Abstract
The engineering of bacterial strains with specific phenotypes frequently requires the use of blocks or “cassettes” of genes that act together to perform a desired function. The potential benefits of utilizing type III secretion systems in this regard are becoming increasingly realized since these systems can be used to direct interactions with host cells for beneficial purposes such as vaccine development, anticancer therapies, and targeted protein delivery. However, convenient methods to clone and transfer type III secretion systems for studies of a range of different types of bacteria are lacking. In addition to functional applications, such methods would also reveal important information about the evolution of a given type III secretion system, such as its ability to be expressed and functional outside of the strain of origin. We describe here the cloning of the Salmonella enterica serovar Typhimurium pathogenicity island 2 (SPI-2) type III secretion system onto a vector that can be easily transferred to a range of gram-negative bacterial genera. We found that expression of the cloned SPI-2 system in different Gammaproteobacteria and Alphaproteobacteria (as monitored by SseB protein levels) is dependent on the bacterial strain and growth medium. We also demonstrate that the cloned system is functional for secretion, can direct interactions with macrophages, and can be used as a novel tool to analyze the predicted interaction of SseB with host cells. This work provides a foundation for future applications where the cloned SPI-2 region (or other cloned type III systems) can provide a desired function to an engineered gram-negative strain.
The organized compilation and applied use of blocks or “cassettes” of genes that can be used to conveniently engineer microbes for specific purposes are being increasingly developed (3, 12, 14). The potential to use bacterial type III secretion systems in this regard is significant and can be illustrated by recent uses of these systems to direct interactions with host cells for beneficial purposes such as vaccine development, anticancer therapies, and targeted protein delivery (13, 28, 32, 33, 35, 36). Type III secretion systems allow secretion of protein substrates to the extracellular milieu and facilitate translocation of effector proteins from bacteria to eukaryotic host cells (7, 15). Originally, these systems were discovered and characterized by their ability to facilitate interactions of pathogens with their host cells, and the list of different gram-negative species that have evolved to use the type III pathway includes Salmonella enterica serovars, Yersinia spp., Shigella spp., Escherichia coli, and Pseudomonas aeruginosa (8). Recently, however, type III systems have been harnessed as tools for targeted beneficial applications. A growing body of work has demonstrated that type III systems can be used to deliver foreign protein antigens, resulting in epitope presentation to the immune system, a vigorous immune response, and subsequent protection against challenge by the organism from which the foreign epitope was derived (6, 21, 35, 36). Other studies have shown that protein delivery via type III systems can be used to elicit cytotoxic effector and memory CD8+ T-cell responses, resulting in the prevention of new tumor growth and regression of established tumors in mice (13, 28, 31). In addition, type III systems have been used in several applications where targeted protein delivery to eukaryotic cells reveals key information about how translocated bacterial effectors serve to alter host cell signaling pathways via a variety of effector/host protein interactions (1, 9, 32, 33, 38).
An improved ability to genetically engineer different kinds of bacteria with type III secretion systems will lead to advances that could have profound effects on our strategies to utilize these systems as beneficial molecular tools. In this study, we report the cloning of the entire Salmonella pathogenicity island 2 (SPI-2) type III secretion system onto a plasmid vector that can be conveniently self-transferred to a range of gram-negative bacterial genera. A previous study has demonstrated the cloning of SPI-2, but this construct was on a narrow-host-range vector and was not self-transmissible, thus limiting its application for studies in an extended range of bacterial genera (17). We transferred SPI-2 to several different nonpathogenic or attenuated gamma- and alphaproteobacterial genera and assayed for SPI-2 expression (by monitoring SseB protein levels) after culture in different growth media. Altered expression of the ssrAB two-component regulatory genes on the cloned SPI-2 was used to obtain SseB expression under a nonpermissive condition, but this result was found to be bacterium specific. The results demonstrate that the cloned SPI-2 system is functional for secretion and can direct interactions with macrophage cells. We also demonstrate that the cloned SPI-2 system can be used as a novel tool to analyze the predicted interaction of the SseB protein with eukaryotic cells. The results provide a foundation for future applications involving the transfer, expression, and function of SPI-2 and other secretion systems in an extended range of bacterial genera.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
All bacterial strains used in this study are as follows: Salmonella enterica serovar Typhimurium χ3339 (16), χ3339 ΔSPI-2 (this study; contains a chloramphenicol marker at the site of the SPI-2 deletion), LT2 (25), and 14028 (ATCC 14028) (generously provided by Yakhya Dieye); Salmonella enterica serovar Typhi TY2 and ISP1820 (generously provided by Roy Curtiss III) (11, 20); E. coli TOP10 (Invitrogen, Carlsbad, CA) and MG1655 (5); Salmonella bongori SARC11 (generously provided by Michael McClelland) (34); Pseudomonas putida ATCC 12633 (40); P. aeruginosa PAK pilA and PAO1 pilA (generously provided by Michael Schurr); Agrobacterium tumefaciens A136 (40); and Rhodobacter sphaeroides 2.4.1 (42). Rifampin derivatives of the above strains were isolated and used as plasmid recipients in conjugative transfers. Bacterial cells were grown in magnesium minimal medium (MgM) (4) containing either 10 mM MgCl2, pH 7.5 (MgM 10), or 8 μM MgCl2, pH 5.0 (MgM 8), and Lennox broth (LB) (2, 24). The growth of strains in MgM 8 was generally not as robust as that in the other media used in this study. However, we found that initial growth in MgM 10 followed by a wash in MgM 8 and subsequent inoculation of the MgM 8 allowed improved growth under this condition in most cases. Antibiotics were used at the indicated concentrations (in μg per milliliter): rifampin, 75; streptomycin, 100; and spectinomycin, 125 or 250.
