Abstract
We report that endogenously synthesized (−)-proto-quercitol (1d-1,3,4/2,5-cyclohexanepentol) and glycine betaine were the principal compatible solutes of Schizochytrium sp. strain S8 (ATCC 20889) and three new osmotolerant isolates of thraustochytrids (strains T65, T66, and T67). The compatible solutes were identified and quantified by use of nuclear magnetic resonance spectroscopy, and their identity was confirmed by mass spectroscopy and measurement of the specific optical rotation. The cellular content of compatible solutes increased with increasing NaCl concentration of a defined medium. (−)-proto-Quercitol was the dominating solute at all NaCl concentrations tested (0.25 to 1.0 M), e.g., cells of S8 and T66 stressed with 1.0 M NaCl accumulated about 500 μmol (−)-proto-quercitol and 100 μmol glycine betaine per g dry weight. To our knowledge, (−)-proto-quercitol has previously been found only in eucalyptus. The 18S rRNA gene sequences of the four (−)-proto-quercitol-producing strains showed 99% identity, and they displayed the same fatty acid profile. The only polyunsaturated fatty acids accumulated were docosahexaenoic acid (78%) and docosapentaenoic acid (22%). A less osmotolerant isolate (strain T29), which was closely phylogenetically related to Thraustochytrium aureum (ATCC 34304), did not contain (−)-proto-quercitol or glycine betaine. Thus, the level of osmotolerance and the osmolyte systems vary among thraustochytrids.
Thraustochytrids are saphrophytic marine protists which can be cultivated on dissolved organic nutrients. Data compiled by Raghukumar (48) show that thraustochytrids have a wide distribution in coastal and oceanic waters and sediments. They prefer sites of high nutrient concentration, such as detritus from mangrove leaf, algae, and phytoplankton. Depending on the nutritional conditions, their numbers in the water column vary from below the detection level to 106 cells liter−1. At their maxima, their biomass in the water column can be similar to the bacterial biomass (reference 48 and references therein).
Many thraustochytrids accumulate large amounts of triacylglycerols containing polyunsaturated fatty acids (PUFA), particularly docosahexaenoic acid (DHA; C22:6 n-3). Thus, thraustochytrids are primary producers of long-chain PUFA for marine food chains (48). Dried products of DHA-containing thraustochytrids have been commercialized as feed for rotifers used in marine aquaculture and feed supplement to egg-laying hens. For humans, consumption of DHA has been linked to a broad range of health benefits, including fetal brain development and cancer treatments. It is believed that thraustochytrids have the potential to become important production organisms for the commercial DHA market (7, 35).
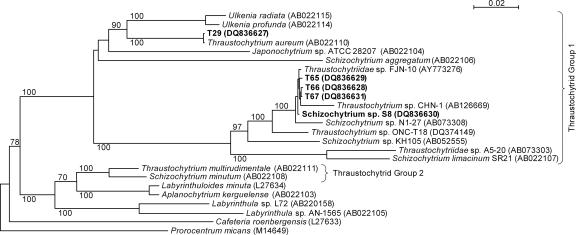
Adl et al. (2) rank thraustochytrids under Labyrinthulomycetes, which are again ranked under Stramenopiles (Chromista). Labyrinthulomycetes are divided into two groups: Thraustochytriaceae, comprising seven genera including Thraustochytrium, Schizochytrium, and Ulkenia, and Labyrinthulaceae, with only one genus, Labyrinthula. However, the taxonomy of thraustochytrids remains confusing because classical taxonomy based on morphology and molecular taxonomy based on 18S rRNA gene sequences do not agree at the genus level (27, 28). Thraustochytrids have recently been divided into two subgroups based on 18S ribosomal DNA taxonomy (33) (see Fig. 1, below). For thraustochytrids isolated from coastal waters of Japan and Fiji, it has been reported that strains with similar PUFA profiles cluster on the phylogenetic tree (28).
FIG. 1.
Neighbor-joining tree of thraustochytrids and labyrinthulids based on 18S rRNA gene sequences with Prorocentrum micans as the outgroup. GenBank accession numbers are shown in parentheses. Bold type indicates the strains for which the 18S rRNA gene sequences were determined in the present study. Bootstrap values equal to and above 70% of 1,000 replicates are shown above branches.
Thraustochytrids have a requirement for sodium ions (20-22, 29). They often prefer salinities corresponding to 40 to 100% seawater (15, 29, 64), but strains that grow well with a wider range of salinities and tolerate at least 150% (11) or 200% (64) seawater have been described. All eukaryotic microorganisms analyzed to date accumulate compatible solutes (low-molecular-weight organic osmolytes) to provide osmotic balance and turgor. This strategy allows organisms to adapt to changes in environmental salinity without undertaking any gross changes of their cytoplasmic macromolecule and cell organelle constituents (63). Osmotically stressed yeasts and fungi often accumulate polyols, such as glycerol, mannitol, arabitol, or sorbitol (9). The known compatible solutes of microalgae belong to several chemical categories: for examples, sugar and sugar derivatives, such as sucrose, polyols, and heterosides; methylated tertiary N compounds, such as glycine betaine and homarine; the methylated S compound dimethylsulphoniopropionate; and amino acids such as proline and glutamate (57, 58). In addition, some microalgae accumulate species of cyclohexanetetrol, e.g., dextrorotary 1d-1,4/2,5-cyclohexanetetrol (14, 18, 19). Wethered and Jennings (60) reported that the proline content of Thraustochytrium aureum and Thraustochytrium roseum increases linearly with the external salt concentration in cells grown in 50 to 100% seawater, but they pointed out that proline and other free amino acids give only a small contribution to the osmotic pressure of the cytoplasm. At first it was proposed that K+ and Na+ ions are major cytoplasmic osmolytes in these organisms (60), but this proposal was later retracted (20). Thus, the major osmolyte systems of thraustochytrids remain unknown.
Quercitol is commonly used as the generic name of cyclohexanepentols, which comprise a family of 16 stereoisomers. Twelve of them are optically active. In the chemical literature, cyclohexanepentols attract attention particularly because of the challenge in synthesizing enantiomerically pure compounds. The isomer of cyclohexanepentol first discovered was isolated from acorn of oak (Quercus sp.) in 1849, hence, the name quercitol. This compound (1l-1,3,4/2,5-cyclohexanepentol) is dextrorotary, and the name (+)-proto-quercitol has come into use. Levorotary (−)-proto-quercitol (1d-1,3,4/2,5) was discovered in leaves of Eucalyptus populnea in 1961 (see Fig. 3, below, for the formula). The only other optically active cyclohexanepentol found in nature is (−)-vibo-quercitol (1l-1,2,4/3,5) of the evergreen shrub Viburnum tinus (references 37 and 51 and references therein). It should be noted that both stereoisomers of proto-quercitol are sometimes simply called quercitol; for example, it has been reported that salt- and drought-tolerant species of eucalyptus accumulate quercitol (1, 39).
