Abstract
A physical and mathematical model for wine fermentation kinetics was adapted to include the influence of temperature, perhaps the most critical factor influencing fermentation kinetics. The model was based on flask-scale white wine fermentations at different temperatures (11 to 35°C) and different initial concentrations of sugar (265 to 300 g/liter) and nitrogen (70 to 350 mg N/liter). The results show that fermentation temperature and inadequate levels of nitrogen will cause stuck or sluggish fermentations. Model parameters representing cell growth rate, sugar utilization rate, and the inactivation rate of cells in the presence of ethanol are highly temperature dependent. All other variables (yield coefficient of cell mass to utilized nitrogen, yield coefficient of ethanol to utilized sugar, Monod constant for nitrogen-limited growth, and Michaelis-Menten-type constant for sugar transport) were determined to vary insignificantly with temperature. The resulting mathematical model accurately predicts the observed wine fermentation kinetics with respect to different temperatures and different initial conditions, including data from fermentations not used for model development. This is the first wine fermentation model that accurately predicts a transition from sluggish to normal to stuck fermentations as temperature increases from 11 to 35°C. Furthermore, this comprehensive model provides insight into combined effects of time, temperature, and ethanol concentration on yeast (Saccharomyces cerevisiae) activity and physiology.
The progress of a wine fermentation is determined by the concentration of residual sugar. Problem fermentations occur when the completion of sugar utilization exceeds 7 to 10 days (sluggish fermentations) or when sugar utilization ceases with more than 0.4% wt/vol residual sugar still present in the wine (stuck fermentations) (5). Sluggish and stuck wine fermentations are often associated with musts that contain inadequate nutrients. The primary nutrient associated with these problem fermentations is nitrogen. The minimum concentration of required nitrogen in order to complete fermentation may be dependent upon other factors, such as temperature and initial sugar concentration. More comprehensive reviews of all known factors that lead to problem fermentations are available (1, 5, 6, 14); however, in this work, to predict problem fermentations, we concentrated on developing a comprehensive model that includes the three main factors of temperature and initial nitrogen and sugar concentrations.
These three variables are well-known critical factors for determining the kinetics of wine fermentations. Ough and Amerine performed extensive studies to characterize the effects of temperature on cell growth, sugar utilization, and ethanol production (25, 26). Others have studied yeast (Saccharomyces cerevisiae) sensitivity to ethanol at various temperatures (22, 27, 28). In addition, insufficient nitrogen has been well documented as an important factor that leads to stuck and sluggish fermentations (1, 2, 5, 14, 15, 20). Ough also studied the combined effects of initial nitrogen concentration and temperature on fermentation rates (23), while still others have studied the combined effects of initial sugar concentration, temperature, and pH on fermentation (9, 24). Overall, these works present a plethora of data that support the importance of understanding the effects of temperature and initial nitrogen and sugar concentrations on wine fermentation and that supply ample qualitative evidence of how each variable affects the overall rate of fermentation (sugar utilization). However, none of these previous studies presented a comprehensive model that includes all three of these variables and that can consistently predict problem fermentations.
Several physical and mathematical models have been developed for predicting wine fermentation kinetics. One of the first mechanistic models of enological fermentations that incorporated the relevant state variables, such as sugar concentration and temperature, was developed by Boulton in 1980 (7). Caro et al. also developed a similar mechanistic model for grape must fermentation which took into account other microbial functions, such as respiration and synthesis of products other than ethanol (8). Nanba et al. developed a kinetic wine fermentation model that incorporated yeast sensitivity to ethanol and temperature effects of yeast cell growth parameters (21). Other researchers have also developed more empirical models within an enological context (4, 13, 16). However, none of these models have been shown to acceptably predict problem fermentations with respect to temperature and initial conditions. A more comprehensive review of these models, along with others, can be found in the work of Marin (19).
One problem with the aforementioned mechanistic models is that the chosen limiting nutrient for cell growth was sugar. Substantial evidence has shown a strong link between nitrogen and yeast growth (2, 14, 20, 23), and a proven method for avoiding certain sluggish or stuck fermentations has been the addition of nitrogen sources (5). Therefore, the inclusion of nitrogen as a component for yeast growth is critical in a model aimed at predicting problem fermentations. The first enological model to include a nitrogen-limited cell growth mechanism was developed by Cramer et al. (11). Malherbe et al. also developed a model that incorporated nitrogen-limited growth (18). However, neither of these models successfully incorporates the effects of temperature in a manner that allows the prediction of sluggish and stuck fermentations.
Here we elaborate on the model previously presented by Cramer et al. (11) by including the effects of temperature. This model consists of five coupled ordinary differential equations (ODEs) that are combined with four one-dimensional regression models (describing the effects of temperature on three temperature-dependent model parameters and initial nitrogen conditions on one model parameter) to accurately predict the variation of sugar utilization for different temperatures and different initial concentrations of sugar and nitrogen. This is the first comprehensive model that predicts a transition from sluggish to normal to stuck wine fermentations with respect to increasing temperature.
MATERIALS AND METHODS
Experimental design.
Fermentations of 400 ml were performed in 500-ml Erlenmeyer flasks with fermentation locks. Fermentations were run at six temperatures (11, 15, 20, 25, 30, and 35°C), and samples were taken at appropriate intervals throughout the fermentation (12 to 18 samples during each fermentation). At each temperature, three different juice conditions were used. The three conditions used in these experiments were normal sugar and normal nitrogen, normal sugar and low nitrogen, and high sugar and normal nitrogen. A fourth juice condition (high sugar and low nitrogen) was used for model validation and was performed at 11 and 35°C. For these experiments, “normal” and “high” sugar correspond to 265 g/liter and 300 g/liter, respectively, while “low” and “normal” nitrogen correspond to 80 mg N/liter and 330 mg N/liter, respectively. Each flask experiment was run in duplicate for a total of 40 fermentations. °Brix (measured with a DMA 35N densitometer; Anton Paar, Ashland, VA), total cell count, and viable cell count were taken for all experiments. In addition, sugar (glucose and fructose), ethanol, ammonia, and α-amino nitrogen data were collected for one of each replicated pair of experiments by using procedures described below.
Fermentations and juice preparation.
For all fermentations, diluted Chardonnay juice was used, with sugar and nitrogen added back to appropriate levels. The purpose of diluting the juice was to create a medium where sugar and nitrogen levels could be manipulated, but all other nutrients were equal throughout the experiments. The juice was from the 2002 vintage and was supplied by Beringer Blass Wine Estates (Napa, CA) from a vineyard in Sonoma County, CA. The juice had the following measurements: °Brix, 25.4; titratable acidity, 7.5 g/liter; pH, 3.35; ammonia, 111 mg N/liter; and α-amino nitrogen concentration, 172 mg N/liter. Sulfur dioxide was added to the juice at 50 mg/liter. A yeast nutritional supplement (Superfood; The Wine Lab, Napa, CA) was also added to the juice at 500 mg/liter just prior to making dilutions.