DNA methods.
DNA manipulations and PCR were performed using standard protocols as described previously (2, 37). All plasmid DNA was isolated using QIAGEN columns as described by the manufacturer (QIAGEN, Valencia, CA).
Cloning and transfer of S. enterica serovar Typhimurium SPI-2 region.
For cloning and transfer of the S. enterica serovar Typhimurium SPI-2 region, please refer to File S1 in the supplemental material.
Protein methods used for analysis of SseB protein expression and secretion.
Analysis of SseB protein expression in strains containing cloned SPI-2 was performed as described previously via Western blotting and probing with rabbit polyclonal anti-SseB antisera (generously provided by Michael Hensel) (4, 41). Protein expression was tested with lysates from cultures inoculated from freshly streaked plates and grown for 16 to 18 h or inoculated by diluting an overnight culture and growing it for an additional 4 to 16 h. All the samples shown in the figures included here are from cultures grown under the former condition; however, similar results were obtained with either culture condition. Culture optical density measurement, Ponceau S staining for total protein, and control antibody probes were used in combination to obtain and verify equivalent sample loading for each strain as described previously (41). Samples from cultures that differed more than twofold in optical density (such cultures were usually slightly lower in density) were normalized for equal loading. In addition, a standard amount of SseB protein from the same batch of total cell lysate was routinely run on each gel as a standardization control for each Western blot assay. Preparation of secreted bacterial proteins, sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), Coomassie blue staining, and Western blot transfers were performed using standard protocols as described previously (2, 27).
Construction of R995 + SPI-2:plac-ssrAB and R995 + SPI-2 ssaV.
The plasmids R995 + SPI-2::plac-ssrAB and R995 + SPI-2 ssaV were constructed using the suicide plasmid pMAK705 via methods described previously (41). In R995 + SPI-2::plac-ssrAB, the suicide plasmid is integrated upstream of ssrAB on the cloned SPI-2 region (genome coordinates 1479612 to 1479987) so that the lac promoter on this plasmid is driving expression of the ssrAB genes. In R995 + SPI-2 ssaV, the suicide plasmid is integrated internally in the reading frame of the ssaV gene on the cloned SPI-2 region (genome coordinates 1494132 to 1494679) such that this gene is disrupted. The structures of each plasmid construct were verified by PCR analysis (data not shown).
Macrophage assays.
The survival assay of bacterial strains in the murine macrophage cell line RAW264.7 (ATCC TIB-71) was performed as described previously using 2-h and 18-h time points and a multiplicity of infection of approximately 20 (26). The survival assays were replicated in four independent experiments using triplicate tissue culture sample wells in each experiment. Detection of SseB protein associated with J774 murine macrophages infected with bacterial strains containing SPI-2 was performed using methods described previously (39) except for the following modifications. To separate intracellular bacteria from the host cell fraction, infected J774 cells were lysed with 0.1% Triton X-100 and the lysate was centrifuged at 27,000 × g to remove intracellular bacteria (which remain intact during this treatment). The supernatant from this centrifugation was filtered with a 0.45-μm filter, precipitated with 10% trichloroacetic acid, and centrifuged at 12,000 × g, and the pellet was resuspended with phosphate-buffered saline and SDS-PAGE sample loading buffer. The J774 macrophages were infected in T-75 flasks with the indicated bacterial strains for 1.5 h at a multiplicity of infection of approximately 50. For this length of infection, all strains infected the macrophages to equivalent levels (data not shown and description of Fig. 7 in text). The results of this assay were replicated in three independent experiments.
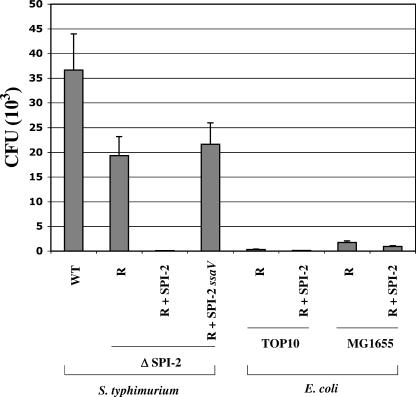
FIG. 7.
Survival of strains containing the cloned SPI-2 region in RAW264.7 macrophages. Equivalent numbers of cells from cultures of the indicated bacterial strains containing either R995 (R), R995 + SPI-2 (R + SPI-2), or R995 + SPI-2 ssaV (R + SPI-2 ssaV) were incubated with RAW264.7 macrophages for 1 h. At this point, the infection medium was changed to contain gentamicin, and the infected-cell cultures were incubated for an additional 17 h (18-h total infection). Intracellular bacteria were obtained at this time and enumerated via plating for CFU. The results were replicated in four independent trials with each trial being performed in triplicate sample wells. WT, wild type.
Biosafety.
All studies were performed in accordance with established biosafety guidelines and have been approved by the investigators' institutional biosafety committee. The cloned SPI-2 region was transferred to bacterial strains that are nonpathogenic or significantly attenuated for virulence.
RESULTS
Cloning of the S. enterica serovar Typhimurium SPI-2 type III secretion system.