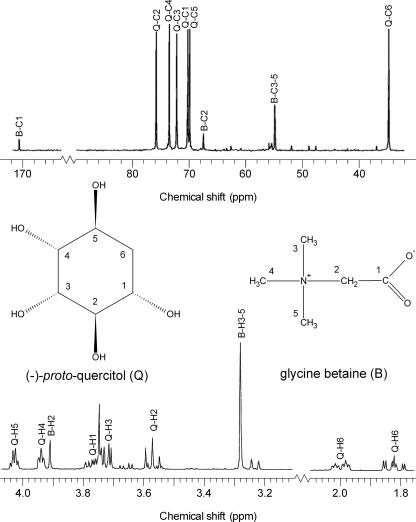
FIG. 3.
13C NMR (top) and 1H NMR (bottom) spectra of (−)-proto-quercitol (Q) and glycine betaine (B) in the ethanolic extract of strain T66. The assignments of the signals are marked on the spectra, i.e., compatible solute (Q or B) and atom (C or H) with the numberings given on the shown formulae of the compatible solutes. The assignments were based on previously published NMR data (3, 16, 37, 47, 51). The cells were grown in medium M1 with 1.0 M NaCl added.
Organic osmolytes comprise a major part of the cytoplasmic pool of metabolites in osmotically stressed microorganisms, and they can therefore be identified and quantified by nuclear magnetic resonance (NMR) spectroscopy of crude cell extracts (34). By performing 1H and 13C NMR analyses together with mass spectroscopy (MS) and measurement of specific optical rotation, we showed that (−)-proto-quercitol and glycine betaine were the principal compatible solutes of four osmotolerant strains of thraustochytrids.
MATERIALS AND METHODS
Isolation procedure and strains.
The pine pollen baiting method (10) was used for isolation of thraustochytrids; that is, the marine samples were mixed with small amounts of γ-irradiated pine pollen and supplemented with ampicillin, penicillin G, streptomycin, and nystatin (0.3 g liter−1 of each). After incubation at 25°C for 3 to 5 days, samples of pollen judged by microscopy to harbor thraustochyrids were streaked onto solid seawater-based medium containing (g liter−1): glucose (20), yeast extract (2), peptone (2), (NH4)2SO4 (2), ampicillin (0.3), streptomycin (0.3), and agar (10) in 60% seawater. Ensuing colonies of thraustochytrids were purified by repeated transfer on the same agar medium. The isolates were stored in 15% glycerol at −80°C. Strains T65, T66, and T67 were isolated from a mixture of marine sediment and seawater sampled from the south coast of Madeira, Portugal, and strain T29 was isolated from seawater sampled from the south coast of Norway near Kristiansand. Schizochytrium sp. strain S8 (ATCC 20889) was obtained from the American Type Culture Collection.
Growth media and growth conditions.
Our growth medium was based on a previously published medium (22) which was modified to give higher growth yields. Medium M1 used for cultivation (28°C) of strains T65, T66, T67, and S8 contained (g liter−1): glycerol (20), sodium glutamate (5), MgCl2 (0.8), Na2SO4 (2.4), KCl (0.4), KH2PO4 (0.5), maleic acid (5.8), Tris base (6.1), ampicillin (0.3), streptomycin (0.3), cyanocobalamin (5 × 10−6), and thiamine-HCl (5 × 10−5) plus 0.5 ml liter−1 trace mineral solution (see below). The medium was supplemented with 0.25 to 2.0 M NaCl as stated in the text. In some instances, 0.47 g glycine betaine liter−1 (4 mM) was added, or the standard amount of sodium glutamate was replaced with 5 g glycine betaine liter−1. pH was adjusted to 7.0 with KOH. Medium M2, which was used to test the Na+ requirement of cells, was identical to medium M1, except that 5 g sodium glutamate was replaced with 2 g glutamic acid and 2.4 g Na2SO4 was replaced with 3.0 g K2SO4. Medium M2 was supplemented with 0.10 M NaCl, 0.25 M NaCl, or 0.10 M NaCl and 0.15 M KCl as stated in the text. The trace mineral solution contained (g liter−1): FeSO4 · 7H2O (50.0), CuSO4 · 5H2O (3.9), ZnSO4 · 7H2O (4.4), MnSO4 · H2O (1.5), Na2MoO4 · 2H2O (0.1), and CoCl2 · 6H2O (0.2) in 0.6 M HCl. Strain T29 was grown at 25°C in medium M1 supplemented with 2 g yeast extract liter−1.
Unless otherwise stated, the organisms were cultured in 500-ml baffled shake flasks containing 100 ml medium. The flasks were incubated on an orbital shaker (200 rpm) in a moisturized chamber. The cells were always pregrown in the same medium as used in the following growth experiment. Cell growth was monitored by measuring the turbidity (optical density) at 660 nm (OD660) after proper dilution of the culture with an isotonic solution of NaCl and breaking of cell aggregates by using a syringe to press and draw the cell suspension three times through a needle of 0.55 by 25 mm. Doubling times in the exponential growth phase were calculated by linear regression of semilogarithmic plots of the OD660 versus time. Standard deviations of the mean doubling time were calculated on the basis of 10 independent parallel cultures. This was performed for three of the salt concentrations tested (0.25, 0.50, and 1.0 M). These values, as a percentage of the mean value, were used as the basis for calculation of standard deviations in other growth experiments. However, all measurements were based on at least two independent growth experiments. Cells used for chemical analyses were harvested by centrifugation at 3,200 × g for 10 min (4°C), washed once with an isotonic solution of NaCl, and then freeze-dried.
Osmolalities of media M1 and M2 with added NaCl were measured by freezing point depression by using an Osmomat Gonotec 030D apparatus (Gonotec). The values for osmotic strength given in the text and below in Fig. 4 and 5 were the measured osmolalities of the glycerol-containing medium minus the previously reported value for the osmolality of 20 g glycerol liter−1, i.e., 0.22 osmol kg−1 (61). This subtraction was made because it is a common experience that glycerol does not exert any osmotic effect in living cells, at least not in bacteria (31, 59).
