify total “Candidatus Accumulibacter” 16S rRNA genes by qPCR, we chose universal primer 518f (reverse complementary to 534r) (26) as the forward primer and PAO-846r as the reverse primer because all the “Candidatus Accumulibacter” 16S rRNA sequences retrieved from this study matched perfectly with probe PAO 846 but had various mismatches with PAO 462 or PAO 651.
qPCR conditions.
qPCR was conducted on an iCycler (Bio-Rad, Hercules, CA) using iQ SYBR green supermix (Bio-Rad, Hercules, CA) with a total reaction volume of 25 μl. All qPCR programs consisted of an initial 3-min denaturation at 95°C, followed by 45 cycles of denaturation at 94°C for 30 s, annealing for 45 s, and extension at 72°C for 30 s. The annealing temperature, primer concentration, and denaturant addition were optimized for PCR efficiency and specificity (Table 2).
Six-point calibration curves for qPCR were produced by 10-fold serial dilution of positive controls in duplicate within each assay, at 103 to 108 target copies per reaction. Controls for ppk1- or 16S rRNA-targeted PCR were generated from appropriate clones from ppk1 or 16S plus internal transcribed spacer rRNA clone libraries. Briefly, plasmid was extracted from the clones by using a QIAGEN plasmid mini kit (QIAGEN, Valencia, CA) and linearized with restriction enzyme ScaI digestion (Promega, Madison, WI). The completion of restriction digestion was checked by gel electrophoresis. The plasmid was purified with a QIAquick PCR purification kit (QIAGEN), and mass concentration was determined by the PicoGreen double-stranded-DNA quantification kit (Invitrogen, Carlsbad, CA). Copy number was calculated based on the mass concentration and the average molecular weight of the plasmid.
For all unknown samples, 5 ng of community-derived genomic DNA was added as the template. In each assay, a no-template control was included to check for contamination and primer-dimer formation. To avoid nonspecific cross-detection, a negative control (a plasmid containing a nontarget ppk1 fragment with the fewest mismatches to the primer set) was also included and applied at 106 copies per reaction.
Data analysis.
Quantification was performed using iCycler iQ optical system software version 3.0a. We found that the quantifications were slightly different when the threshold was set at different relative fluorescence unit values. Therefore, for all assays, the cycle threshold was determined by setting the threshold at a relative fluorescence unit value of 50, where the fluorescence was above the background noise and in early exponential phase. PCR amplification efficiency was estimated from the slope of the standard curve by the formula 10−1/slope − 1. To account for intra- and inter-qPCR assay variation, both standard curve and samples were run on triplicate plates, with duplicate reactions within each plate. Standard deviations were calculated from the averages for the triplicate runs.
Descriptors of “Candidatus Accumulibacter” lineage diversity.
Measurements of clade diversity and evenness within the “Candidatus Accumulibacter” lineage were calculated for individual samples by using the Shannon index (H = −∑Pi ln Pi) (19) and the Pielou regularity index (R = H/lnS) (17), respectively. The relative abundance of each clade as determined by qPCR (normalized to total “Candidatus Accumulibacter” ppk1 abundance) was used as a proxy for relative species abundance (Pi), and the number of clades that were detected by qPCR was used as the richness value (S) in the above equations.
Nucleotide sequence accession numbers.
The GenBank accession numbers for the nucleotide sequences determined in this study are EF565147 to EF565162 and EF559317 to EF559355.
RESULTS
Phylogeny reconstructed by 16S rRNA and ppk1 genes.