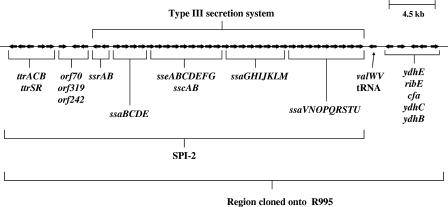
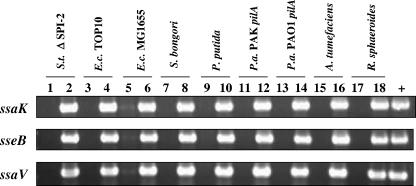
To obtain the type III secretion system encoded by S. enterica serovar Typhimurium SPI-2 as a single cloned fragment, we used the VEX-capture technique for the targeted excision and cloning of large bacterial chromosomal sections (40). The VEX-capture technique allows precise Cre/lox-mediated excision of the desired chromosomal fragment as a nonreplicating, covalently closed circular molecule and simultaneous cloning (or “capture”) of the excised circular molecule by homologous recombination. The vector used to capture the excised chromosomal region was the self-transmissible, broad-host-range plasmid R995 (30) containing a DNA fragment from the orf70-319-242 region of SPI-2 (18, 25). A map of S. enterica serovar Typhimurium SPI-2 and adjacent genes that are present in the R995 + SPI-2 clone is presented in Fig. 1. We transferred this plasmid to an S. enterica serovar Typhimurium strain containing a deletion of SPI-2 as well as to a range of other gram-negative proteobacterial genera, and we confirmed the presence of the cloned SPI-2 region in these strains via PCR analysis as shown in Fig. 2.
FIG. 1.
S. enterica serovar Typhimurium SPI-2 region. A map of the S. enterica serovar Typhimurium SPI-2 region is depicted. The arrows indicate the genes of the region that have been cloned onto R995 using VEX-capture. The genes that comprise the SPI-2 genomic island and the type III secretion system within that island are also indicated.
FIG. 2.
Transfer of R995 + SPI-2 to different bacterial hosts as shown via PCR analysis. Plasmid DNA isolated from the indicated transconjugant strains containing either R995 or R995 + SPI-2 was used as a template in PCR analysis with primers hybridizing to portions of the indicated SPI-2 genes. The PCR products were run on DNA agarose gels and stained with ethidium bromide. Odd-numbered and even-numbered lanes indicate samples from strains containing R995 or R995 + SPI-2, respectively. The lane marked “+” indicates PCRs where chromosomal DNA from wild-type S. enterica serovar Typhimurium strain χ3339 was used as a template. Abbreviations: S.t., S. enterica serovar Typhimurium; E.c., E. coli; P.a., P. aeruginosa.
Analysis of SseB expression from R995 + SPI-2 in different bacterial genera.
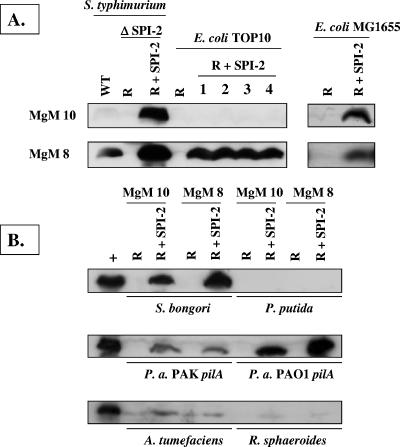
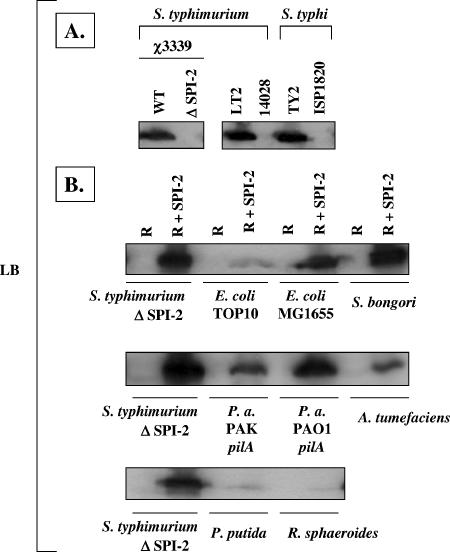
As a measure of SPI-2 gene expression from the R995 + SPI-2 construct, we probed Western blots of different bacterial cell lysates with antisera to the SseB protein (Fig. 3). SseB is a substrate of the SPI-2 type III secretion system that is essential for the translocation of other effector proteins (19, 27). We grew strains in two types of MgM that have previously been shown to be either noninducing (MgM 10) or inducing (MgM 8) for SPI-2 gene expression and secretion (4, 10). While the wild-type S. enterica serovar Typhimurium strain χ3339 exhibited the expected medium-specific regulation of SseB expression (i.e., expression only in MgM 8), the cloned SPI-2 island in the S. enterica serovar Typhimurium ΔSPI-2 background was expressed in both MgM 10 and MgM 8. The observed expression of SseB from cloned SPI-2 under the “noninducing” condition is likely due to the fact that multiple copies of the R995 + SPI-2 plasmid are present (about 10 copies per chromosome) (43). In the E. coli strains, the medium-specific expression of SseB from R995 + SPI-2 was found to be strain specific: SseB expression in TOP10 (R995 + SPI-2) was MgM specific while SseB in MG1655 (R995 + SPI-2) was expressed in both MgMs. In the other genera, SseB was expressed from R995 + SPI-2 at different levels depending on the strain, but the expression in each strain was observed to be similar in either MgM. Expression of SseB was undetectable under these conditions in the P. putida strain background.
FIG. 3.
Western blot analysis of SseB expression in different bacterial strains containing R995 + SPI-2 grown in MgM 10 and MgM 8. (A) Total cell lysates were obtained from S. enterica serovar Typhimurium and E. coli strains containing either R995 (R) or R995 + SPI-2 (R + SPI-2) grown in the indicated medium, and equivalent amounts of these lysates were analyzed via SDS-PAGE, Western blotting, and probing with anti-SseB antisera. Lanes numbered 1 to 4 indicate four separate isolates of the strain TOP10 (R + SPI-2). MgM 10 and MgM 8 indicate media used for SPI-2 noninducing and inducing conditions, respectively. WT, wild type. (B) An analysis identical to that described for panel A was performed for other gram-negative strains containing R995 (R) or R995 + SPI-2 (R + SPI-2). The lane marked “+” contains a sample from the same batch of lysate obtained from the strain S. enterica serovar Typhimurium ΔSPI-2 (R + SPI-2) as a positive control. P.a., P. aeruginosa.