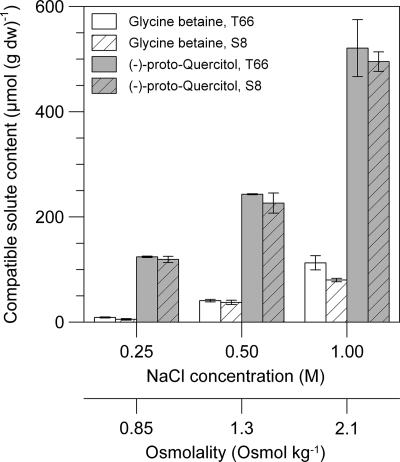
FIG. 4.
Osmotic stress-dependent accumulation of (−)-proto-quercitol and glycine betaine in Schizochytrium sp. strain S8 (ATCC 20889) and strain T66. The cells were grown in medium M1 with 0.25 to 1.0 M NaCl added. The resulting osmolalities are shown on the second x axis. The cell contents of the compatible solutes (μmol per g dry weight) were quantified by 1H NMR spectroscopy of ethanolic extracts. The values given are the means ± standard deviations of measurements from four separate cell cultures.
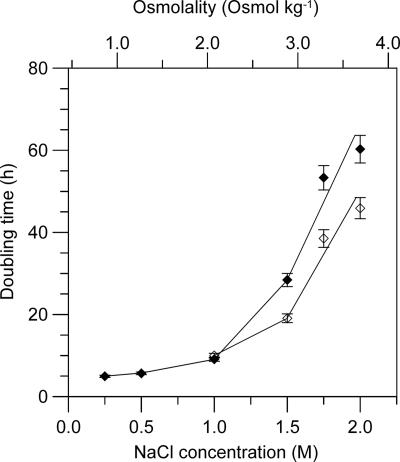
FIG. 5.
Influence of osmotic stress generated with NaCl and externally added glycine betaine on the doubling time of strain T66. Filled diamonds, cells grown in medium M1 with 0.25 to 2.0 M NaCl added; open diamonds, cells grown in medium M1 with 1.0 to 2.0 M NaCl plus 4 mM glycine betaine added. The resulting medium osmolalities are shown on the upper x axis. Cell growth was measured turbidimetrically at 660 nm. The values given for the doubling times are the means ± standard deviations of measurements from 2 to 10 separate cell cultures (see Materials and Methods).
Cultivations in bioreactors.
Cells used for analysis of fatty acids were cultivated in a 1.8-liter working volume stirred reactor (Applicon) equipped to maintain the pH at 7.0 and dissolved oxygen at 20% of saturation. Medium M1 was used with the following modifications (g liter−1): sodium glutamate (25), KH2PO4 (4.0), Na2SO4 (3.0), CaCl2 · 2H2O (0.5), plus 2.5 ml trace mineral solution (see above). The initial concentration of glycerol was 30 g liter−1 (strain T29) or 145 g liter−1 (other strains), and the concentration was always kept above 20 g liter−1 by addition of batches of glycerol. The medium was supplemented with 0.25 M NaCl. Antifoam (Clerol FBA 622) was added as required. The growth medium of T29 was additionally supplemented with 2 g yeast extract liter−1.
Classification based on 18S rRNA gene sequence. (i) DNA extraction.
The cell pellet of a 2-ml culture was suspended in 200 μl lysis buffer (0.25 M Tris-HCl, 0.1 M Na2-EDTA, 2% [wt/vol] sodium dodecyl sulfate, and 0.1 M NaCl, pH 8.2), and 20 μl proteinase K solution (DNeasy tissue kit; QIAGEN) was added. The mixture was incubated for 30 min at 70°C. One hundred μl of the cell suspension was diluted with 360 μl deionized water, and DNA was extracted using the DNeasy tissue kit, according to the manufacturer's instructions. Purified DNA was dissolved in 100 μl water and stored at −20°C.
(ii) PCR amplification and DNA sequencing.
Five μl of diluted DNA solution (1:10) was added to 35 μl PCR solution containing 0.3 mM deoxynucleoside triphosphate solution, 1.25 μM of primers FA1 and RA3 designed by Mo et al. (41), 3.5 U DNA polymerase, and 12.5% (vol/vol) buffer 1 from the Expand High-Fidelity PCR system (Roche Diagnostics). PCR amplifications were performed as described previously (41). PCR products were purified from agarose gels using a QIAEX II kit (QIAGEN) and sequenced by MWG Biotech (Germany) using primers FA1, FA2, FA3, RA1, RA2, and RA3 (41).
(iii) Phylogenetic analysis.
The obtained 18S rRNA gene sequences were aligned with corresponding sequences of related microorganisms available from the National Center for Biotechnology Information using ClustalX version 1.83 (56). Positions with gaps and undetermined and ambiguous bases were removed before the subsequent phylogenetic analyses. A phylogenetic tree was constructed by the neighbor-joining method (50) with Prorocentrum micans as the outgroup. The statistical significance of the tree branches was assessed by 1,000 bootstrap resamplings (17).
Fatty acid analysis.
Lipids were extracted from freeze-dried cells with chloroform-methanol-water according to the method of Bligh and Dyer (8), as modified by Hardy and Keay (24). The lipids were then hydrolyzed in 0.5 M NaOH, and free fatty acids were methylated with 14% BF3 in methanol (40). Analysis of the methyl esters was carried out using an Agilent 6890 series gas chromatograph equipped with an autoinjector (injection volume of 1 μl, split ratio of 1:100) operated at 260°C and a flame ionization detector operated at 310°C. An SPB PUFA (Supelco) fused silica capillary column (30 m by 0.32 mm inner diameter) coated with a polyalkylene glycol stationary phase was used. The oven was operated at a constant temperature of 210°C for 60 min. Helium was used as the carrier gas. The signals were identified by comparison with known standards (mixture ME 81; Larodan Fine Chemicals) and quantified according to the response factor of the internal fatty acid standard C21:0 (Larodan Fine Chemicals). The fatty acid contents and profiles presented were based on analyses of samples from three independent bioreactor cultures.
Ethanol extraction of cells.
Freeze-dried cells (0.5 to 1.0 g) were suspended in 15 ml of 90% (vol/vol) ethanol in water and incubated in rotating tubes at room temperature for 30 min. After centrifugation, the supernatant was removed and the extraction was repeated three times using 15 ml of 80% (vol/vol) ethanol. The combined supernatants were filtered through a 0.2-μm membrane filter, and the solvents were removed under reduced pressure.