In order to obtain the previously mentioned juice conditions, juice was diluted 3:1 (3 parts water or stock solutions to 1 part juice) to a total of 400 ml in a 500-ml Erlenmeyer flask. Sugar was added back by means of a 550-g/liter stock solution composed of 50% glucose (Sigma Chemical Company, St. Louis, MO) and 50% fructose (Sigma Chemical Company, St. Louis, MO) dissolved in sterile, filtered, deionized water. Nitrogen was added by means of a stock solution consisting of 10 g/liter diammonium phosphate (DAP; Sigma Chemical Company, St. Louis, MO). Fermentation flasks were made up to 400 ml with sterile deionized water.
The yeast used in all fermentations was Premier Cuvee (Red Star, Milwaukee, WI). Fermentations were performed in temperature-controlled incubators with shaker tables set at a constant 120 rpm. Temperature and agitation levels were maintained throughout the fermentation. Fermentations were monitored until the normal sugar and normal N flasks reached a constant sugar level for at least 2 days (300 to 800 h, depending on the temperature).
Fermentation sampling procedures.
Samples (approximately 3.5 ml/sample) were taken at appropriate times. Three milliliters of the sample was filtered through a 0.45-μm syringe filter into a 5.0-ml sterile test tube with a sealing cap and was frozen for later analysis. The remainder of the sample was used immediately for obtaining total cell count and viable cell count as well as for obtaining an optical density reading.
Viable and total cell concentration procedures.
For determining total and viable cell counts, a Bright-Line hemacytometer (Hausser Scientific, Horsham, PA) was used under a Zeiss light microscope at ×400 magnification. A 100-μl sample was diluted with water such that when a final dilution with methylene blue at a 1:1 ratio was made, the final dilution would give a minimum of 100 cells and a maximum of 400 cells in the counting area of the hemacytometer. Samples were mixed on a vortex mixer at each stage of the dilution. The methylene blue used was 0.02% wt/vol in a citrate buffer. Cells were allowed to be in contact with methylene blue for at least 1 min, but not more than 5 min. Blue cells were counted as dead, and noncolored cells were counted as live. Five of the 25 squares in the hemacytometer grid were counted, and the result was multiplied by 5 to give a total count. Except for early in the fermentation when the cell number was very low, a minimum of 100 cells were counted. When time permitted, replicate counts were made using the chamber on the opposite side of the hemacytometer. Each cell count was converted to grams per liter of cell mass, assuming that each cell weighs 4 × 10−11 g and that the 25 squares in the hemacytometer grid were covered with 10 μl. Based upon replicate flask experiments, the standard deviations of total cell mass and viable cell mass estimates were a maximum of 0.63 g/liter and 0.48 g/liter, respectively, with a significantly smaller replication error found during the exponential growth phase (approximately the first 100 h), closer to 0.005 g/liter.
High-pressure liquid chromatography analysis for sugars and ethanol.
Samples were analyzed by high-pressure liquid chromatography for glucose, fructose, and ethanol (30). The system used was a Hewlett-Packard 1100 series with two HPX-87H columns (Bio-Rad; catalog no. 125-0140) in series, preceded by a cation H+ guard column (Bio-Rad; catalog no. 125-0140). The detector used was an HP 407A refractive index detector. Elution was isocratic using a mobile phase of 1.5 mM sulfuric acid at a flow rate of 0.6 ml/min. The column temperature was maintained at 50°C. The injection volume used was 20 μl. The columns were flushed with water for at least 1 h after every 18 to 24 samples.
Determination of total nitrogen concentration.
Before and during fermentations, total nitrogen was determined by separately measuring ammonia concentration and alpha amino acid concentration and summing the two values. Alpha amino acid concentration was measured using the procedure described previously by Dukes and Butzke (12). Ammonia was determined via an enzymatic kit produced by R-Biopharm (South Marshall, MI).
Determination of model parameters and generation of simulated fermentations.
Sugar, nitrogen, ethanol, total biomass, and viable biomass concentrations were used for parameter estimation. However, only the first seven observed time points of total and viable biomass were used. These time points were used because the model is thought to represent cell activity and not cell viability. However, cell activity is not yet easily measured (11). MATLAB software (version 7.0; The MathWorks, Inc., Natick, MA) was then used to find values for the seven model parameters. Markov chain Monte Carlo integration (MCMC) in conjunction with Bayesian parameter estimation (10) was used to determine the expectation and credible regions of all seven model parameters for each fermentation (see “Parameter estimation” below). All parameter regression surfaces were performed using Bayesian robust linear regression and MCMC methods (see “Regression modeling of model parameters” below). A function called “metrop2” in a free toolbox from the Laboratory of Computational Engineering (http://www.lce.hut.fi/research/mm/mcmcstuff/) was adapted to perform all MCMC procedures.
Fermentation model.
The nitrogen-limited model utilized in this paper was first proposed by Cramer et al. (11). It consists of five coupled ODEs:
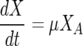 |
(1) |
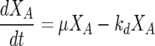 |
(2) |
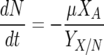 |
(3) |
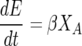 |
(4) |
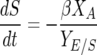 |
(5) |
where the five state variables are total biomass (X [g/liter]), active biomass (XA [g/liter]), nitrogen (N [mg/liter]), ethanol (E [g/liter]), and sugar (S [g/liter]). The specific growth rate for cell mass (μ) is a function of nitrogen concentration
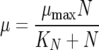 |
(6) |
where μmax and KN are the maximum specific growth rate and the Monod constant for nitrogen (N)-limited growth, respectively. The death rate or rate of cell inactivation (kd) is a function of total ethanol concentration
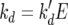 |
(7) |
where k′d is a parameter that describes the sensitivity of yeast to ethanol. The rate of sugar utilization per cell is a function of sugar concentration
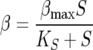 |
(8) |
where βmax and KS are the maximum specific rate of sugar utilization and the Michaelis-Menten-type constant for sugar utilization, respectively. In total, there are seven model parameters: μmax, the maximum specific growth rate; KN, the Monod constant for nitrogen-limited growth; k′d, a death or inactivation parameter describing the sensitivity of the cells to ethanol; βmax, the maximum rate of sugar utilization; KS, the Michaelis-Menten-type constant for sugar transport across the cell membrane; YX/N, the yield coefficient for cell mass grown per mass of nitrogen utilized; and YE/S, the yield coefficient for ethanol produced per sugar consumed.