The large majority of experiments involving in vitro expression of SPI-2 genes have involved the use of media containing specific nutritional and environmental signals to induce SPI-2 gene expression, including a shift to low pH, low magnesium levels, and phosphate limitation (10, 23). We examined whether the SPI-2 protein SseB is expressed in a rich medium (LB) since bacterial growth in this medium is more robust and it can be used to grow a broader range of bacterial species than the other specialized minimal media frequently used in previous studies. We found that expression of SseB in the S. enterica serovar Typhimurium strains χ3339 and LT2 was robust and easily detectable (Fig. 4). However, this observation was found to be strain specific, as the S. enterica serovar Typhimurium strain 14028 did not express SseB under the same conditions. We also examined SseB expression in LB medium for two S. enterica serovar Typhi strains, TY2 and ISP1820. We found a strain-specific expression pattern in this species as well, as TY2 expressed SseB but ISP1820 did not under these conditions. These results indicate that LB medium can be used to obtain significant expression of chromosomally encoded SPI-2 SseB in strains of S. enterica serovar Typhimurium and S. enterica serovar Typhi. The expression of SseB from cloned SPI-2 in LB medium was readily observed in most of the bacterial host genera and generally followed a strain-specific pattern similar to that found in the MgM (Fig. 4). The low expression of SseB in LB in the bacterial hosts P. putida and R. sphaeroides was consistent with the low SseB expression in MgM in these species. Taken together, the results indicate that the cloned SPI-2 can be successfully established in a range of bacterial genera and SseB protein expression can be detected in most of these strains in a range of growth media.
FIG. 4.
Western blot analysis of SseB expression in different bacterial strains containing R995 + SPI-2 grown in LB medium. (A) Equivalent amounts of total cell lysates from the indicated strains grown in LB medium were analyzed via Western blotting and probing with anti-SseB antisera. WT, wild type. (B) Equivalent amounts of total cell lysates from the indicated strains containing R995 (R) or R995 + SPI-2 (R + SPI-2) grown in LB medium were analyzed as described for panel A. P.a., P. aeruginosa.
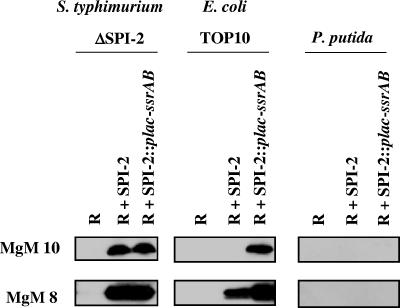
The ssrAB genes located in SPI-2 encode a two-component regulatory system that is essential for expression of several SPI-2 genes including sseB (4). In strains containing cloned SPI-2 where SseB expression is low or undetectable, manipulation of ssrAB expression could allow increased SseB (and likely other SPI-2 gene) expression levels to be obtained. To test this hypothesis, we inserted a heterologous promoter upstream of the ssrAB genes on the cloned SPI-2 (R995 + SPI-2::plac-ssrAB) and tested SseB expression from this construct in three different bacterial genera (Fig. 5). In these strains, this promoter (plac) is active due to low or absent LacI repressor activity. Since SseB expression from cloned SPI-2 in S. enterica serovar Typhimurium ΔSPI-2 was already at significant levels in all media tested, the plac-ssrAB construct had little effect on SseB expression in this strain. However, in the E. coli strain TOP10, the R995 + SPI-2::plac-ssrAB plasmid produced detectable SseB expression under conditions where the R995 + SPI-2 plasmid did not. Under the same conditions in P. putida, R995 + SPI-2::plac-ssrAB did not allow detectable SseB expression to be observed. These results indicate that the R995 + SPI-2::plac-ssrAB construct can allow SseB expression under certain “nonpermissive” conditions but that this result is strain specific.
FIG. 5.
Expression of SseB from the R995 + SPI-2::plac-ssrAB construct. Strains containing the plasmids R995 (R), R995 + SPI-2 (R + SPI-2), and R995 + SPI-2::plac-ssrAB (R + SPI-2::plac-ssrAB) were grown in the indicated media and assayed for SseB protein expression via Western blotting of equivalent amounts of cell lysate. Please refer to the text for additional details.
Protein secretion via the cloned SPI-2 type III system.
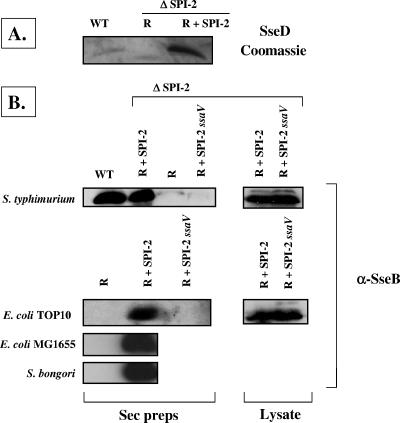
To demonstrate that the cloned SPI-2 type III secretion system is functional for protein secretion, we made protein preparations from culture supernatants harvested from strains containing cloned SPI-2. The R995 + SPI-2 plasmid complemented the S. enterica serovar Typhimurium ΔSPI-2 strain for secretion as shown using Coomassie blue staining and Western blot analysis (Fig. 6). We constructed a mutation in the ssaV gene in the cloned SPI-2 region (resulting in plasmid R995 + SPI-2 ssaV) to demonstrate that the observed secretion was dependent on the activity of the type III secretion system. The ssaV gene encodes an essential component of the SPI-2 secretion apparatus, and the secretion of SseB has been previously shown to be dependent on a functional ssaV gene (4). Accordingly, secretion of SseB from strains containing the R995 + SPI-2 ssaV construct was not observed. We tested SseB secretion from E. coli TOP10, E. coli MG1655, and S. bongori SARC11 containing cloned SPI-2 and observed secretion of SseB from these strains. This indicates that R995 + SPI-2 is functional for secretion in bacterial backgrounds other than S. enterica serovar Typhimurium.