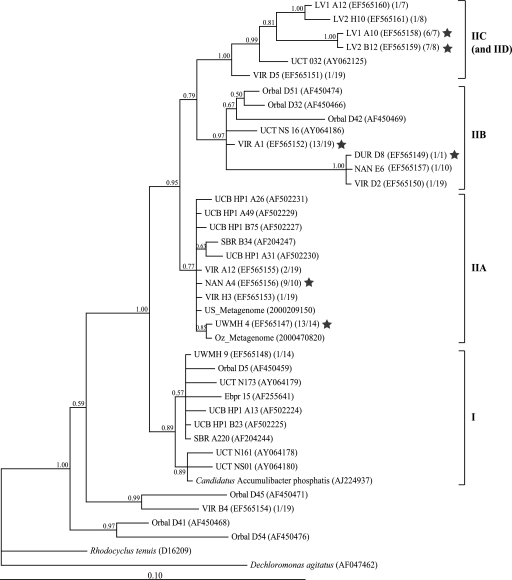
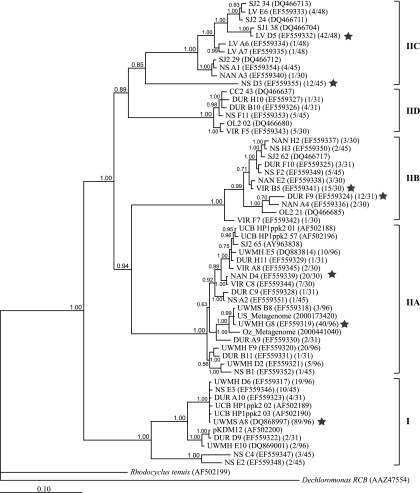
We were able to retrieve “Candidatus Accumulibacter” 16S rRNA and ppk1 genes from each EBPR sample with which clone libraries were constructed. Figure 1 shows a phylogram of 16S rRNA genes from this study and some relevant sequences from other EBPR studies. Figure 2 shows phylogenies of “Candidatus Accumulibacter” ppk1 from UWMH, UWMS, NAN, VIR, DUR, LV1, LV2, and NS (this study); CC, OL, and SJ (13); and UCB (12). The 16S rRNA and ppk1 sequences from metagenomic analysis of “Candidatus Accumulibacter”-enriched laboratory-scale reactors in the United States and Australia (4) (designated as US_Metagenome and Oz_Metagenome, respectively) were also included in the phylogenies presented in Fig. 1 and 2. At least four distinct 16S rRNA clades and five distinct ppk1 clades appeared to comprise the “Candidatus Accumulibacter” lineage, as shown in Fig. 1 and 2. Previously, McMahon and coworkers defined type I and type II “Candidatus Accumulibacter” clades by their ppk1 sequences (12, 13). Because no pure-culture representatives are available from the “Candidatus Accumulibacter” lineage, it is difficult to link 16S rRNA phylogeny to that of ppk1. However, due to the following similar traits shared by both phylogenies, we were able to relate the 16S rRNA clades to ppk1 clades. (i) In agreement with 16S rRNA phylogeny, ppk1 genes from laboratory-scale reactors were affiliated only with clades I and IIA (previously called types I and II). (ii) Both 16S rRNA and ppk1 sequences from metagenomic libraries (US_Metagenome and Oz_Metagenome) were affiliated with clade IIA. (iii) When clone frequencies were available, it was observed that the OTU with the highest 16S rRNA clone frequency in a sample affiliated with the clade that contained the OTU with the highest ppk1 clone frequency in that sample. (iv) Specifically, 16S rRNA and ppk1 sequences from samples LV1 and LV2 were found only in clade IIC. Therefore, we proposed the same clade names for 16S rRNA and ppk1 phylogenies and determined that the trees were largely congruent.
FIG. 1.
Unrooted phylogenetic tree for “Candidatus Accumulibacter”-related 16S rRNA sequences. A total of 1,022 bp (E. coli 16S rRNA positions 439 to 1459) were included for phylogenetic analysis. The posterior probabilities are shown next to the nodes. The first number in the second set of parentheses indicates the number of clones belonging to this OTU, and the second number indicates the total number of “Candidatus Accumulibacter” clones in that sample. The star indicates the OTU with the highest clone frequency in that library. Sequence names starting with “UCT” were recovered from plant NS in the study by Zilles et al. (29). The scale bar indicates the number of changes per site.
FIG. 2.
Phylogram indicating inferred relatedness of ppk1 genes from the “Candidatus Accumulibacter” lineage. A total of 1,080 bp (“Candidatus Accumulibacter” ppk1 [GenBank accession number AF502200] positions 265 to 1344) were included for Bayesian analysis. The posterior probabilities are shown next to the nodes. The first number in the second set of parentheses indicates the number of clones belonging to this OTU, and the second number is the total number of “Candidatus Accumulibacter” ppk1 clones retrieved from that sample. The star indicates the OTU with the highest clone frequency in that library. Sequences from LV1 and LV2 are combined as OTU LV because all ppk1 sequences from LV2 were >99% identical to those from LV1. The scale bar indicates the number of changes per site.
It is noteworthy that about one-quarter of the ppk1 sequences recovered from sample NS belonged to OTU NS D3 in clade IIC. Interestingly, NS D3 was only distantly related to other sequences in this clade and had regions (usually with lengths of about 20 to 50 bp) similar to those of other clades at various locations along the sequence, such that it appeared to be a mosaic combination of all five clades. This sequence was unlikely to be a PCR artifact, as discussed below.
qPCR specificity and accuracy.