There are two minor differences between the previous model (11) and the model presented here. First, total cell mass has been included in the here-presented model. This inclusion does not change the model at all. Total cell mass is included in this work to later illustrate the difference between total and active cell mass. The second difference is that we no longer refer to active cell mass as viable cell mass. Cramer et al. (11) provided significant evidence that suggested live cells may not be completely active in terms of growth, utilization of sugar and nitrogen, and production of ethanol. Thus, we have formally acknowledged this in the model by renaming viable cell mass as active cell mass.
To adapt this model for temperature dependence, we first assume that the basic structure of the coupled ODEs does not vary with temperature. Only the parameters within the ODEs (μmax, k′d, βmax, KN, KS, YX/N, and YE/S) are considered to be potential functions of temperature. Here, we find the temperature dependence of these parameters and then we model this temperature dependence by using robust polynomial regression (see “Regression modeling of model parameters” below). We developed the polynomial model simply to facilitate interpolation between data values from the temperatures studied. For this reason, it would be possible to utilize other regression models, lookup tables, neural networks, nonlinear regression of Arrhenius-type equations, or any other expression that describes the experimentally derived temperature dependence of the model parameters.
RESULTS
Sugar utilization.
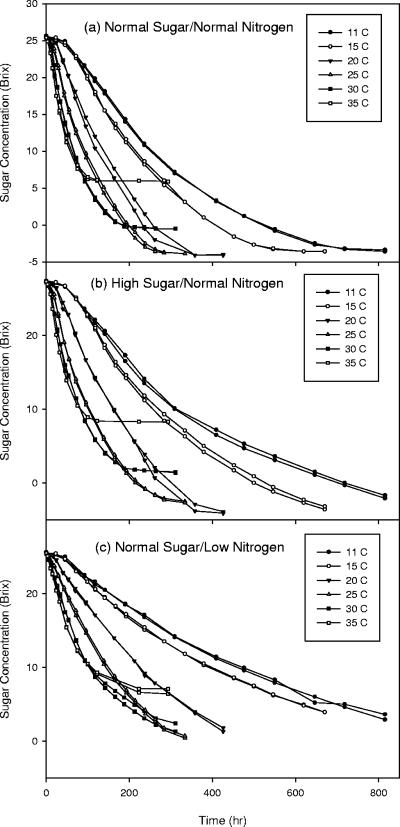
°Brix curves were measured for all fermentations and are presented in Fig. 1. First it should be noted that the replicate experiments demonstrate the highly reproducible nature of the observations. Several key features can be seen with respect to changing temperature and initial conditions. At high temperatures, regardless of initial conditions, fermentations have an initial period of rapid sugar utilization, followed by a sudden cessation as the fermentation becomes stuck. At midrange temperatures, fermentations reach dryness in the minimum amount of time (i.e., normal fermentation activity). At low temperatures, fermentations were very sluggish. Normal nitrogen and sugar conditions at low temperatures result in fermentations that reach completion. However, for high-sugar or low-nitrogen conditions, fermentations become stuck or at least are extremely sluggish.
FIG. 1.
Experimental °Brix curves for three different initial conditions and six different temperatures. (a) Normal sugar and normal nitrogen. (b) High sugar and normal nitrogen. (c) Normal sugar and low nitrogen. Each different temperature is shown with a different symbol, and the fermentation for each temperature was performed in duplicate. The data from these duplicate fermentations give an indication of the small inherent experimental error observed for these measurements.
Initial sugar conditions seem to have a smaller effect on the overall rate of sugar utilization. For example, at 11°C, the normal sugar and normal nitrogen fermentations dropped an average of 29.1 °Brix in 815 h, while the high-sugar fermentations and normal nitrogen fermentations dropped an average of 29.2 °Brix in 815 h. While there are definitely visual differences between normal and high initial sugar concentrations, there do not seem to be drastic changes in the overall rate of sugar utilization.
Initial nitrogen conditions had a much stronger effect on the overall rate of sugar utilization. For example, at 11°C, the normal sugar and low-nitrogen fermentations dropped an average of 22.3 °Brix in 815 h. Likewise, normal sugar and low-nitrogen fermentations at all other temperatures had a significantly lower overall change in °Brix compared to normal sugar and nitrogen and high-sugar and low-nitrogen fermentations given the same change in time of fermentation.
Model fit.
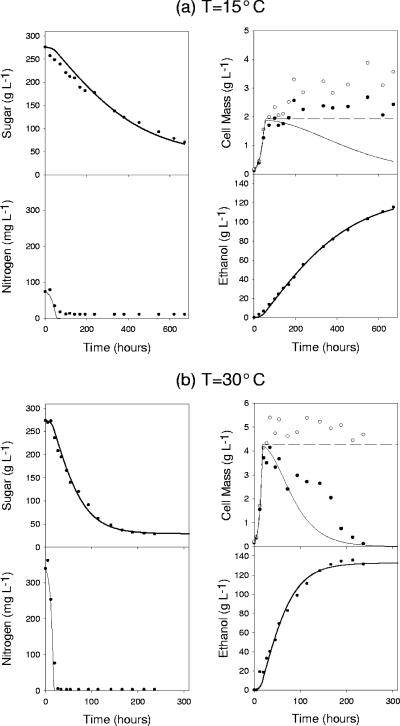
All 18 fermentations (three initial conditions at six temperatures) were fit to the model, and each resulted in a corresponding set of model parameters. Figure 2 shows two plots of typical model fits for various fermentations. Notice that sugar, ethanol, and nitrogen are accurately fit to the model. More discrepancies exist between the fits for cell mass. At lower temperatures, total cell mass seems to gradually increase even after the exponential growth phase. The observed viable cell mass at lower temperatures gradually decreases; however, in order to more accurately fit the other state variables (sugar and ethanol), the model predicts that cell activity decreases more rapidly. In general, total cell mass is more accurately predicted for higher temperature fermentations and cell activity more closely follows the measured cell viability (Fig. 2). In Fig. 2b, we see that the experimental total cell mass does not increase after the exponential growth phase and that the viable cell mass more rapidly decreases. These model fit results are consistent with the results of Cramer et al. (11).
FIG. 2.
Typical model fits for two different fermentations. (a) Normal sugar and low-nitrogen fermentation at 15°C. (b) Normal sugar and normal nitrogen fermentation at 30°C. Both panel a and panel b show all five state variables: sugar, viable cell mass (filled circles), total cell mass (open circles and broken line), nitrogen, and ethanol. It should be noted that the model does not predict viable cell mass but rather active cell mass (solid line).
Effects of temperature and initial conditions on model parameters.