FIG. 6.
Protein secretion via the cloned SPI-2 type III secretion system. Preparations of secreted proteins were obtained from culture supernatants of the indicated strains containing either R995 (R), R995 + SPI-2 (R + SPI-2), or R995 + SPI-2 ssaV (R + SPI-2 ssaV) grown in MgM 8. (A) Coomassie blue staining of secretion preparations obtained from the indicated S. enterica serovar Typhimurium strains and run on an SDS-polyacrylamide gel. In this assay, the SPI-2-encoded SseD protein is detected as a prominent band running at approximately 20 kDa. (B) Samples of secreted proteins from the indicated strains were Western blotted and probed with anti-SseB antisera. Samples from secretion preparations (“Sec preps”) and cell lysates are indicated. WT, wild type.
Use of R995 + SPI-2 to mediate interactions with macrophages.
One of the phenotypes associated with the SPI-2 secretion system is its role in the survival of S. enterica serovar Typhimurium in macrophages after entry into these cells (22, 29, 39). Since mutants in SPI-2 genes have been shown to be defective in macrophage intracellular survival, we tested whether the R995 + SPI-2 construct could complement this phenotype in the S. enterica serovar Typhimurium ΔSPI-2 background (Fig. 7). However, we found that the presence of cloned SPI-2 in this strain had the opposite effect: the survival of the S. enterica serovar Typhimurium ΔSPI-2 (R995 + SPI-2) strain was much less than that of both the wild-type and ΔSPI-2 strains. The S. enterica serovar Typhimurium ΔSPI-2 (R995 + SPI-2 ssaV) strain survived at a level equal to that of the ΔSPI-2 (R995) strain, thus indicating that it is likely that overactivity of the cloned SPI-2 secretion system is causing the enhanced survival defect of the S. enterica serovar Typhimurium ΔSPI-2 (R995 + SPI-2) strain. These strains all infected the macrophages at almost equal levels at a 2-h time point (the survival defect of the R995 + SPI-2 strain was at 18 h, per Fig. 7), indicating that decreased initial entry into the host cells was not the cause of the enhanced survival defect (data not shown). When E. coli strains TOP10 and MG1655 containing cloned SPI-2 were tested in the same assay, only very slight differences were observed between SPI-2-containing strains and the R995-only controls. Also, these strains were much less able to survive in macrophages than was wild-type S. enterica serovar Typhimurium. Both of these observations with the E. coli strains are most likely due to the fact that other substrates which are secreted by the SPI-2 system and which are encoded elsewhere on the S. enterica serovar Typhimurium chromosome are absent in these backgrounds. The severe survival defect observed in the S. enterica serovar Typhimurium ΔSPI-2 (R995 + SPI-2) strain could be potentially useful in cases where clearance of a SPI-2-containing strain is desirable after it has performed its desired function. Consequently, this phenotype is being further characterized in our laboratory for future applications.
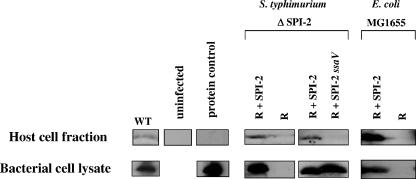
To further characterize the ability of cloned SPI-2 to direct bacterial cell/host cell interactions, we tested for an association between the SseB protein and host cell macrophages. The SseB protein has been shown to be localized to the surface of S. enterica serovar Typhimurium cells and to function as a translocator protein that acts to allow movement of SPI-2 substrates from the bacterial cell to eukaryotic host cells (4, 27). This activity of the SseB protein predicts that SseB would interact with eukaryotic cells and possibly be found in the eukaryotic host cell fraction of an infection assay. After infection of J774 macrophage cells with strains containing cloned SPI-2 (and appropriate control strains), bacterium-free J774 total host cell fractions were obtained, Western blotted, and probed for the presence of the SseB protein (Fig. 8). Although detection of SseB association with the host cell fraction from the wild-type strain was somewhat variable and weak (Fig. 8 and data not shown), we readily detected SseB in the host cell fraction from infections using cloned SPI-2 in strains S. enterica serovar Typhimurium ΔSPI-2 and E. coli MG1655. This result was found to be dependent on the normal function of the cloned SPI-2 type III system since SseB was not detected in the host cell fraction from infections with S. enterica serovar Typhimurium ΔSPI-2 (R995 + SPI-2 ssaV). As an additional control, we incubated SseB protein from an amount of bacterial cells equivalent to the inoculum used in the infection asssays (approximately 5 × 108 cells) with the J774 cells. We did not observe SseB to be present in the host cell fractions in this control assay, indicating that the extensive washing of cells postinfection was sufficient to remove any extracellular SseB that may be present during the assays. Since our assay does not discriminate between host cell membrane and cytosolic fractions, the host cell-associated SseB could be targeted to either compartment in the host cells.
FIG. 8.