For accurate quantification by qPCR, high specificity and good amplification efficiency are necessary, as is an absence of inhibition by the DNA template. To avoid nonspecific cross-detection of ppk1 from other “Candidatus Accumulibacter” clades, we included an appropriate negative control for each clade-specific primer set. This control was a plasmid containing the nontarget “Candidatus Accumulibacter” ppk1 fragment that had the fewest mismatches with the forward and reverse primers (Table 2). The negative control was applied at 106 copies per reaction. The annealing temperature, primer concentration, and denaturant addition were tested in order to achieve high PCR efficiency and specificity. We found that increasing PCR stringency increased specificity, as indicated by decreased amplification from the negative control, but it also resulted in lower PCR efficiency. During qPCR, the reaction efficiency should range from 90% to 110% for accurate quantification. Therefore, we chose the reaction conditions providing the best specificity when the PCR efficiency was ≥90%. Specificity was controlled by setting a “specific fractioning limit,” which we defined as the specific detection limit for the fraction of a “Candidatus Accumulibacter” clade relative to total “Candidatus Accumulibacter” bacteria. This limit was determined by dividing the qPCR “quantified” ppk1 copy number in the negative control due to nonspecific amplification by the actual ppk1 copy number in the negative control (106). This gave an estimation of the chance that a nontarget template would be nonspecifically amplified if it were present in very high abundance. Therefore, when a measured relative clade abundance was lower than the specific fractioning limit, we could not distinguish whether the clade was truly present, albeit at a low abundance, or if the PCR primers were nonspecifically amplifying a related nontarget. This limit was conservative because it assumes that the nontarget with the fewest mismatches was always present in the sample. For most primer sets, the specific fractioning limit was ≤1% (Table 2). Therefore, we generally could specifically detect these clades when their relative abundances were higher than 1% of the total “Candidatus Accumulibacter” lineage abundance.
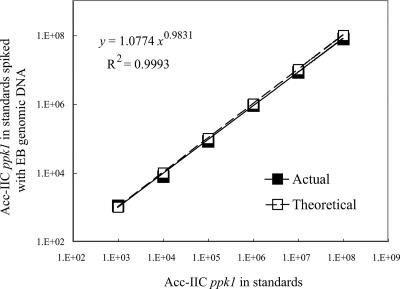
To determine whether or not the extracted genomic DNA contained contaminants that inhibit qPCR, a series of ppk1 standards were spiked with 5 ng of genomic DNA extracted from sample EB, which was a non-EBPR sample in which “Candidatus Accumulibacter” ppk1 was not detected by agarose gel electrophoresis or later qPCR results. Because all genomic DNA templates were extracted and further purified in the same way, EB DNA was representative of DNA templates from all other samples in this study. qPCR was conducted on the standards with and without the EB DNA spike. ppk1 abundance in the spiked standards was determined using the standard curve generated from the nonspiked standards. The ppk1 copy numbers quantified in the spiked standards were plotted against those in their nonspiked counterparts (Fig. 3). Both the slope and the exponent were close to 1.0, indicating that the assay was accurate and the inhibition of PCR from genomic DNA template was minimal (27).
FIG. 3.
Validation curve plotting the clade IIC ppk1 copy numbers quantified in the standards spiked with 5 ng of genomic DNA from sample EB against those of their nonspiked counterparts. The slope and exponent value are close to 1, indicating that the inhibitory effect on PCR from the genomic DNA template was negligible.
qPCR results.
The relative distribution of each “Candidatus Accumulibacter” clade was assessed by qPCR using the clade-specific “Candidatus Accumulibacter” ppk1 primer sets that we had designed (Table 3). Total “Candidatus Accumulibacter” ppk1 abundance was calculated as the sum of the ppk1 abundances detected from the five “Candidatus Accumulibacter” clades. The “detected” and “not detected” information presented in Table 3 was consistent with the qualitative PCR using clade-specific ppk1 primers with 30 cycles of amplification, visualized by agarose gel electrophoresis (data not shown). No clades of “Candidatus Accumulibacter” ppk1 were detected from the two non-EBPR samples by qPCR, in agreement with a qualitative PCR using clade-specific ppk1 primers or the total-“Candidatus Accumulibacter”-targeted ppk1 or 16S rRNA primers.
TABLE 3.