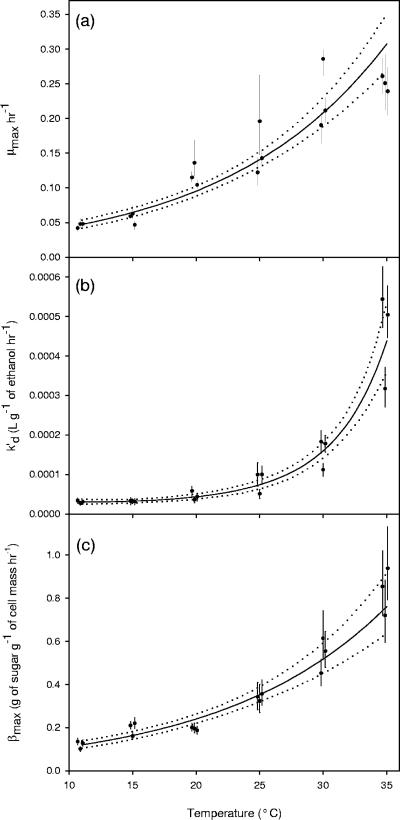
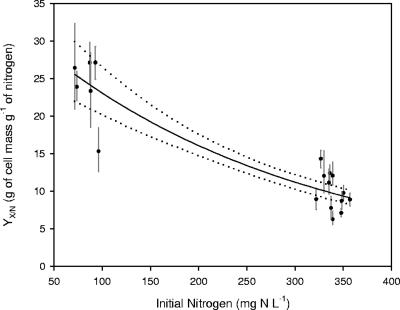
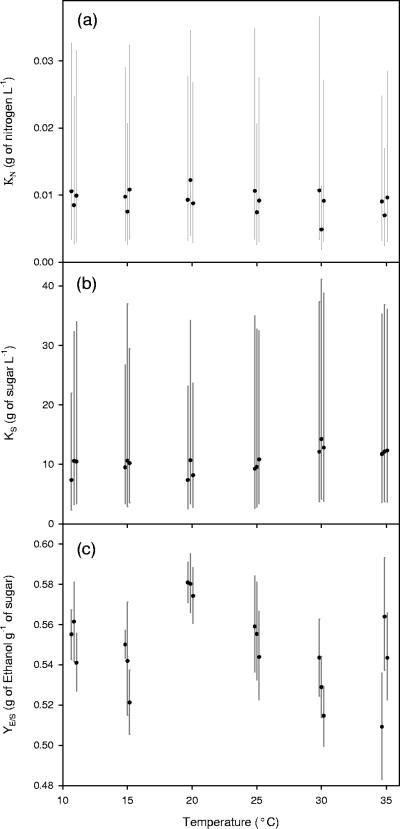
The resulting model parameter values were then tested for significant relationships with temperature and initial conditions. The maximum specific growth rate (μmax), the inactivation constant (k′d), and the maximum specific sugar utilization rate (βmax) were all shown to have significant relationships with respect to temperature (Fig. 3). Both μmax and βmax gradually increase by a factor of six as the temperature shifts from 11 to 35°C. The inactivation constant k′d also increases as temperature shifts from 11 to 35°C; however, once the temperature increases higher than 25°C, a more drastic increase occurs. It should be noted that k′d at 35°C is approximately 13 times greater than k′d at 11°C. The yield coefficient between cell mass production and nitrogen utilization (YX/N) was found to significantly change with respect to the initial nitrogen concentration (Fig. 4). All other parameters (YE/S, KN, and KS) were found to not change significantly with respect to temperature or initial conditions. Figure 5 shows these variables with respect to temperature.
FIG. 3.
Three model parameters from equations 1 through 8 were shown to have a significant relationship with respect to temperature. Each parameter estimate shows the mean value from the MCMC integration (•) along with its 95% credible region (vertical lines). Regression surfaces (solid lines) also include a 95% credible region (dotted lines). The maximum specific growth rate (a) increases by a factor of 6 as temperature increases from 11 to 35°C. The ethanol sensitivity constant which causes cell inactivation (b) increases by a factor of 13 as temperature increases from 11 to 35°C. The maximum rate of sugar utilization (c) increases by a factor of 6 as temperature increases from 11 to 35°C. Also note that some of the data points were plotted slightly off-center so as to prevent the overlap of error bars.
FIG. 4.
One model parameter from equations 1 through 8 was found to have a significant relationship with respect to initial nitrogen concentration. The yield coefficient between cell mass and nitrogen was about two times greater at lower initial nitrogen concentrations. The mean value from the MCMC parameter estimation is shown (•) along with the 95% credible regions (vertical lines). Regression surfaces (solid lines) also include a 95% credible region (dotted lines). Also note that some of the data points were plotted slightly off-center so as to prevent the overlap of error bars.
FIG. 5.
Three model parameters from equations 1 through 8 were shown to not have a significant relationship with respect to temperature or initial conditions. These parameters (Monod constant for nitrogen-limited growth [a], Monod constant for sugar transport [b], and the yield coefficient between ethanol and sugar [c]) are plotted against temperature. Each parameter estimate shows the mean value from the MCMC integration (•) along with its 95% credible region (vertical lines). Notice that the error bars are typically larger than the variation between the estimated mean values. Also note that some of the data points were plotted slightly off-center so as to prevent the overlap of error bars. It should be noted that the error bars are the dominant features of these plots.
Comprehensive model.
A comprehensive model was then formed by combining the relationships shown in Fig. 3 and 4 with the fermentation model shown in equations 1 through 8. For any given temperature and initial nitrogen concentration, the parameters μmax, k′d, βmax, and YX/N can be estimated from the relationships shown in Fig. 3 and 4. The parameters determined to not have a significant effect with temperature or initial conditions (YE/S, KN, and KS) are set to the mean value of all fermentations (e.g., KS would be set to approximately 10 g/liter). Estimates for all seven parameters are then combined and placed into equations 1 through 8 to estimate the time profiles of all five state variables (X, XV, N, E, and S).