Association of SseB with host macrophage cells. The eukaryotic host cell protein fraction from infections of J774 macrophages with the indicated bacterial strains was obtained as described in Materials and Methods and analyzed for the presence of SseB via Western blotting and probing with anti-SseB antisera. Samples of cell lysate from cultures of the bacterial strains used in the infections are also indicated. The protein control was performed by adding SseB protein from a cell lysate (equivalent to the number of bacterial cells used in the infection) to demonstrate that any extracellular SseB is washed away before macrophage lysis. WT, wild type.
Taken together, the above results indicate that the cloned SPI-2 type III secretion system can function to direct interactions with host cells.
DISCUSSION
The ability to obtain entire type III secretion systems as single cloned DNA fragments that could be moved between bacterial cells would greatly enhance our ability to genetically engineer different bacteria for certain beneficial purposes. In this study, we report the cloning of the entire SPI-2 type III secretion system as a single DNA fragment contained on a plasmid vector that can be conveniently transferred to and established in a range of gram-negative proteobacteria. We demonstrate that the SPI-2 protein SseB is detectably expressed from the R995 + SPI-2 construct in strains of S. enterica serovar Typhimurium, S. bongori, E. coli, P. aeruginosa, P. putida, A. tumefaciens, and R. sphaeroides. However, SseB expression in the P. putida and R. sphaeroides backgrounds ranges from very low to undetectable. We attempted to obtain elevated SseB expression in the P. putida background by manipulating the expression of the SPI-2 ssrAB regulator genes, but this did not result in increased SseB expression in this strain. However, this approach did result in detectable SseB expression under a “noninducing” condition in the strain E. coli TOP10. Further study will be required to determine if low levels of SseB expression in a particular strain are due to defects at the transcriptional, posttranscriptional, translational, or posttranslational levels. Interestingly, our laboratory has previously cloned the S. enterica serovar Typhimurium SPI-1 type III secretion system in a similar fashion onto the R995 vector, but we could not detect SPI-1 protein expression in strains other than those of S. enterica serovar Typhimurium upon its transfer to and establishment in other gram-negative genera (41). This difference in results between the cloned SPI-1 and SPI-2 systems highlights two important points: (i) the different evolution of the two type III systems such that SPI-2 appears to be able to be expressed and to function outside of the host bacterial species of origin while SPI-1 appears to have barriers that prevent this under the conditions that we have examined, and (ii) the potential advantage in obtaining and studying clones of different type III systems, since the activities and specificities of these different systems may vary, and this variability may provide flexibility such that a given type III system could be targeted for a specific application or environmental condition.
Our studies demonstrate that the cloned SPI-2 region is functional to secrete cognate substrates and direct interactions between bacterial strains containing SPI-2 and eukaryotic host cells. However, we observed an enhanced survival defect in macrophages with the strain S. enterica serovar Typhimurium ΔSPI-2 (R995 + SPI-2) that is dependent on the function of the cloned type III system. The most likely reason for this result is that the cloned SPI-2 system in the S. enterica serovar Typhimurium background is secreting and/or translocating substrates in an unregulated manner that is detrimental to the survival of this strain inside the macrophages. This observation may indicate a potentially beneficial phenomenon that can be used in particular applications where clearance of a strain containing SPI-2 can be obtained after the strain has performed its desired function. Regulating SPI-2 (or its substrate proteins) to function at different levels such that a desired result can be obtained at one level of activity and strain clearance can be obtained at another activity level could be very useful in vaccine delivery and other such applications. Further study will be needed to determine the specific SPI-2 substrates or specific aspects of SPI-2 type III secretion activity that are involved in causing the enhanced survival defect. We also demonstrate that the R995 + SPI-2 construct can be used to study interactions between SseB (and potentially other SPI-2 translocation proteins) and eukaryotic host cells. To our knowledge, this study provides the first results indicating that SseB does indeed associate with host cells as predicted by previous studies that document its activity as a translocation protein (4, 27).
The use of type III secretion systems to deliver antigens for vaccines is currently being developed, and recent results indicate much promise in utilizing type III systems for this purpose (6, 21, 35, 36). The potential to use cloned type III secretion systems to design particular bacterial vaccine delivery strains in this regard is significant. First, epitopes can be specifically designed and delivered by the type III system to induce a desired immune response and obtain potential protection against the source organism of the epitope. Second, since the design of polyvalent vaccines is frequently advantageous, the ability to use cloned type III systems and cognate substrates provides the potential for a range of different epitopes to be expressed from a single bacterial strain carrying the type III system. For example, a bacterial background strain that expresses endogenous antigens from its chromosome (either against the strain itself or another organism) could carry a cloned type III system that expresses a different set of antigen epitopes. The different set of epitopes could be directed toward the background organism itself (to provide a more potent polyvalent response against this strain) or toward a heterologous pathogenic organism. In the latter case, protection could be obtained against multiple organisms with a single vaccine. However, since the plasmid constructs described here are self-transmissible, the likelihood of their use in approved vaccines is low, though their convenient use for research studies in this area has much potential. Future studies aimed at permanently knocking out the R995 transfer system upon establishment of the cloned type III system in a given bacterial strain would help to solve this problem. Alternatively, a method to irreversibly integrate a cloned type III system into a given host chromosome can be developed to circumvent this issue as well.
Supplementary Material
Acknowledgments
We thank Michael Hensel for generously providing anti-SseB antisera for use in this study. We thank the following individuals for generously providing bacterial strains: Michael Schurr for P. aeruginosa strains PAK pilA and PAO1 pilA, Michael McClelland and Steffen Porwollik for S. bongori SARC11, Roy Curtiss III for S. enterica serovar Typhi strains TY2 and ISP1820, and Yakhya Dieye for S. enterica serovar Typhimurium 10428.