Relative distributions of “Candidatus Accumulibacter” clades and estimated abundance of the total “Candidatus Accumulibacter” lineage relative to the bacterial community
| Sample | Relative distribution (%) of indicated clade within the “Candidatus Accumulibacter” lineagea
|
% of total “Candidatus Accumulibacter” lineage within the bacterial community as estimated by:
|
||||||
|---|---|---|---|---|---|---|---|---|
| I | IIA | IIB | IIC | IID | qPCR-16Sb | qPCR-ppk1c | FISHd | |
| CC | 1.5 ± 0.2 | 83 ± 3 | 16 ± 3 | 6 ± 2 | 7 ± 2 | ND | ||
| OL | 2 ± 1 | 89 ± 1 | 9 ± 1 | 6.1 ± 0.8 | 9 ± 1 | ND | ||
| SJ | 12 ± 1 | 38 ± 2 | 43 ± 1 | 8 ± 2 | 4.8 ± 0.1 | 6.9 ± 0.4 | ND | |
| NAN | 65 ± 2 | 27 ± 2 | 6 ± 1 | 2 ± 1 | 24 ± 6 | 26 ± 3 | 20 ± 5 | |
| VIR | 25 ± 5 | 60 ± 3 | 1.0 ± 0.2 | 14 ± 2 | 15 ± 3 | 19 ± 2 | 24 ± 7 | |
| DUR | 20 ± 1 | 21 ± 3 | 44 ± 5 | 16 ± 2 | 4.4 ± 0.1 | 4.1 ± 0.6 | 11 ± 1 | |
| LV1 | 6 ± 1 | 94 ± 1 | 9 ± 2 | 3.4 ± 0.3 | 11 ± 3 | |||
| LV2 | 4 ± 2 | 96 ± 2 | 7 ± 2 | 4 ± 1 | 9 ± 2 | |||
| NS | 35 ± 4 | 5 ± 2 | 13 ± 4 | 27.0 ± 0.3 | 19 ± 2 | 9.4 ± 0.3 | 6.3 ± 0.6 | 20 ± 3 |
| UCB | 98 ± 1 | 2 ± 1 | 67 ± 7 | 74 ± 10 | 80 ± 5 | |||
| UWMH | 7 ± 1 | 93 ± 1 | 73 ± 2 | 73 ± 8 | 80 ± 10 | |||
| UWMS | 76 ± 3 | 24 ± 3 | 51 ± 3 | 58 ± 1 | 55 ± 10 | |||
The percentage was obtained by dividing the ppk1 copy number from each clade by the sum of the ppk1 copy numbers for the five clades. The numbers in bold indicate the dominant clade in each sample. Blank cells in the table represent amplification that was below the detection limits of both the qPCR assay and visualization by agarose gel electrophoresis.
“Candidatus Accumulibacter” percentage estimated by “Candidatus Accumulibacter” 16S rRNA genes and bacterial 16S rRNA genes.
“Candidatus Accumulibacter” percentage estimated by total “Candidatus Accumulibacter” ppk1 genes and bacterial 16S rRNA genes.
Two different pairs of primers were applied to quantify clade IIC. For sample NS, the primer set including OTU NS D3 (Acc-ppk1-254f and Acc-ppk1-460r) detected higher copy numbers than the primer set excluding OUT NS D3 (Acc-ppk1-1123f and Acc-ppk1-1376r). However, for other samples there was no significant difference between the two quantifications acquired from the two primer sets. This suggests that OTU NS D3 was not likely a PCR artifact, despite its long branch length in the phylogenetic tree (Fig. 2) and its apparent mosaic combination of sequences from other clades. In Table 3, for all samples except NS, the reported relative abundance of IIC ppk1 was determined using the primer set excluding OTU NS D3.
The fractional abundance of the total “Candidatus Accumulibacter” lineage relative to the bacterial community in many of the samples included in this study was previously measured using 16S rRNA-targeted FISH (5, 12), and these quantifications are included in Table 3. To validate quantifications derived from qPCR, we also estimated the percentage of the total “Candidatus Accumulibacter” lineage relative to the bacteria by using qPCR-based methods. For this purpose, qPCR with general bacterial 16S rRNA primers was performed to estimate total bacterial abundance. As ppk1 is a single-copy gene in “Candidatus Accumulibacter” (4), its abundance can represent the cell abundance of this organism. However, since many bacteria possess multiple copies of the rrn operon, it is difficult to accurately determine cell number using qPCR with 16S rRNA primers. The metagenomic analysis of “Candidatus Accumulibacter”-dominated EBPR reactors indicated that “Candidatus Accumulibacter” had at least 2 copies of the rrn operon (4). Therefore, we estimated the fraction of the total “Candidatus Accumulibacter” lineage by assuming that the “Candidatus Accumulibacter” genome had 2 copies of the rrn operon and that the other bacterial genomes in activated sludge had an average of 4.1 copies of the rrn operon, as has been assumed previously (9). This assumption provided an approximate estimation of the “Candidatus Accumulibacter” fraction by using the abundance of “Candidatus Accumulibacter” ppk1 and bacterial 16S rRNA genes, although we acknowledge that it does not ensure accurate determination. As a comparison, under the same assumption, the “Candidatus Accumulibacter” fraction was also estimated using 16S rRNA gene-targeted qPCR with general bacterial and PAO-846r primers since all “Candidatus Accumulibacter” 16S rRNA sequences retrieved in this study matched the PAO-846r primer. These two qPCR-based estimations are also shown in Table 3.