Identification and quantification of organic osmolytes by NMR spectroscopy.
Dried ethanolic extracts of cells were dissolved in deuterated water. NMR spectra were recorded at 25°C on a Bruker Avance DPX 400 spectrometer. 1H NMR spectra were acquired using a 90° pulse, a spectral width of 4,789 Hz, acquisition time of 4.0 s, and a repetition delay of 16.0 s. The repetition delay was five times the longest relaxation time (T1) of the compounds measured, which was 3.2 s, obtained for the internal standard sodium-3′-trimethylsilylpropionate-2,2,3,3-d4 (TSP). The number of scans was 16. Exponential line broadening of 0.3 Hz was used. Chemical shifts and peak integrals for the metabolites were measured relative to TSP at 0 ppm. The concentration of (−)-proto-quercitol was quantified by calculating the average integral of four signals of the spectrum at 1.82, 2.00, 3.94, and 4.03 ppm, and the concentration of glycine betaine was calculated on basis of the signal at 3.28 ppm (see Fig. 3, below). The values reported are mean values of four separate experiments. 13C NMR spectra were acquired using a 30° pulse, a spectral width of 31,847 Hz, acquisition time of 1.0 s, and a repetition delay of 3.0 s. The number of scans was 3,722.
Purification of proto-quercitol and determination of optical rotation.
Dried ethanolic extract of T66 cells was dissolved in pyridine, and proto-quercitol was acetylated as described previously (51). The resulting quercitol pentaacetate was fractionated from impurities over a silica gel using pentane and ethylacetate (3:1) as solvents. The solvents were removed by evaporation under reduced pressure, and quercitol pentaacetate was confirmed by 1H NMR analysis. Deacetylation with ammonia in methanol was performed as described previously (51), and the resulting proto-quercitol was crystallized from hot ethanol and confirmed by 1H NMR and MS analyses. Specific optical rotation was measured by using a Perkin-Elmer 241 polarimeter at 20°C.
MS analyses. (i) Identification of glycine betaine.
Dried ethanolic extract of T66 cells was dissolved in 70% (vol/vol) methanol. Ammonium acetate was added to a final concentration of 10 mM, and direct infusion MS analysis was performed on an Agilent SL ion-trap MS apparatus using electrospray ionization in positive mode. An injection speed of 10 μl min−1 was used.
(ii) Identification of proto-quercitol.
Purified proto-quercitol from strain T66 (see above) was dissolved in water. An accurate mass determination was performed by flow injection MS analysis performed on an Agilent 1100 high-pressure liquid chromatograph connected to an Agilent mass selective detector time-of-flight MS apparatus with atmospheric pressure chemical ionization in negative mode. The mobile phase consisted of water and methanol (9:1). A flow rate of 0.2 ml min−1 and an injection of 2 μl were used.
Nucleotide sequence accession numbers.
The 18S rRNA gene sequences of Schizochytrium sp. strain S8 (ATCC 20889) and the new isolates T65, T66, T67, and T29 have been submitted to GenBank with the accession numbers that are stated in Fig. 1.
RESULTS
Characterization of test organisms.
Strains T65, T66, and T67, isolated by us from the coast of Madeira, Portugal, and Schizochytrium sp. strain S8 (ATCC 20889), previously isolated from the coast of California, were the main test organisms used in this study. These strains grew well in defined liquid medium containing glutamate and glycerol as the only sources of carbon, nitrogen, and energy and with vitamins B1 and B12 as the only growth factors (medium M1). They tolerated a wide range of NaCl concentrations (0.1 to 2.0 M; see below). All four strains formed biflagellated zoospores. Based on their 18S rRNA gene sequences, they belonged to thraustochytrid group 1, and they formed a tight cluster together with Schizochytrium sp. strain N1-27 (28, 55) and Thraustochytriidae sp. strain FJN-10 (accession number AY773276) (Fig. 1). As originally noted by Honda et al. (27), 18S rRNA genes of thraustochytrids carry a variable insert of approximately 14 to 28 bp in the region between nucleotides 1712 and 1738. Strains T66, T67, S8, FJN-10, and N1-27 carried the same insert (CTTTTCGCTGCTCGCAG), whereas T65 carried a unique insert (ATTATCGTTGTTTTGCAG). Pair-wise comparisons of the 18S rRNA gene sequences of T65, T66, T67, and S8 showed 99% identity. They displayed 89% identity with Schizochytrium limacinum SR21 (42), which appeared on the same phylogenetic branch (Fig. 1).
In order to induce a high level of lipid accumulation, T65, T66, T67, and S8 were grown in bioreactors in a medium with a high C-to-N ratio (excess of glycerol) and 0.25 M NaCl. After an incubation period of approximately 130 to 140 h (28°C), DHA content reached its maximum value of about 29% of total fatty acids (Fig. 2), and the lipid content was 55% of dry weight. At this time, DHA comprised 78%, and docosapentaenoic acid (DPA; C22:5 n-6) comprised 22% of total PUFA. The content of other PUFA did not exceed the cutoff level, which was set at 1% of total fatty acids. Within the limits of the experimental errors, strains T65, T67, and S8 had the same fatty acid composition as T66 (data not shown).
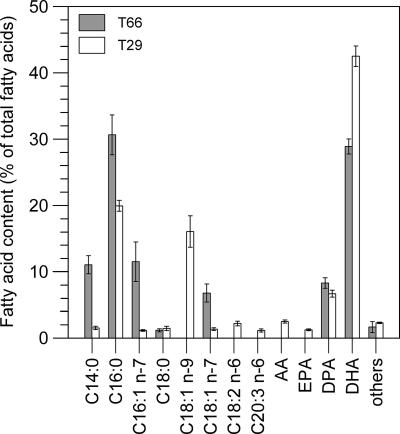
FIG. 2.
Fatty acid profiles of strains T29 and T66. The cells were grown in medium M1 with a high C-to-N ratio (excess glycerol) and assayed at the time the percentage of DHA was at its maximum value (see text). The values given are the means ± standard deviations of measurements of cells from three separate bioreactor cultures. Abbreviations: AA, arachidonic acid (C20:4 n-6); EPA, eicosapentaenoic acid (C20:5 n-3); DPA, docosapentaenoic acid (C22:5 n-6); DHA, docosahexaenoic acid (C22:6 n-3).