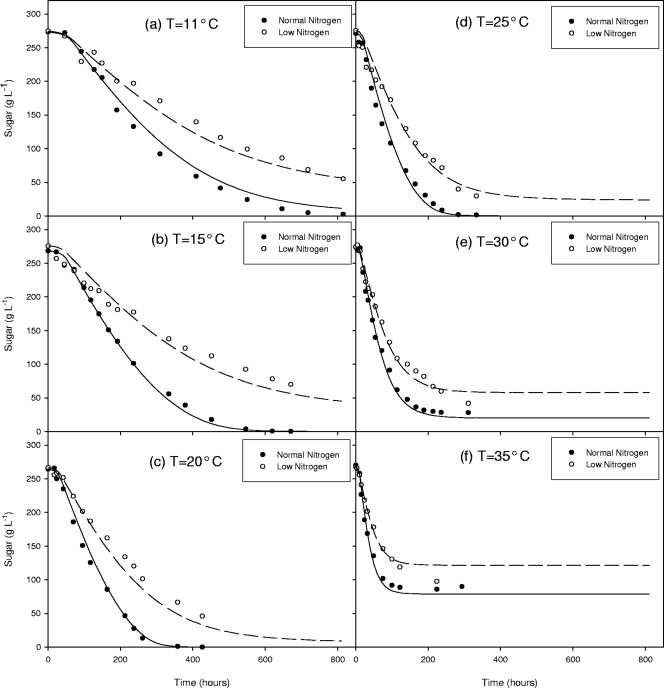
This comprehensive model was then shown to accurately predict a transition from sluggish to normal to stuck fermentations as the temperature rose from 11 to 35°C. Figure 6 shows this transition for both normal sugar/normal nitrogen and normal sugar/low-nitrogen fermentations. In general, model fits are best near 20°C; at extreme temperatures, model fits are slightly less accurate. The underlying feature to recognize in Fig. 6 is that at low temperatures, the sugar utilization rate is low; at the midrange temperatures, fermentations seem more normal; and at high temperatures, sugar utilization starts out rapidly and is then followed by a sudden cessation of fermentation activity. Experimental data and model fits are a close match and both show these underlying phenomena, unlike previous models for wine fermentation kinetics.
FIG. 6.
The comprehensive model fits of sugar utilization for normal sugar/normal nitrogen and normal sugar/low nitrogen fermentations ranging from 11 to 35°C. Notice that the model is able to explain the experimental transition from sluggish fermentation activity at low temperatures to normal fermentation activity at midrange temperatures to stuck fermentation activity at high temperatures.
Model predictions at extreme conditions.
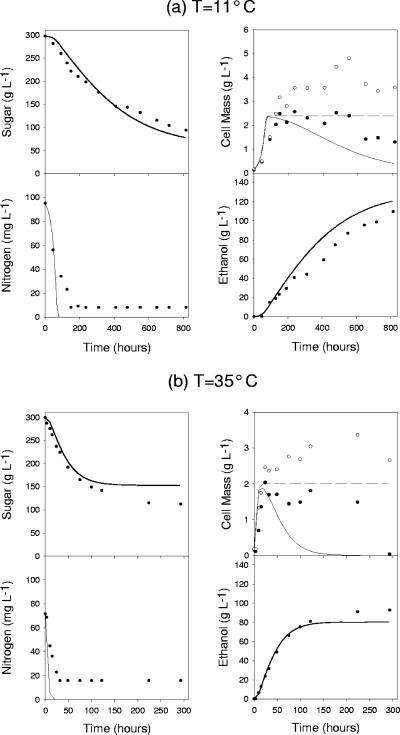
To test the predictive capabilities of the comprehensive model, two additional fermentations were performed at extreme conditions. One experiment was at 11°C, and the other was at 35°C. Both of these fermentations had high levels of sugar and low levels of nitrogen (an initial condition set that had not been tested). Figure 7 shows the model predictions and experimental data of all five state variables for both experiments. Both sugar utilization and ethanol production are accurately predicted by the model, with some minor discrepancies. For example, the residual sugar left in the fermentation at 35°C is slightly lower than what was predicted by the model. Predicted nitrogen concentrations are relatively accurate, with some minor discrepancies. For example, the model predicts that both fermentations will utilize all available nitrogen; however, some residual amounts remain at the end of fermentation. The model predictions of total cell mass concentration are relatively accurate; however, in both cases, the experimental data increase slightly after the exponential growth phase. The least accurately predicted state variable is the viable cell mass. This result is expected because the model predicts active cell mass (a currently immeasurable variable) and not viable cell mass. In both cases, the predictions of active cell mass decrease more rapidly than does the experimentally observed viable cell mass.
FIG. 7.
Predictions made by the comprehensive model for two different fermentations at extreme conditions. Experiments for both panel a and panel b start from high-sugar/low-nitrogen conditions; however, the experiment for panel a was run at a low temperature and that for panel b was carried out at a high temperature. The activity of both fermentations is relatively well predicted. The main differences between experimental and simulated predictions are the underestimations of viable (filled circles) and total (open circles and broken line) cell mass. However, sugar utilization is the most critical state variable to predict. Furthermore, it should be noted that the model predicts active cell mass and not viable cell mass. Thus, a difference between model prediction and observed data is expected.
DISCUSSION
Here we demonstrate that the model first proposed by Cramer et al. (11) has been successfully adapted to wine fermentations at various temperatures and initial conditions. This success is shown through the model's ability to fit various types of fermentation activity with respect to temperature and initial conditions (Fig. 2 and 7). Furthermore, the changes in the types of fermentation activity (i.e., sluggish, normal, stuck) can be accurately predicted by the model (Fig. 6). The reason behind this transition can be understood by examining the relationship each parameter has with respect to temperature. For example, the overall sluggish behavior of the fermentations at low temperatures can be understood by observing that the maximum sugar utilization rate (βmax) is lower at low temperatures (Fig. 3c). The stuck fermentations at higher temperatures can be explained by the significantly higher cell inactivation constant (k′d) sharply increasing above 25°C (Fig. 3b). While the cells may prefer a higher temperature for growth and sugar utilization, they become increasingly more sensitive to ethanol at higher temperatures. One potential explanation for this could be an increased physical response of the cell lipid bilayer to ethanol at elevated temperatures (17, 29). Furthermore, the quantitative predictions made by the model accurately resemble the observed experimental data at all temperatures studied, which span the normal range for industrial wine fermentations. While previous mechanistic wine models have been successful at predicting fermentation kinetics at room temperature (11) or for midrange temperatures (18), this is the first time that a wine model has been able to predict transitions to problem fermentations caused by high or low temperatures.
The model was also able to accurately predict fermentation activity at extreme conditions that had not been previously tested. From Fig. 6, we can see that model fits are typically less accurate at 11 or 35°C. However, data from fermentations using novel initial conditions (high sugar and low nitrogen) at these temperatures were shown to be accurately predicted by the model (Fig. 7). This shows that the model is robust across extreme conditions and could be used to predict problem fermentations at temperatures and initial conditions not yet observed, making it an interesting candidate for use in a practical winery setting.