DISCUSSION
In this study, the fine-scale population structure of the “Candidatus Accumulibacter” lineage was investigated, using ppk1 as a genetic marker. Since the function of ppk1 in EBPR as the exclusive poly(P) generation mechanism has not been confirmed and not all “Candidatus Accumulibacter” bacteria accumulate poly(P) at all times, we were not using ppk1 as a functional marker to indicate EBPR activity. Instead, ppk1 was used only as an alternative phylogenetic marker to study the fine-scale population structure of “Candidatus Accumulibacter” that cannot be distinguished by 16S rRNA genes. To do this, we examined the structure of the “Candidatus Accumulibacter” lineage by using two different genetic loci, 16S rRNA and ppk1. Some 16S rRNA sequences from geographically and operationally distinct systems were almost identical, exhibiting unresolved branching patterns, especially in clades I and IIA (Fig. 1). For example, the dominant 16S rRNA sequence from the UWM reactor (OTU UWMH 4) was 100% identical to the sequence obtained from the metagenomic analysis of a laboratory-scale reactor in Australia (4). The close affiliation may suggest the wide dispersal of “Candidatus Accumulibacter” among different EBPR systems (Kunin et al., submitted). But because of the limited phylogenetic resolution provided by 16S rRNA sequences, it is not possible to determine if these sequences represent essentially identical populations of “Candidatus Accumulibacter.” The ppk1 gene exhibits higher interspecies variation than 16S rRNA but is conserved enough to be well suited for phylogenetic analysis (13; Kunin et al., submitted). As expected, ppk1-based tree topology was more resolved at most but not all nodes (Fig. 2). The ppk1 sequences in these unresolved clades share very high levels of sequence identity, even though they were recovered from geographically and operationally distinct EBPR systems. For instance, the dominant ppk1 sequence from the UWM reactor (OTU UWMH G8) shared 98.6% DNA sequence identity with the ppk1 sequence recovered during metagenomic analysis of the reactor in Australia, and the dominant ppk1 sequence retrieved from UWM at another sampling time (OTU UWMS A8) was more than 99% identical to OTU DUR A10, retrieved from a full-scale system in Oregon. This high degree of sequence identity supports the hypothesis that “Candidatus Accumulibacter” is less restricted in its global dispersal than some organisms (e.g., hot spring Archaea [25]) and that its ecological niche within EBPR systems is quite broad (Kunin et al., submitted). We note that the newly described diversity within the “Candidatus Accumulibacter” lineage is not surprising; many bacterial groups originally identified by 16S rRNA-targeted techniques have been subsequently shown to be quite diverse at the genome level (11, 23). We propose that efforts to determine the ecological significance of this diversity relative to the EPBR process would be a fruitful direction for future research.
It was previously suggested that 16S rRNA- and ppk1-based phylogenies were partially incongruent, possibly implying horizontal transfer of ppk1 genes (13). In this study, we tried to relate phylogenies reconstructed from 16S rRNA and ppk1 genes by using the common characteristics shared by these two phylogenetic trees since there are no pure culture representatives available within the “Candidatus Accumulibacter” lineage. In most cases, 16S rRNA- and ppk1-based “Candidatus Accumulibacter” phylogenetic trees were largely congruent, suggesting that ppk1 has not been horizontally transferred and is therefore a suitable locus for studying population structure within the “Candidatus Accumulibacter” lineage. However, it is noteworthy that clade IID was underrepresented in the 16S rRNA phylogenetic tree. This was probably caused by differences in the gene retrieval procedures used. For “Candidatus Accumulibacter” 16S rRNA, PCR was first conducted with general bacterial primers and the resulting clones were screened using “Candidatus Accumulibacter”-specific primers (5), whereas ppk1 sequences were obtained directly by amplification with the ppk1 primer set specific to “Candidatus Accumulibacter.” Therefore, if a “Candidatus Accumulibacter” clade was present at a relatively low abundance relative to the total bacterial population (such as clade IID, as indicated by both clone frequencies and qPCR results), few such 16S rRNA sequences would have been captured by our screening method. Therefore, more 16S rRNA sequences belonging to clade IID and a higher degree of congruence between 16S rRNA and ppk1 phylogenies would have been expected if more 16S rRNA clones had been screened.
The qPCR-based estimation of total “Candidatus Accumulibacter” abundance determined using the ppk1 primer sets developed in this study was not significantly different from that derived by qPCR using 16S rRNA gene-targeted primer PAO-846f (paired Student t test, P = 0.32) but was different from that determined by 16S rRNA-based FISH (paired Student t test, P = 0.04). The fact that the two qPCR-based estimates were comparable suggests that ppk1 provides as adequate a measurement of total “Candidatus Accumulibacter” population size as does 16S rRNA, in spite of uncertainties about rrn operon copy number per genome. The differences between qPCR-based estimation and FISH quantification are possibly due to the following two reasons: (i) the assumption about the rrn operon copy number per genome in activated sludge was not precise enough, although it provided a rough estimation, and (ii) the FISH results, except those for sample UCB, were obtained using a probe mixture including probe RHC 439, which may overestimate “Candidatus Accumulibacter” abundance because RHC 439 also targets other members in Rhodocyclaceae. Specifically, 16S rRNA sequences matching probe RHC 439 but not probe PAO 462(b), PAO 651, or PAO 846(b) had been recovered from sample DUR (OTU DUR A10 [GenBank accession no. EF559323]) and sample NS (26). Therefore, within the sample subset excluding DUR and NS, qPCR-based estimation was more comparable to FISH quantifications (paired Student t test, P = 0.18).