This work was supported by NASA-Ames grant NAG 2-1378.
Footnotes
Published ahead of print on 3 August 2007.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Angot, A., A. Vergunst, S. Genin, and N. Peeters. 2007. Exploitation of eukaryotic ubiquitin signaling pathways by effectors translocated by bacterial type III and type IV secretion systems. PLoS Pathog. 3:e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.). 1996. Current protocols in molecular biology. John Wiley & Sons, Inc., New York, NY.
- 3.Baker, D., G. Church, J. Collins, D. Endy, J. Jacobson, J. Keasling, P. Modrich, C. Smolke, and R. Weiss. 2006. Engineering life: building a fab for biology. Sci. Am. 294:44-51. [DOI] [PubMed] [Google Scholar]
- 4.Beuzon, C. R., G. Banks, J. Deiwick, M. Hensel, and D. W. Holden. 1999. pH-dependent secretion of SseB, a product of the SPI-2 type III secretion system of Salmonella typhimurium. Mol. Microbiol. 33:806-816. [DOI] [PubMed] [Google Scholar]
- 5.Blattner, F. R., G. Plunkett III, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453-1474. [DOI] [PubMed] [Google Scholar]
- 6.Chen, L. M., G. Briones, R. O. Donis, and J. E. Galan. 2006. Optimization of the delivery of heterologous proteins by the Salmonella enterica serovar Typhimurium type III secretion system for vaccine development. Infect. Immun. 74:5826-5833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cornelis, G. R. 2006. The type III secretion injectisome. Nat. Rev. Microbiol. 4:811-825. [DOI] [PubMed] [Google Scholar]
- 8.Cornelis, G. R., and F. Van Gijsegem. 2000. Assembly and function of type III secretory systems. Annu. Rev. Microbiol. 54:735-774. [DOI] [PubMed] [Google Scholar]
- 9.Cuzick, A., F. R. Stirling, S. L. Lindsay, and T. J. Evans. 2006. The type III pseudomonal exotoxin U activates the c-Jun NH2-terminal kinase pathway and increases human epithelial interleukin-8 production. Infect. Immun. 74:4104-4113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deiwick, J., T. Nikolaus, S. Erdogan, and M. Hensel. 1999. Environmental regulation of Salmonella pathogenicity island 2 gene expression. Mol. Microbiol. 31:1759-1773. [DOI] [PubMed] [Google Scholar]
- 11.Deng, W., S. R. Liou, G. Plunkett III, G. F. Mayhew, D. J. Rose, V. Burland, V. Kodoyianni, D. C. Schwartz, and F. R. Blattner. 2003. Comparative genomics of Salmonella enterica serovar Typhi strains Ty2 and CT18. J. Bacteriol. 185:2330-2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Endy, D. 2005. Foundations for engineering biology. Nature 438:449-453. [DOI] [PubMed] [Google Scholar]
- 13.Epaulard, O., B. Toussaint, L. Quenee, M. Derouazi, N. Bosco, C. Villiers, R. Le Berre, B. Guery, D. Filopon, L. Crombez, P. N. Marche, and B. Polack. 2006. Anti-tumor immunotherapy via antigen delivery from a live attenuated genetically engineered Pseudomonas aeruginosa type III secretion system-based vector. Mol. Ther. 14:656-661. [DOI] [PubMed] [Google Scholar]
- 14.Fu, P. 2006. A perspective of synthetic biology: assembling building blocks for novel functions. Biotechnol. J. 1:690-699. [DOI] [PubMed] [Google Scholar]
- 15.Galan, J. E., and H. Wolf-Watz. 2006. Protein delivery into eukaryotic cells by type III secretion machines. Nature 444:567-573. [DOI] [PubMed] [Google Scholar]
- 16.Gulig, P. A., and R. Curtiss III. 1987. Plasmid-associated virulence of Salmonella typhimurium. Infect. Immun. 55:2891-2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hansen-Wester, I., D. Chakravortty, and M. Hensel. 2004. Functional transfer of Salmonella pathogenicity island 2 to Salmonella bongori and Escherichia coli. Infect. Immun. 72:2879-2888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hensel, M., T. Nikolaus, and C. Egelseer. 1999. Molecular and functional analysis indicates a mosaic structure of Salmonella pathogenicity island 2. Mol. Microbiol. 31:489-498. [DOI] [PubMed] [Google Scholar]
- 19.Hensel, M., J. E. Shea, S. R. Waterman, R. Mundy, T. Nikolaus, G. Banks, A. Vazquez-Torres, C. Gleeson, F. C. Fang, and D. W. Holden. 1998. Genes encoding putative effector proteins of the type III secretion system of Salmonella pathogenicity island 2 are required for bacterial virulence and proliferation in macrophages. Mol. Microbiol. 30:163-174. [DOI] [PubMed] [Google Scholar]
- 20.Hone, D. M., A. M. Harris, S. Chatfield, G. Dougan, and M. M. Levine. 1991. Construction of genetically defined double aro mutants of Salmonella typhi. Vaccine 9:810-816. [DOI] [PubMed] [Google Scholar]
- 21.Husseiny, M. I., F. Wartha, and M. Hensel. 2007. Recombinant vaccines based on translocated effector proteins of Salmonella pathogenicity island 2. Vaccine 25:185-193. [DOI] [PubMed] [Google Scholar]
- 22.Kuhle, V., and M. Hensel. 2002. SseF and SseG are translocated effectors of the type III secretion system of Salmonella pathogenicity island 2 that modulate aggregation of endosomal compartments. Cell. Microbiol. 4:813-824. [DOI] [PubMed] [Google Scholar]