The main discrepancy between the model fit and the data is the difference between experimental and model predictions of cell mass after the exponential growth phase. The difference between model predictions of cell activity and the experimentally observed cell viability is to be expected and is consistent with the results of Cramer et al. (11). This difference is likely due to cells that are inactive rather than dead. For example, ethanol may be preventing sugar transport into the cell, thus reducing sugar utilization even though the cells are not yet dead. Regardless of the reason, it seems certain that the cell viability measurements made are not a good measure of cell activity. This is the main reason that only the first seven data points from total and viable cell mass are included into the parameter estimation routine. This approach is well justified by the model's excellent prediction capabilities of sugar and ethanol (the most important state variables from an enologist's point of view). The model was also unable to predict that the total cell mass for many fermentations gradually increased after the exponential phase. This result may be due to the gradual degradation of nitrogen sources being generated from dead yeast cells. This source of nitrogen could then potentially be utilized by the active cells for cell reproduction. Regardless, not accounting for such additional cell growth had little to any effect on the final quantitative predictions of the other state variables.
Another small discrepancy between the model and the data is that the nitrogen measurements suggest that not all nitrogen is utilized during fermentation. Some small amount of nitrogen is typically left over in the fermentation. This leftover amount may be due to the inability of the yeast to utilize some types of nitrogen sources. However, fermentations at higher temperatures consistently resulted in higher residual nitrogen levels at the end of the fermentations (Fig. 7). This result could be due to cell growth inhibition at higher temperatures due to ethanol. Due to this added sensitivity of ethanol at higher temperatures, the cells may be unable to fully utilize all of the available nitrogen.
One interesting finding for the presented model is that the yield coefficient of cell mass on nitrogen turned out to significantly vary with respect to the initial nitrogen concentration (Fig. 4). The simplest and most likely explanation for this observation is that at higher concentrations of initial nitrogen, a different nutrient component could be growth limiting. This explanation would be consistent with the findings and data of Cramer et al. (11) and Malherbe et al. (18). A related potential explanation is that the cell mass yield is different for various forms of nitrogen. The juices in this work were first diluted, and then appropriate additions of DAP were added back to achieve the desired levels of nitrogen. The original juice contained 111 mg N/liter (ammonia) and 172 mg N/liter (α-amino nitrogen). After the dilution and addition of DAP, the low-nitrogen juices contained about equal parts nitrogen from ammonia and α-amino nitrogen. However, over 85% of the nitrogen in high-nitrogen juices was from ammonia. This difference in initial nitrogen composition could lead to differences in observed yield. As the temperature dependence of the fermentation is well predicted, regardless of the initial nitrogen level and composition, it is apparent that these two issues can be separated, leaving further explanation of the changes in biomass yield on nitrogen for further detailed study.
Despite these model discrepancies, the presented model accurately predicts the most important state variable: sugar concentration. Sugar utilization is of the utmost importance for any model that aims to aid in the prediction of stuck and sluggish fermentations. The accurate prediction of ethanol concentration, cell growth, cell activity, or nitrogen consumption gives valuable insight into microbial physiology; however, for the purposes of winemaking, accurate predictions of these variables are primarily important because they lead to the accurate prediction of sugar utilization. This model may be of value to winemakers because it always yields reasonable predictions of sugar utilization. Winemakers could potentially use this model to make decisions in the winery. For example, if a must is known to have a low level of nitrogen, a winemaker may typically supplement the natural level of available nitrogen with DAP to avoid a stuck or sluggish fermentation. However, certain winemakers may prefer to avoid the use of DAP because of the expense or because they believe it affects wine quality. The model presented in this paper could then be used to predict when the addition of DAP is critical and when it can be avoided.
Another important result to be stressed is that low-nitrogen fermentations are more sensitive to extremes in temperature. For example, Fig. 6 shows that for normal nitrogen conditions, any temperature between 11 and 25°C could be used to complete the fermentations (although 25°C would be the fastest). For low-nitrogen initial conditions, fermentation activity more easily becomes problematic at higher or lower temperatures. The optimal temperature range to complete low-nitrogen fermentations becomes smaller. While the experimental data at low-nitrogen levels were not allowed to go to completion, the model suggests that a temperature of 20°C would yield the greatest opportunity to finish a low-nitrogen-level fermentation.
While low levels of nitrogen have long been linked with problem fermentations, what the relationship between low levels of nitrogen and problem fermentations is has not been precisely determined. Some authors have linked low nitrogen to low cellular activity (2, 3), while others associated this condition with resulting low levels of biomass (11, 20). The data presented in this paper support the latter of these two hypotheses. Only one of the model parameters was shown to vary with respect to the initial nitrogen concentration. In addition, this one parameter (YX/N) only determines the amount of cell mass produced from a given quantity of nitrogen and does not directly affect the metabolic rates of reaction within a given cell. In the context of the fermentations used in this study, we can conclude that low levels of nitrogen contribute to the problematic fermentations because they result in fewer cells. However, this conclusion may not hold for other juices or strains of yeast which may react differently to low levels of nitrogen (15).
The adaptation of the presented model to industrial fermentations will be dependent upon collecting samples and determining the key state variables. These tasks must be carried out for several fermentations at different temperatures and conditions before predictions can be made. The empirical modeling methods presented below in “Parameter estimation” and “Regression modeling of model parameters” are carefully constructed to fully leverage all of the available information from the observed data and make optimal decisions with respect to parameter estimates and regression surfaces. However, these methods are not necessary for constructing a working model capable of predicting problematic fermentations. For example, a simple lookup table could be used to estimate model parameters that are temperature dependent (i.e., a regression model is not necessary to determine that fermentations at 11°C will result in a maximum specific growth rate of approximately 0.05 h−1). Future versions of this model may implement more-informative regression models for temperature dependence (e.g., mixtures of Arrhenius-type equations); however, these regression models will still be dependent upon data collected and parameters estimated for new systems.
In conclusion, we have shown that the presented model can accurately predict fermentation behavior across a wide variety of temperatures and initial conditions. This model is the first wine fermentation model that predicts a transition from sluggish to normal to stuck fermentations with respect to increasing temperature. Furthermore, the model could be used in a winery setting to determine the length of time required to finish a fermentation based on just the concentration of sugar and nitrogen in the juice, the optimal temperature to reach a minimum in sugar concentration, or whether the addition of a supplementary nitrogen source is critical. In order to adapt this model to various systems, it would be necessary for a winery to first collect data on the various state variables during the course of different fermentations. Once this collection of data has been achieved, a comprehensive model could be used to make predictions for new systems in different winery settings.
Acknowledgments
We thank the American Vineyard Foundation and the California Competitive Grant Program for Research in Viticulture and Enology for funding of this research.
APPENDIX
Parameter estimation.