The clustering patterns in both 16S rRNA and ppk1 phylogenies clearly reveal the distinct clades within the “Candidatus Accumulibacter” lineage. It was difficult to design specific 16S rRNA gene-targeted primer sets to distinguish each “Candidatus Accumulibacter” clade because the 16S rRNA gene is too conserved among clades. However, we were able to design specific ppk1 primers and develop qPCR assays to quantitatively study the distribution of “Candidatus Accumulibacter” clades among EBPR systems. In agreement with 16S rRNA and ppk1 phylogenies, only “Candidatus Accumulibacter” ppk1 clades I and IIA were detected in the laboratory-scale reactors by qPCR, whereas “Candidatus Accumulibacter” bacteria in most full-scale WWTPs were represented by at least three clades. This likely reflects the higher level of complexity and temporal fluctuations associated with wastewater composition and operational conditions in the full-scale WWTPs, which create more niches for these closely related groups. The relative distributions of “Candidatus Accumulibacter” clades were generally different among these full-scale EBPR systems, with the exception of pairs LV1-LV2 and CC-OL. It is expected that LV1 and LV2 had almost identical wastewater compositions, as they were parallel treatment trains in the same WWTP, with the only difference being their SRTs 5 days prior to sample collection. CC and OL also harbored very similar “Candidatus Accumulibacter” population structures. These two WWTPs were geographically very close (less than 30 miles apart), both were configured with high-rate anaerobic selectors, and both had very short SRTs (2 to 3 days) compared to other EBPR plants in this study (8 to 11 days). The similar population structures likely reflect these similar operational conditions, which could define similar ecological niches for “Candidatus Accumulibacter.”
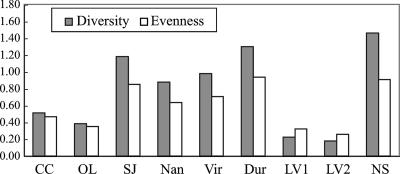
To compare the “Candidatus Accumulibacter” clade diversity and evenness among the full-scale EBPR samples, the Shannon index (diversity) and Pielou regularity index (evenness) of the “Candidatus Accumulibacter” lineage were calculated using the relative abundances of the five clades as determined by qPCR (Fig. 4). The least diversity and evenness were found in LV1 and LV2, with more than 90% of “Candidatus Accumulibacter” in clade IIC. Notably, sample NS showed the highest level of diversity, with all five clades detected and more evenly distributed than in other full-scale WWTP samples. This may be related to the complex configuration in the NS WWTP, where activated sludges from trains configured as University of Cape Town and anaerobic-oxic process reactors are combined in the secondary clarifier, with the combined sludge being recycled back to each train. Sample DUR exhibited the second-highest level of diversity. It is noteworthy that “Candidatus Accumulibacter” 16S rRNA sequences recovered from this sample affiliated only with clade IIB (Fig. 1), while ppk1 sequences of DUR were distributed among four clades (Fig. 2), with qPCR results showing dominance in clade IIB. The underestimation of diversity by 16S rRNA analysis in this case was probably caused by differences in the gene retrieval procedures applied, as discussed earlier. When “Candidatus Accumulibacter” was present at a low abundance, such as in sample DUR, only the 16S rRNA sequences from the numerically dominant clades were recovered by the screening procedure, leading to underestimated 16S rRNA sequence diversity.
FIG. 4.
Shannon index (diversity) and Pielou regularity index (evenness) of the “Candidatus Accumulibacter” lineage in the full-scale EBPR treatment plants, calculated from the relative abundances of the five “Candidatus Accumulibacter” clades (normalized to total “Candidatus Accumulibacter” ppk1 genes) as shown in Table 3.
It is noteworthy that the laboratory-scale UWM reactor was inoculated with activated sludge from the NS plant. It was previously estimated that all “Candidatus Accumulibacter” bacteria in the NS plant were involved in poly(P) accumulation (6). Therefore, if there were no distinguishable physiological difference among these five clades, all of them could be enriched in the laboratory-scale reactor where EBPR conditions were optimized and well controlled. However, after 10 months of operation (sample UWMH), three out of five “Candidatus Accumulibacter” clades could not be detected. Compared with the NS plant, the UWM reactor was fed with acetate as the sole carbon source, had no dissolved oxygen in the anaerobic phase caused by sparging with nitrogen gas, had well-controlled pH and temperature, and had only slight variations in carbon and nutrient levels in the influent. The well-defined operational conditions likely reduced the number of ecological niches available to “Candidatus Accumulibacter.” This environment acted as a strong selective force to favor the members that fit this constrained niche best, allowing them to out-compete not only non-“Candidatus Accumulibacter” but also other “Candidatus Accumulibacter” clades. We also examined “Candidatus Accumulibacter” population structure in the UWM reactor after the reactor had been operated for 2 years (UWMS). The dominant “Candidatus Accumulibacter” clade had switched, with the predominance of clade I over clade IIA determined by qPCR, consistent with their occurrence frequencies in the clone libraries (Fig. 2). Several additional samples were collected from this reactor at different time points, and dramatic changes in the relative abundances of clades I and IIA over time were observed (data not shown), indicating that the population structure varies significantly over time. This dynamic may be caused by subtle fluctuation of operational conditions, periodic selection of one clade over the other, or strain-specific viral invasion since high viral activity was suspected in this system (Kunin et al., submitted).