For comparison, we performed some experiments with strain T29 isolated from the south coast of Norway. This strain did not grow in the defined medium M1 but grew well when this medium was fortified with yeast extract. Furthermore, T29 tolerated only about 0.75 M NaCl. It formed biflagellated zoospores and belonged phylogenetically to the thraustochytrid group 1. It clustered (Fig. 1) with Thraustochytrium aureum (ATCC 34304) (5, 22), i.e., their 18S rRNA gene sequences had nearly 100% identity and they carried the same insert (ATTTGTTTTTTTCTCATTTTGGGAGGGGG). The 18S rRNA gene sequence of T29 had approximately 85% identity with the four other strains examined by us.
In T29 cells grown in yeast extract-fortified medium M1 (0.25 M NaCl) with a high C-to-N ratio, the DHA content reached its maximum value of 43% of total fatty acids after an incubation period of approximately 220 to 230 h (25°C). The lipid content was then 45% of dry weight. In terms of percentage of total PUFA, DHA accounted for 76% and DPA for 12%. PUFA that occurred in amounts of 2 to 4% were arachidonic acid (AA; C20:4 n-6), eicosapentaenoic acid (EPA; C20:5 n-3), eicosatrienoic acid (C20:3 n-6), and linoleic acid (C18:2 n-6) (Fig. 2).
Identification of (−)-proto-quercitol and glycine betaine in osmotically stressed cells.
For identification of putative compatible solutes, T65, T66, T67, and S8 were grown in medium M1 with a low C-to-N ratio (20 g glycerol and 5 g sodium glutamate liter−1), and 1.0 M NaCl was added. The cells were harvested in late exponential growth phase. Under these growth conditions, the lipid content of the cells was below 15% of dry weight when harvested. 1H and 13C NMR spectra of the ethanolic extracts of all four strains gave exactly the same signal pattern. To ensure that the compatible solutes were not modified by the ethanol-extraction procedure, we showed that T66 cells extracted with 5% (wt/vol) trichloroacetic acid gave rise to the same 1H and 13C NMR signal patterns (data not shown). Therefore, only ethanolic extract of T66 was analyzed in more detail.
As shown in Fig. 3, the 13C NMR spectrum of the T66 extract contained nine strong signals; other signals were barely visible. Three signals of different intensities (54.8, 67.4, and 170.7 ppm) corresponded to the published chemical shifts at neutral pH of glycine betaine (16), which is a common compatible solute in many organisms (34, 52, 58, 63). The six remaining 13C NMR signals (34.6, 69.9, 70.2, 72.2, 73.5, and 75.8 ppm) were stronger and had similar intensities. The signal at 34.6 ppm was in the resonance region typical of CH2 groups, whereas the five other signals mentioned were in the region typical of carbons bound to an electronegative atom, e.g., a halogen, O, or N (16). We therefore suspected that the extract contained a polyol, since such compounds are common osmolytes in eukaryotic microorganisms (9, 57, 58).
The 1H NMR spectrum (Fig. 3) of T66 extract showed two singlets at 3.28 and 3.91 ppm with a quantitative ratio of 9 to 2. From the positions (16) and ratio of these signals and from analysis of extract with added authentic glycine betaine, we concluded that they represented the three identical CH3 groups and the single CH2 group of glycine betaine, respectively. The remaining spectrum comprised seven signals with splitting. Two of them (1.82 and 2.00 ppm) were in the region typical for signals of CH2 groups. Five (3.57, 3.71, 3.75, 3.94, and 4.03 ppm) were in the region typical for signals of protons attached to carbons that are linked to an electronegative atom, conceivably a hydroxyl group in this case.
A 1H-1H correlated spectroscopy two-dimensional spectrum of the T66 extract showed couplings between the two singlets (3.28 and 3.91 ppm) found to represent glycine betaine and between the seven remaining signals with splitting (data not shown). This indicated that the extracts contained only one additional major compound besides glycine betaine. Based on the data and considerations given above, we inferred that the unidentified signals of the 1H and 13C NMR spectra represented a species of cyclohexanepentol. Through a literature search, we found that these signals were in accordance with the NMR spectra of synthetic proto-quercitol (3, 37, 51) and extracted proto-quercitol from leaves of cork oak (47).
Since it is not possible to distinguish between the two optical isomers of proto-quercitol by NMR spectroscopy, we purified the proto-quercitol species of strain T66 (grown with 1.0 M NaCl) and showed that it was levorotatory, [α]D20 of −21.3 (c 1.0; H2O). This value for specific optical rotation was in good agreement with previously reported values for synthetic (−)- and (+)-proto-quercitols, which range from ±18.0 to 25.3 (23, 37, 46). Accurate mass determination of the purified compound, performed with a time-of-flight MS operated with atmospheric pressure chemical ionization in negative mode, displayed an m/z value of 163.06, consistent with a deprotonated species of cyclohexanepentol, such as proto-quercitol. Furthermore, analysis of the crude extract of T66 with an ion trap-MS system operated with electrospray ionization in positive mode revealed one major signal with an m/z value of 118.4, consistent with protonated glycine betaine.
Osmotic stress-dependent accumulation of compatible solutes.
Strains T66 and S8 were cultivated as described for the identification procedure of compatible solutes (see above), except that the osmolality of medium M1 was varied by addition of 0.25, 0.5, or 1.0 M NaCl. (−)-proto-Quercitol and glycine betaine were measured by quantitative 1H NMR spectroscopy of ethanolic extracts. As illustrated in Fig. 4, the cell contents of both metabolites increased nearly linearly with increasing external NaCl concentration, showing that they acted as compatible solutes. T66 and S8 accumulated similar amounts of these solutes, and (−)-proto-quercitol dominated at all salt concentrations tested (Fig. 4). No other solutes were detected that displayed osmotic stress-dependent accumulation.
Osmotic tolerance and NaCl requirement of strain T66.
When grown in medium M1, T66 tolerated at least 2.0 M NaCl, but faster growth was observed with the lower salt concentration tested, e.g., a doubling time of 5 h with 0.25 M NaCl added (Fig. 5). Particularly with NaCl additions above 1.0 M, the doubling time considerably increased. Since the osmotic tolerance of many bacteria is enhanced by externally supplied glycine betaine (references 6, 32, and 52 and references therein), we tested this effect for T66. Addition of 4 mM glycine betaine gave a moderate growth stimulation of T66 in medium with 1.5 M or higher amounts of NaCl added (Fig. 5).