Bayesian parameter estimation was used to estimate the seven model parameters for each set of fermentation data. The likelihood (L) for a given set of fermentation data was defined as
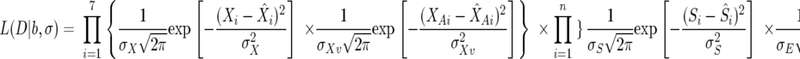 |
(A1) |
where D is the entire set of data collected for one fermentation, b is the vector of all model parameters (μmax, KN, k′d, βmax, KS, YX/N, and YE/S), σ is a vector of the noise parameters for each state variable (σX, σXA, σS, σE, and σN), n is the total number of points collected for each state variable, Xi is the experimental observed total cell mass at the ith observation, and X̂i is the model prediction for the total cell mass at the ith observation. Similarly, the circumflex marks the model prediction as opposed to the experimental value (no circumflex) for each of the other state variables as well. These model predictions are calculated using the model parameter values in b and equations 1 through 8.
Prior probabilities were then defined for each of the seven model parameters and each of the five noise parameters. For example, the prior probability over the maximum specific growth rate was
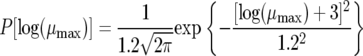 |
(A2) |
This is essentially using a normal distribution to state that all fermentations will have a μmax value between 0.0001 h−1 and 1.8 h−1 and most likely be closer to 0.05 h−1. Notice that we define the prior probability of μmax over the log(μmax) because it is a positive constant. Similar prior probabilities are defined over all model and noise parameters in the form of a normal distribution. The mean and standard deviation for each of these priors is shown in Table A1. These priors are then combined with the likelihood function (equation A1) to form the posterior probability.
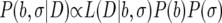 |
(A3) |
where P(b) and P(σ) are the combined priors for all model and noise parameters and P(b,σ|D) is the posterior distribution for all model and noise parameters given a set of data, D. This posterior is evaluated via MCMC integration to find the mean and credible regions shown for each model parameter shown in Fig. 3, 4, and 5 (10).
TABLE A1.
Values used to define the prior probabilities over all model and noise parametersa
| Parameter | Mean | Standard deviation |
|---|---|---|
| Model | ||
| Log(μmax) | −3 | 1.2 |
| Log(KN) | −4.61 | 0.7 |
| Log(k′d) | −9.21 | 1 |
| Log(YX/N) | 3.43 | 5 |
| Log(YE/S) | −0.755 | 2 |
| Log(βmax) | −1.2 | 2 |
| Log(KS) | 2.3 | 0.7 |
| Noise | ||
| Log(σX) | −2.3 | 0.05 |
| Log(σXv) | −4.6 | 1 |
| Log(σS) | 0 | 2.24 |
| Log(σE) | 0 | 2.24 |
| Log(σN) | −2.3 | 0.05 |
The mean and standard deviation values are for normal distributions defined over the log of each parameter.
Regression modeling of model parameters.
A robust method of regression was used to determine the regression surfaces shown in Fig. 3 and 4. The model for each surface was linear and in the form of the equation
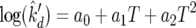 |
(A4) |
where a0, a1, and a2 are parameters to be estimated, and T is temperature in Celsius. As an example for all three parameters, the likelihood function to estimate k′d is shown by equation A5
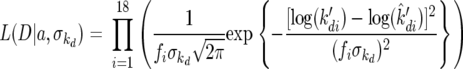 |
(A5) |
where D represents the data for the model parameter estimates of k′d, a represents the regression parameters in equation A4, 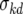 is the noise parameter associated with k′d, log(k̂′di) is the estimation of k′d for the ith fermentation from the regression surface of equation A4, and fi is the standard deviation of the credible region for the ith parameter estimate. Essentially, all the fi values do is adjust the importance of the fit for each parameter value based upon the size of its credible regions. For example, the credible regions at high temperatures are larger in Fig. 3b. Thus, fi when T is 35°C is greater than fi when T is 11°C. No prior distributions were used for the regression surface parameters (a) or the noise parameter
is the noise parameter associated with k′d, log(k̂′di) is the estimation of k′d for the ith fermentation from the regression surface of equation A4, and fi is the standard deviation of the credible region for the ith parameter estimate. Essentially, all the fi values do is adjust the importance of the fit for each parameter value based upon the size of its credible regions. For example, the credible regions at high temperatures are larger in Fig. 3b. Thus, fi when T is 35°C is greater than fi when T is 11°C. No prior distributions were used for the regression surface parameters (a) or the noise parameter 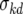 . Thus, MCMC integration was used to evaluate likelihood in equation A5 and to estimate mean values and credible regions for the regression surface parameters (10). The values for all regression surfaces are summarized in Table A2.
. Thus, MCMC integration was used to evaluate likelihood in equation A5 and to estimate mean values and credible regions for the regression surface parameters (10). The values for all regression surfaces are summarized in Table A2.
TABLE A2.
Estimated parameter values for each of the regression surfacesa
| Model parameter | Value for regression surface
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
|
a0
|
a1
|
a2
|
|||||||
| Mean | 5% CR | 95% CR | Mean | 5% CR | 95% CR | Mean | 5% CR | 95% CR | |
| Log(μmax) | −3.92 | −4.14 | −3.70 | 7.82 × 10−2 | 6.98 × 10−2 | 8.79 × 10−2 | NA | NA | NA |
| Log(KN) | −4.73 | −5.02 | −4.44 | NA | NA | NA | NA | NA | NA |
| Log(k′d) | −9.81 | −10.6 | −8.94 | −1.08 × 10−3 | −1.94 × 10−1 | −3.30 × 10−2 | 4.78 × 10−3 | 3.16 × 10−3 | 6.58 × 10−3 |
| Log(YX/N) | 3.50 | 3.31 | 3.70 | −3.61 | −4.35 × 10−3 | −2.93 × 10−3 | NA | NA | NA |
| Log(YE/S) | −5.98 × 10−1 | −6.36 × 10−1 | −5.58 × 10−1 | NA | NA | NA | NA | NA | NA |
| Log(βmax) | −2.30 | −3.22 | −2.74 | 7.71 × 10−2 | 6.66 × 10−2 | 8.85 × 10−2 | NA | NA | NA |
| Log(KS) | 2.33 | 1.99 | 2.65 | NA | NA | NA | NA | NA | NA |
Estimated parameter values for each of the regression surfaces shown in Fig. 3, 4, and 5. When regression surfaces did not utilize all linear parameters, the symbol NA (not applicable) is put in place of the estimated value. For example, log(k′d) utilizes a0, a1, and a2 (a quadratic surface); however, log(KN) utilizes only a0 [thus, all values of log(KN) are estimated by the mean value of a0]. CR, credible region.
Footnotes
Published ahead of print on 6 July 2007.