The possibility that some “Candidatus Accumulibacter” clades can out-compete other clades in certain EBPR systems supports the assertion that these clades were distinct not only in their ppk1 sequences but also in their physiological characteristics. Unique clades may occupy ecological niches with subtle differences, leading to the observed distribution patterns in different EBPR systems or at different times within a single system. Cohan has suggested using the ecotype concept to describe a set of strains occupying the same ecological niche (2). From the definition and properties of an ecotype, each bacterial ecotype is expected to be identifiable as a sequence cluster, where the average sequence divergence between clusters is greater than the average sequence divergence within the cluster (2). The five clades revealed by ppk1 genes meet this criterion and thus may represent five putative ecotypes of “Candidatus Accumulibacter.” Further investigation is needed to confirm whether these five clades are truly ecologically distinct.
Acknowledgments
This project was funded by the National Science Foundation (BES 0332136).
Special thanks to Daniel Noguera, Brook Peterson, Jason Flowers, and Philip Hugenholtz for thoughtful discussions; Andy Torkelson for operating the laboratory-scale reactor; April Gu for assistance with acquiring WWTP samples; and Ryan Newton for help with phylogenetic analysis. The assistance of personnel in the WWTPs included in this study is greatly appreciated.
Footnotes
Published ahead of print on 3 August 2007.
REFERENCES
- 1.Ahn, K. H., and A. Kornberg. 1990. Polyphosphate kinase from Escherichia coli—purification and demonstration of a phosphoenzyme intermediate. J. Biol. Chem. 265:11734-11739. [PubMed] [Google Scholar]
- 2.Cohan, F. M. 2002. What are bacterial species? Annu. Rev. Microbiol. 56:457-487. [DOI] [PubMed] [Google Scholar]
- 3.Crocetti, G. R., P. Hugenholtz, P. L. Bond, A. Schuler, J. Keller, D. Jenkins, and L. L. Blackall. 2000. Identification of polyphosphate accumulating-organisms and the design of 16S rRNA-directed probes for their detection and quantitation. Appl. Environ. Microbiol. 66:1175-1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Garcia Martin, H., N. Ivanova, V. Kunin, F. Warnecke, K. W. Barry, A. C. McHardy, C. Yeates, S. He, A. A. Salamov, E. Szeto, E. Dalin, N. H. Putnam, H. J. Shapiro, J. L. Pangilinan, I. Rigoutsos, N. C. Kyrpides, L. L. Blackall, K. D. McMahon, and P. Hugenholtz. 2006. Metagenomic analysis of two enhanced biological phosphorus removal (EBPR) sludge communities. Nat. Biotechnol. 24:1263-1269. [DOI] [PubMed] [Google Scholar]
- 5.He, S., A. Z. Gu, and K. D. McMahon. 2006. Fine-scale differences between Accumulibacter-related bacteria in enhanced biological phosphorus removal activated sludge Water Sci. Technol. 54:111-117. [DOI] [PubMed] [Google Scholar]
- 6.He, S., A. Z. Gu, and K. D. McMahon. 29 June 2007, posting date. Progress toward understanding the distribution of Accumulibacter among full-scale enhanced biological phosphorus removal systems. Microb. Ecol. doi: 10.1007/s00248-007-9270-x. [DOI] [PubMed]
- 7.Hesselmann, R. P. X., C. Werlen, D. Hahn, J. R. van der Meer, and A. J. B. Zehnder. 1999. Enrichment, phylogenetic analysis and detection of a bacterium that performs enhanced biological phosphate removal in activated sludge. Syst. Appl. Microbiol. 22:454-465. [DOI] [PubMed] [Google Scholar]
- 8.Huelsenbeck, J. P., and F. Ronquist. 2001. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17:754-755. [DOI] [PubMed] [Google Scholar]
- 9.Kaetzke, A., D. Jentzsch, and K. Eschrich. 2005. Quantification of Microthrix parvicella in activated sludge bacterial communities by real-time PCR. Lett. Appl. Microbiol. 40:207-211. [DOI] [PubMed] [Google Scholar]
- 10.Kong, Y., J. L. Nielsen, and P. H. Nielsen. 2004. Microautoradiographic study of Rhodocyclus-related polyphosphate-accumulating bacteria in full-scale enhanced biological phosphorus removal plants. Appl. Environ. Microbiol. 70:5383-5390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Martiny, A. C., M. L. Coleman, and S. W. Chisholm. 2006. Phosphate acquisition genes in Prochlorococcus ecotypes: Evidence for genome-wide adaptation. Proc. Natl. Acad. Sci. USA 103:12552-12557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McMahon, K. D., M. A. Dojka, N. R. Pace, D. Jenkins, and J. D. Keasling. 2002. Polyphosphate kinase from activated sludge performing enhanced biological phosphorus removal. Appl. Environ. Microbiol. 68:4971-4978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McMahon, K. D., S. Yilmaz, S. He, D. L. Gall, D. Jenkins, and J. D. Keasling. 2 August 2007, posting date. Polyphosphate kinase genes from full-scale activated sludge plants. Appl. Microbiol. Biotechnol. doi: 10.1007/s00253-007-1122-6. [DOI] [PubMed]
- 14.Mino, T., M. C. M. Van Loosdrecht, and J. J. Heijnen. 1998. Microbiology and biochemistry of the enhanced biological phosphate removal process. Water Res. 32:3193-3207. [Google Scholar]
- 15.Neethling, J., B. Bakke, M. Benisch, A. Gu, and H. Stephens. 2005. Factors influencing the reliability of enhanced biological phosphorus removal. Report 01CTS3. Water Environment Research Foundation, Alexandria, VA.