It is also noteworthy that T66 was able to utilize glycine betaine as the single source of N in the glycerol-containing medium (doubling time of 5 h with 0.25 M NaCl added), but it was unable to grow in medium M1 with glycine betaine as the sole source of energy, C, and N.
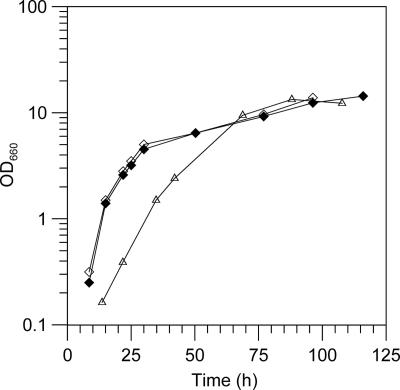
Besides the stated amounts of added NaCl, medium M1 contained 0.06 M Na+, which was contributed by other medium components. In order to test the dependency of T66 for NaCl, we used medium M2, which did not contain any other sodium salts. T66 did not grow in M2 without any NaCl added but grew with an initial doubling time of 9 h in M2 with 0.10 M NaCl added (0.45 osmol kg−1). Presumably, the ionic or osmotic strength, rather than the NaCl concentration, was too low, since a higher initial growth rate was observed in M2 containing either 0.25 M NaCl or 0.10 M NaCl and 0.15 M KCl (0.75 osmol kg−1). Typical growth curves of T66 under these conditions are depicted in Fig. 6.
FIG. 6.
Influences of added NaCl and KCl on growth of strain T66 in medium of low osmotic strength. The cells were grown in medium M2 with NaCl and/or KCl added as follows: open triangles, 0.10 M NaCl (0.45 osmol kg−1); filled diamonds, 0.10 M NaCl and 0.15 M KCl; open diamonds, 0.25 M NaCl (0.75 osmol kg−1). The added NaCl was the only source of Na+. Representative growth curves are presented.
Lack of quercitol and glycine betaine in strain T29.
1H and 13C NMR spectra of ethanolic extracts of T29 grown in medium M1 (fortified with yeast extract) with 0.25 to 0.75 M NaCl added showed signals that differed from those observed for the other strains (data not shown). Though we were unable to interpret the spectra, we could conclude that T29 did not accumulate glycine betaine or any species of quercitol. Since this strain showed a weak osmotolerance and did not grow in our defined media, it was not examined in more detail.
DISCUSSION
The present data show that Schizochytrium sp. strain S8 (ATCC 20889) and three phylogenetically related strains isolated by us (T65, T66, and T67) displayed osmotic stress-dependent accumulation of (−)-proto-quercitol and glycine betaine. At the salt concentrations tested (0.25 to 1.0 M NaCl), the ratio between (−)-proto-quercitol and glycine betaine in the cells was about 5 to 1. To our knowledge, (−)-proto-quercitol has previously been found only in species of eucalyptus (1, 37, 39, 51), whereas glycine betaine is a common compatible solute in many prokaryotes and eukaryotic microorganisms and multicellular organisms (34, 52, 58, 63). Since the present analyses were performed on thraustochytrids grown in a simple defined medium, it is evident that their compatible solutes were synthesized endogenously without the need of any particular precursors.
The previous finding that the related organism S. limacinum SR21 (Fig. 1) contains phosphatidylinositol and phosphatidylcholine (62) indicates that (−)-proto-quercitol and glycine betaine were made de novo from myo-inositol and choline, respectively, in these eukaryotic microorganisms. In plants, myo-inositol is synthesized from glucose-6-phosphate, and it can be processed into other isomeric inositols (36). Conversions of inositols to quercitols (deoxyinositols) have not been described for eukaryotes, but bacterial bioconversion of myo-inositol into several optically active quercitols, including (+)-proto-quercitol, has been reported previously (54), and a mechanism for such bioconversion has recently been proposed (44). Depending on the type of organism, eukaryotes can synthesize choline by a three-step methylation of ethanolamine and phosphatidylethanolamine (38, 49), and free choline can be converted to glycine betaine via betaine aldehyde by a two-step oxidation (25, 49). Enzymes that oxidize choline to betaine aldehyde are localized in cell organelles of glycine betaine-producing eukaryotes, e.g., choline dehydrogenase of mammalian mitochondria (25) and choline monooxygenase of plant chloroplasts (49). Betaine-aldehyde dehydrogenase activity is found mainly in the cytoplasm of mammals (13) and in chloroplasts of plants (49). In heterotrophic bacteria, synthesis of glycine betaine occurs usually by a two-step oxidation of externally supplied choline, but certain halophilic actinomycetes and photothropic bacteria utilize a three-step methylation of glycine or both pathways (reference 43 and references therein).
Our growth experiments performed with strain T66 showed that it also had an uptake system(s) which allowed the use of glycine betaine as a source of nitrogen and which conferred some osmoprotection, as is common in many bacteria (6, 32, 52). The finding that T66 grew in media with osmotic strengths ranging from 0.45 to 3.7 osmol kg−1 (containing 0.1 to 2.0 M NaCl) showed that (−)-proto-quercitol and glycine betaine constitute a potent osmolyte system.
Yancey et al. (63) pointed out that osmotolerant organisms have through evolution selected compatible solutes that stabilize intracellular proteins. The protein-stabilizing effect of compatible solutes is generally explained in terms of the preferential exclusion model, i.e., the solutes are preferentially excluded from the surface of proteins and thereby push the equilibrium between unfolded and native protein toward the native state (4, 53). Because of this ability, compatible solutes can protect proteins and organisms against many adverse conditions. Glycine betaine has been shown to protect enzymes against thermal and salt inactivation in vitro (12, 63) and to protect bacteria against stress caused by high external osmolality (6, 32, 34), cold (6, 32), or heat (12, 26). In vitro experiments have shown that (+)-proto-quercitol isolated from oak protects glutamine synthetase against heat inactivation in a concentration-dependent manner (30) and protects the cyclic photophosphorylation activity of isolated thylakoid membranes of spinach during a freeze and thaw cycle (45). Both stereoisomers of proto-quercitol have been linked to protection against cold-induced stress in cork oak (47) and drought- and salt-induced stress in species of eucalyptus (1, 39).