REFERENCES
- 1.Alexandre, H., and C. Charpentier. 1998. Biochemical aspects of stuck and sluggish fermentation in grape must. J. Ind. Microbiol. Biotechnol. 20:20-27. [Google Scholar]
- 2.Bely, M., J. M. Sablayrolles, and P. Barre. 1990. Automatic detection of assimilable nitrogen deficiencies during alcoholic fermentation in enological conditions. J. Ferment. Bioeng. 70:246-252. [Google Scholar]
- 3.Bely, M., J. M. Salmon, and P. Barre. 1994. Assimilable nitrogen addition and hexose-transport system activity during enological fermentation. J. Inst. Brew. 100:279-282. [Google Scholar]
- 4.Bezenger, M. C., and J. M. Navarro. 1987. Grape juice fermentation—effects of initial nitrogen concentration on culture parameters. Sci. Aliments 7:41-60. [Google Scholar]
- 5.Bisson, L. F. 1999. Stuck and sluggish fermentations. Am. J. Enol. Vitic. 50:107-119. [Google Scholar]
- 6.Bisson, L. F., and D. E. Block. 2002. Ethanol tolerance in Saccharomyces, p. 86-98. In M. Ciani (ed.), Biodiversity and biotechnology of wine yeast. Research Signpost, Kerala, India.
- 7.Boulton, R. 1980. Prediction of fermentation behavior by a kinetic-model. Am. J. Enol. Vitic. 31:40-45. [Google Scholar]
- 8.Caro, I., L. Perez, and D. Cantero. 1991. Development of a kinetic-model for the alcoholic fermentation of must. Biotechnol. Bioeng. 38:742-748. [DOI] [PubMed] [Google Scholar]
- 9.Charoenchai, C., G. H. Fleet, and P. A. Henschke. 1998. Effects of temperature, pH, and sugar concentration on the growth rates and cell biomass of wine yeasts. Am. J. Enol. Vitic. 49:283-288. [Google Scholar]
- 10.Coleman, M. C., and D. E. Block. 2006. Bayesian parameter estimation with informative priors for nonlinear systems. AICHE J. 52:651-667. [Google Scholar]
- 11.Cramer, A. C., S. Vlassides, and D. E. Block. 2002. Kinetic model for nitrogen-limited wine fermentations. Biotechnol. Bioeng. 77:49-60. [DOI] [PubMed] [Google Scholar]
- 12.Dukes, B. C., and C. E. Butzke. 1998. Rapid determination of primary amino acids in grape juice using an o-phthaldialdehyde/N-acetyl-L-cysteine spectrophotometric assay. Am. J. Enol. Vitic. 49:125-134. [Google Scholar]
- 13.Giovanelli, G., C. Peri, and E. Parravicini. 1996. Kinetics of grape juice fermentation under aerobic and anaerobic conditions. Am. J. Enol. Vitic. 47:429-434. [Google Scholar]
- 14.Ingledew, W. M., and R. E. Kunkee. 1985. Factors influencing sluggish fermentations of grape juice. Am. J. Enol. Vitic. 36:65-76. [Google Scholar]
- 15.Julien, A., J. L. Roustan, L. Dulau, and J. M. Sablayrolles. 2000. Comparison of nitrogen and oxygen demands of enological yeasts: technological consequences. Am. J. Enol. Vitic. 51:215-222. [Google Scholar]
- 16.López, A., and P. Secanell. 1992. A simple mathematical empirical-model for estimating the rate of heat-generation during fermentation in white-wine making. Int. J. Refrig. 15:276-280. [Google Scholar]
- 17.Ly, H. V., D. E. Block, and M. L. Longo. 2002. Interfacial tension effect of ethanol on lipid bilayer rigidity, stability, and area/molecule: a micropipet aspiration approach. Langmuir 18:8988-8995. [Google Scholar]
- 18.Malherbe, S., V. Fromion, N. Hilgert, and J. M. Sablayrolles. 2004. Modeling the effects of assimilable nitrogen and temperature on fermentation kinetics in enological conditions. Biotechnol. Bioeng. 86:261-272. [DOI] [PubMed] [Google Scholar]
- 19.Marin, M. R. 1999. Alcoholic fermentation modelling: current state and perspectives. Am. J. Enol. Vitic. 50:166-178. [Google Scholar]
- 20.Monteiro, F. F., and L. F. Bisson. 1992. Nitrogen supplementation of grape juice. 1. Effect on amino-acid utilization during fermentation. Am. J. Enol. Vitic. 43:1-10. [Google Scholar]
- 21.Nanba, A., Y. Nishizawa, Y. Tsuchiya, and S. Nagai. 1987. Kinetic-analysis for batch ethanol fermentation of Saccharomyces cerevisiae. J. Ferment. Technol. 65:277-283. [Google Scholar]
- 22.Ough, C. S. 1966. Fermentation rates of grape juice. III. Effects of initial ethyl alcohol, pH, and fermentation temperature. Am. J. Enol. Vitic. 17:74-81. [Google Scholar]
- 23.Ough, C. S. 1964. Fermentation rates of grape juice. I. Effects of temperature and composition on white juice fermentation rates. Am. J. Enol. Vitic. 15:167-177. [Google Scholar]
- 24.Ough, C. S. 1966. Fermentation rates of grape juice. II. Effect of initial °Brix, pH, and fermentation temperature. Am. J. Enol. Vitic. 17:20-26. [Google Scholar]
- 25.Ough, C. S., and M. A. Amerine. 1966. Fermentation rates of grape juice. IV. Compositional changes affecting prediction equations. Am. J. Enol. Vitic. 17:163-173. [Google Scholar]
- 26.Ough, C. S., and M. A. Amerine. 1961. Studies with controlled fermentation. VI. Effects of temperature and handling on rates, composition, and quality of wines. Am. J. Enol. Vitic. 12:117-128. [Google Scholar]
- 27.Piper, P. W. 1995. The heat-shock and ethanol stress responses of yeast exhibit extensive similarity and functional overlap. FEMS Microbiol. Lett. 134:121-127. [DOI] [PubMed] [Google Scholar]
- 28.Sa-Correia, I., and N. Van Uden. 1983. Temperature profiles of ethanol tolerance—effects of ethanol on the minimum and the maximum temperatures for growth of the yeasts Saccharomyces cerevisiae and Kluyveromyces fragilis. Biotechnol. Bioeng. 25:1665-1667. [DOI] [PubMed] [Google Scholar]
- 29.Tierney, K. J., D. E. Block, and M. L. Longo. 2005. Elasticity and phase behavior of DPPC membrane modulated by cholesterol, ergosterol, and ethanol. Biophys. J. 89:2481-2493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vlassides, S., J. G. Ferrier, and D. E. Block. 2001. Using historical data for bioprocess optimization: modeling wine characteristics using artificial neural networks and archived process information. Biotechnol. Bioeng. 73:55-68. [DOI] [PubMed] [Google Scholar]