- 16.Newton, R. J., A. D. Kent, E. W. Triplett, and K. D. McMahon. 2006. Microbial community dynamics in a humic lake: differential persistence of common freshwater phylotypes. Environ. Microbiol. 8:956-970. [DOI] [PubMed] [Google Scholar]
- 17.Pielou, E. C. 1975. Ecological diversity. Wiley, New York, NY.
- 18.Purkhold, U., A. Pommerening-Roser, S. Juretschko, M. C. Schmid, H.-P. Koops, and M. Wagner. 2000. Phylogeny of all recognized species of ammonia oxidizers based on comparative 16S rRNA and amoA sequence analysis: implications for molecular diversity surveys. Appl. Environ. Microbiol. 66:5368-5382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shannon, C. E., and W. Weaver. 1949. The mathematical theory of communication. The University of Illinois Press, Urbana.
- 20.Stackebrandt, E., and J. Ebers. 2006. Taxonomic parameters revisited: tarnished gold standards. Microbiol. Today 33:152-155. [Google Scholar]
- 21.Stackebrandt, E., and B. M. Goebel. 1994. A place for DNA-DNA reassociation and 16S ribosomal-RNA sequence-analysis in the present species definition in bacteriology. Int. J. Syst. Bacteriol. 44:846-849. [Google Scholar]
- 22.Thompson, J. R., L. A. Marcelino, and M. F. Polz. 2002. Heteroduplexes in mixed-template amplifications: formation, consequence and elimination by ‘reconditioning PCR’. Nucleic Acids Res. 30:2083-2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thompson, J. R., S. Pacocha, C. Pharino, V. Klepac-Ceraj, D. E. Hunt, J. Benoit, R. Sarma-Rupavtarm, D. L. Distel, and M. F. Polz. 2005. Genotypic diversity within a natural coastal bacterioplankton population. Science 307:1311-1313. [DOI] [PubMed] [Google Scholar]
- 24.Warnecke, F., R. Amann, and J. Pernthaler. 2004. Actinobacterial 16S rRNA genes from freshwater habitats cluster in four distinct lineages. Environ. Microbiol. 6:242-253. [DOI] [PubMed] [Google Scholar]
- 25.Whitaker, R. J., D. W. Grogan, and J. W. Taylor. 2003. Geographic barriers isolate endemic populations of hyperthermophilic archaea. Science 301:976-978. [DOI] [PubMed] [Google Scholar]
- 26.Yoshida, N., N. Takahashi, and A. Hiraishi. 2005. Phylogenetic characterization of a polychlorinated-dioxin-dechlorinating microbial community by use of microcosm studies. Appl. Environ. Microbiol. 71:4325-4334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yu, Z., F. C. Michel, Jr., G. Hansen, T. Wittum, and M. Morrison. 2005. Development and application of real-time PCR assays for quantification of genes encoding tetracycline resistance. Appl. Environ. Microbiol. 71:6926-6933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang, H., K. Ishige, and A. Kornberg. 2002. A polyphosphate kinase (PPK2) widely conserved in bacteria. Proc. Natl. Acad. Sci. USA 99:16678-16683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zilles, J. L., J. Peccia, M.-W. Kim, C.-H. Hung, and D. R. Noguera. 2002. Involvement of Rhodocyclus-related organisms in phosphorus removal in full-scale wastewater treatment plants. Appl. Environ. Microbiol. 68:2763-2769. [DOI] [PMC free article] [PubMed] [Google Scholar]