Our description of the present strains included their fatty acid profile. Huang et al. (28) carried out an examination of the PUFA profile of DHA-producing thraustochytrids and identified five different C20 to C22 PUFA profiles (called types A to E). They reported that strains with the same profile cluster on the phylogenetic tree, but strain N1-27, which was their only strain with a type B profile (DHA, DPA, and EPA), appears to be closely related to strains A5-20, KH105, and S. limacinum SR21 displaying the simpler type A profile (DHA and DPA). The phylogenetic tree presented in Fig. 1 includes the strains mentioned above plus strain ONC-TN18, which has a type B profile (11), and strains T65, T66, T67, and S8, which have a type A profile. The scattering of these strains on the same phylogenetic branch shows that for use in taxonomy of thraustochytrids the type A and type B profiles should be joined. We also showed that strains with a PUFA profile of type A or type B may carry the same insert in their 18S rRNA gene (see above). It has been reported previously that strain SR21 (56) displays the same growth yield in medium containing from 50 to 200% seawater (corresponding to about 18 to 70 g sea salt liter−1) and that strain ONC-T18 (11) displays practically the same growth yield in medium containing from 2 to 50 g sea salt liter−1. Thus, similarly to our strains of the same phylogenetic branch, SR21 and ONC-T18 appear to adapt well to environments of elevated osmotic strength.
Strain T29, which tolerated only 0.75 M NaCl, did not accumulate (−)-proto-quercitol or glycine betaine. This strain was phylogenetically related to T. aureum (ATCC 34304), which has previously been reported to be rather salt sensitive, i.e., it prefers a medium with 50% seawater and shows strongly reduced growth yield with 100% seawater (29). The close relationship between these thraustochytrids is also reflected in their PUFA profiles. Both accumulate DHA, DPA, AA, EPA, and linoleic acid, but strain T29 accumulated a small amount of eicosatrienoic acid (Fig. 2), which has not been reported for T. aureum (5, 29). Furthermore, by growing their strains of T. aureum and T. roseum in media with 50 to 100% seawater, Wethered and Jennings (60) did not detect osmotic stress-induced accumulation of any Dragendorff-positive quaternary ammonium compound, such as glycine betaine. Apparently, this group of thraustochytrids utilizes osmolytes that give less osmoprotection than (−)-proto-quercitol and glycine betaine.
Acknowledgments
We thank P. Bruheim and S. E. F. Borgos for performing the liquid chromatography-MS analysis and Ø. M. Jakobsen for help with the phylogenetic analysis.
This work was supported by a grant from the Research Council of Norway.
Footnotes
Published ahead of print on 27 July 2007.
REFERENCES
- 1.Adams, M. A., A. Richter, A. K. Hill, and T. D. Colmer. 2005. Salt tolerance in Eucalyptus spp.: identity and response of putative osmolytes. Plant Cell Environ. 28:772-787. [Google Scholar]
- 2.Adl, S. M., A. G. B. Simpson, M. A. Farmer, R. A. Andersen, O. R. Anderson, J. R. Barta, S. S. Browser, G. Brugerolle, R. A. Fensome, S. Fredericq, T. Y. James, S. Karpov, P. Kugrens, J. Krug, C. E. Lane, L. A. Lewis, J. Lodge, D. H. Lynn, D. G. Mann, R. M. McCourt, L. Mendoza, Ø. Moestrup, S. E. Mozley-Standridge, T. A. Nerad, C. A. Shearer, A. V. Smirnov, F. W. Spiegel, and M. F. J. R. Taylor. 2005. The new higher level classification of eukaryotes with emphasis on the taxonomy of protists. J. Eukaryot. Microbiol. 52:399-451. [DOI] [PubMed] [Google Scholar]
- 3.Angyal, S. J., and L. Odier. 1982. The 13C-NMR spectra of inositols and cyclohexanepentols: the validity of rules correlating chemical shifts with configuration. Carbohydr. Res. 100:43-54. [Google Scholar]
- 4.Arakawa, T., and S. N. Timasheff. 1985. The stabilization of proteins by osmolytes. Biophys. J. 47:411-414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bajpai, P. K., P. Bajpai, and O. P. Ward. 1991. Optimization of production of docosahexaenoic acid (DHA) by Thraustochytrium aureum ATCC 34304. J. Assoc. Oil Chem. Soc. 68:509-514. [Google Scholar]
- 6.Bayles, D. O., and B. J. Wilkinson. 2000. Osmoprotectants and cryoprotectants for Listeria monocytogenes. Lett. Appl. Microbiol. 30:23-27. [DOI] [PubMed] [Google Scholar]
- 7.Bergé, J. P., and G. Barnathan. 2005. Fatty acids from lipids of marine organisms: molecular biodiversity, roles as biomarkers, biologically active compounds, and economical aspects. Adv. Biochem. Eng. Biotechnol. 96:49-125. [DOI] [PubMed] [Google Scholar]
- 8.Bligh, E. G., and W. J. Dyer. 1959. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37:911-917. [DOI] [PubMed] [Google Scholar]
- 9.Blomberg, A., and L. Adler. 1992. Physiology of osmotolerance in fungi. Adv. Microbiol. Physiol. 33:145-212. [DOI] [PubMed] [Google Scholar]
- 10.Bremer, G. B. 2000. Isolation and culture of Thraustochytrids, p. 49-61. In K. D. Hyde and S. B. Pointing (ed.), Marine mycology: a practical approach. Fungal Diversity Press, Hong Kong.