- 23.Lee, A. K., C. S. Detweiler, and S. Falkow. 2000. OmpR regulates the two-component system SsrA-SsrB in Salmonella pathogenicity island 2. J. Bacteriol. 182:771-781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lennox, E. S. 1955. Transduction of linked genetic characters of the host by bacteriophage P1. Virology 1:190-206. [DOI] [PubMed] [Google Scholar]
- 25.McClelland, M., K. E. Sanderson, J. Spieth, S. W. Clifton, P. Latreille, L. Courtney, S. Porwollik, J. Ali, M. Dante, F. Du, S. Hou, D. Layman, S. Leonard, C. Nguyen, K. Scott, A. Holmes, N. Grewal, E. Mulvaney, E. Ryan, H. Sun, L. Florea, W. Miller, T. Stoneking, M. Nhan, R. Waterston, and R. K. Wilson. 2001. Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nature 413:852-856. [DOI] [PubMed] [Google Scholar]
- 26.Nickerson, C. A., and R. Curtiss III. 1997. Role of sigma factor RpoS in initial stages of Salmonella typhimurium infection. Infect. Immun. 65:1814-1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nikolaus, T., J. Deiwick, C. Rappl, J. A. Freeman, W. Schroder, S. I. Miller, and M. Hensel. 2001. SseBCD proteins are secreted by the type III secretion system of Salmonella pathogenicity island 2 and function as a translocon. J. Bacteriol. 183:6036-6045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nishikawa, H., E. Sato, G. Briones, L. M. Chen, M. Matsuo, Y. Nagata, G. Ritter, E. Jager, H. Nomura, S. Kondo, I. Tawara, T. Kato, H. Shiku, L. J. Old, J. E. Galan, and S. Gnjatic. 2006. In vivo antigen delivery by a Salmonella typhimurium type III secretion system for therapeutic cancer vaccines. J. Clin. Investig. 116:1946-1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ochman, H., F. C. Soncini, F. Solomon, and E. A. Groisman. 1996. Identification of a pathogenicity island required for Salmonella survival in host cells. Proc. Natl. Acad. Sci. USA 93:7800-7804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pansegrau, W., E. Lanka, P. T. Barth, D. H. Figurski, D. G. Guiney, D. Haas, D. R. Helinski, H. Schwab, V. A. Stanisich, and C. M. Thomas. 1994. Complete nucleotide sequence of Birmingham IncP alpha plasmids. Compilation and comparative analysis. J. Mol. Biol. 239:623-663. [DOI] [PubMed] [Google Scholar]
- 31.Panthel, K., K. M. Meinel, V. E. Sevil Domenech, G. Geginat, K. Linkemann, D. H. Busch, and H. Russmann. 2006. Prophylactic anti-tumor immunity against a murine fibrosarcoma triggered by the Salmonella type III secretion system. Microbes Infect. 8:2539-2546. [DOI] [PubMed] [Google Scholar]
- 32.Patel, J. C., and J. E. Galan. 2006. Differential activation and function of Rho GTPases during Salmonella-host cell interactions. J. Cell Biol. 175:453-463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Patel, J. C., and J. E. Galan. 2005. Manipulation of the host actin cytoskeleton by Salmonella—all in the name of entry. Curr. Opin. Microbiol. 8:10-15. [DOI] [PubMed] [Google Scholar]
- 34.Porwollik, S., R. M. Wong, and M. McClelland. 2002. Evolutionary genomics of Salmonella: gene acquisitions revealed by microarray analysis. Proc. Natl. Acad. Sci. USA 99:8956-8961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Russmann, H. 2003. Bacterial type III translocation: a unique mechanism for cytosolic display of heterologous antigens by attenuated Salmonella. Int. J. Med. Microbiol. 293:107-112. [DOI] [PubMed] [Google Scholar]
- 36.Russmann, H. 2004. Inverted pathogenicity: the use of pathogen-specific molecular mechanisms for prevention or therapy of disease. Int. J. Med. Microbiol. 293:565-569. [DOI] [PubMed] [Google Scholar]
- 37.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Press, Plainview, NY.
- 38.Schlumberger, M. C., A. J. Muller, K. Ehrbar, B. Winnen, I. Duss, B. Stecher, and W. D. Hardt. 2005. Real-time imaging of type III secretion: Salmonella SipA injection into host cells. Proc. Natl. Acad. Sci. USA 102:12548-12553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Uchiya, K., M. A. Barbieri, K. Funato, A. H. Shah, P. D. Stahl, and E. A. Groisman. 1999. A Salmonella virulence protein that inhibits cellular trafficking. EMBO J. 18:3924-3933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wilson, J. W., D. H. Figurski, and C. A. Nickerson. 2004. VEX-capture: a new technique that allows in vivo excision, cloning, and broad-host-range transfer of large bacterial genomic DNA segments. J. Microbiol. Methods 57:297-308. [DOI] [PubMed] [Google Scholar]
- 41.Wilson, J. W., and C. A. Nickerson. 2006. Cloning of a functional Salmonella SPI-1 type III secretion system and development of a method to create mutations and epitope fusions in the cloned genes. J. Biotechnol. 122:147-160. [DOI] [PubMed] [Google Scholar]
- 42.Wilson, J. W., and C. A. Nickerson. 2006. A new experimental approach for studying bacterial genomic island evolution identifies island genes with bacterial host-specific expression patterns. BMC Evol. Biol. 6:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wilson, J. W., E. A. Sia, and D. H. Figurski. 1997. The kilE locus of promiscuous IncP alpha plasmid RK2 is required for stable maintenance in Pseudomonas aeruginosa. J. Bacteriol. 179:2339-2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.