- 11.Burja, A. M., H. Radianingtyas, A. Windust, and C. J. Barrow. 2006. Isolation and characterization of polyunsaturated fatty acid producing Thraustochytrium species: screening of strains and optimization of omega-3 production. Appl. Microbiol. Biotechnol. 72:1161-1169. [DOI] [PubMed] [Google Scholar]
- 12.Caldas, T., N. Demont-Caulet, A. Ghazi, and G. Richarme. 1999. Thermoprotection by glycine betaine and choline. Microbiology 145:2543-2548. [DOI] [PubMed] [Google Scholar]
- 13.Chern, M. K., and R. Pietruszko. 1999. Evidence for mitochondrial localization of betaine aldehyde dehydrogenase in rat liver: purification, characterization, and comparison with human cytoplasmic E3 isozyme. Biochem. Cell Biol. 77:179-187. [PubMed] [Google Scholar]
- 14.Craigie, J. S. 1969. Some salinity-induced changes in growth, pigments, and cyclohexanetetrol content of Monochrysis lutheri. J. Fish. Res. Board Can. 26:2959-2967. [Google Scholar]
- 15.Fan, K. W., L. L. P. Vrijmoed, and E. B. G. Jones. 2002. Physiological studies of subtropical mangrove thraustochytrids. Bot. Mar. 45:50-57. [Google Scholar]
- 16.Fan, T. W. M. 1996. Metabolite profiling by one- and two-dimensional NMR analysis of complex mixtures. Prog. Nucl. Magn. Reson. Spectrosc. 28:161-299. [Google Scholar]
- 17.Felsenstein, J. 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783-791. [DOI] [PubMed] [Google Scholar]
- 18.Fujii, S., and J. A. Hellebust. 1994. Growth and osmoregulation of Boekelovia hooglandii in relation to salinity. Can. J. Bot. 72:823-828. [Google Scholar]
- 19.Fujii, S., N. Nishimoto, A. Notoya, and J. A. Hellebust. 1995. Growth and osmoregulation of Chaetoceros muelleri in relation to salinity. Plant Cell Physiol. 36:759-764. [Google Scholar]
- 20.Garrill, A., N. J. W. Clipson, and D. H. Jennings. 1992. Preliminary observations on the monovalent cation relations of Thraustochytrium aureum, a fungus requiring sodium for growth. Mycol. Res. 96:295-304. [Google Scholar]
- 21.Goldstein, S. 1963. Development and nutrition of new species of Thraustochytrium. Am. J. Bot. 50:271-279. [Google Scholar]
- 22.Goldstein, S. 1963. Morphological variation and nutrition of a new monocentric marine fungus. Arch. Mikrobiol. 45:101-110. [DOI] [PubMed] [Google Scholar]
- 23.Gültekin, M. S., M. Celik, E. Turkut, C. Tanyeli, and M. Balci. 2004. Resolution of (+/−)-anti-2,3-dioxabicyclo[2.2.2]oct-7-en-5-ol via Candida cylindracea lipase: synthesis of (−)- and (+)-proto-quercitol. Tetrahedron Asymm. 15:453-456. [Google Scholar]
- 24.Hardy, R., and N. J. Keay. 1972. Seasonal variations in the chemical composition of Cornish mackerel, Scomber scombrus (L), with detailed reference to the lipids. J. Food Technol. 7:125-137. [Google Scholar]
- 25.Haubrich, D. R., and N. H. Gerber. 1981. Choline dehydrogenase. Assay, properties and inhibitors. Biochem. Pharmacol. 30:2993-3000. [DOI] [PubMed] [Google Scholar]
- 26.Holtmann, G., and E. Bremer. 2004. Thermoprotection of Bacillus subtilis by exogenously provided glycine betaine and structurally related compatible solutes: involvement of Opu transporters. J. Bacteriol. 186:1683-1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Honda, D., T. Yokochi, T. Nakahara, S. Raghukumar, A. Nakagiri, K. Schaumann, and T. Higashihara. 1999. Molecular phylogeny of labyrinthulids and thraustochytrids based on the sequencing of 18S ribosomal RNA gene. J. Eukaryot. Microbiol. 46:637-647. [DOI] [PubMed] [Google Scholar]
- 28.Huang, J., T. Aki, T. Yokochi, T. Nakahara, D. Honda, S. Kawamoto, S. Shigeta, K. Ono, and O. Suzuki. 2003. Grouping newly isolated docosahexaenoic acid-producing thraustochytrids based on their polyunsaturated fatty acid profiles and comparative analysis of 18S rRNA genes. Mar. Biotechnol. 5:450-457. [DOI] [PubMed] [Google Scholar]
- 29.Iida, I., T. Nakahara, T. Yokochi, Y. Kamisaka, H. Yagi, M. Yamaoka, and O. Suzuki. 1996. Improvement of docosahexaenoic acid production in a culture of Thraustochytrium aureum by medium optimization. J. Ferment. Bioeng. 81:76-78. [Google Scholar]
- 30.Jaindl, M., and M. Popp. 2006. Cyclitols protect glutamine synthetase and malate dehydrogenase against heat induced deactivation and thermal denaturation. Biochem. Biophys. Res. Commun. 345:761-765. [DOI] [PubMed] [Google Scholar]
- 31.Kets, E. P. W., J. A. M. deBont, and H. J. Heipieper. 1996. Physiological response of Pseudomonas putida S12 subjected to reduced water activity. FEMS Microbiol. Lett. 139:133-137. [Google Scholar]
- 32.Ko, R., L. T. Smith, and G. M. Smith. 1994. Glycine betaine confers enhanced osmotolerance and cryotolerance on Listeria monocytogenes. J. Bacteriol. 176:426-431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kumon, Y., R. Yokoyama, T. Yokochi, D. Honda, and T. Nakahara. 2003. A new labyrinthulid isolate, which solely produces n-6 docosapentaenoic acid. Appl. Microbiol. Biotechnol. 63:22-28. [DOI] [PubMed] [Google Scholar]
- 34.Larsen, P. I., L. K. Sydnes, B. Landfald, and A. R. Strøm. 1987. Osmoregulation in Escherichia coli by accumulation of organic osmolytes: betaines, glutamic acid, and trehalose. Arch. Microbiol. 147:1-7. [DOI] [PubMed] [Google Scholar]
- 35.Lewis, T. E., P. D. Nichols, and T. A. McMeekin. 1999. The biotechnological potential of thraustochytrids. Mar. Biotechnol. 1:580-587. [DOI] [PubMed] [Google Scholar]
- 36.Loewus, F. A., and P. P. N. Murthy. 2000. myo-Inositol metabolism in plants. Plant Sci. 150:1-19. [Google Scholar]
- 37.McCasland, G. E., M. O. Naumann, and L. J. Durham. 1968. Alicyclic carbohydrates. XXXV. The synthesis of proto-quercitol. 220-MHz proton spectrum with superconducting solenoid. J. Org. Chem. 33:4220-4227. [Google Scholar]
- 38.McNeil, S. D., M. L. Nuccio, M. J. Ziemak, and A. D. Hanson. 2001. Enhanced synthesis of choline and glycine betaine in transgenic tobacco plants that overexpress phosphoethanolamine N-methyltransferase. Proc. Natl. Acad. Sci. USA 98:10001-10005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Merchant, A., M. Tausz, S. K. Arndt, and M. A. Adams. 2006. Cyclitols and carbohydrates in leaves and roots of 13 Eucalyptus species suggest contrasting physiological responses to water deficit. Plant Cell Environ. 29:2017-2029. [DOI] [PubMed] [Google Scholar]
- 40.Metcalfe, L. D., A. A. Schmitz, and J. R. Pelka. 1966. Rapid preparation of fatty acid esters from lipids for gas chromatographic analysis. Anal. Chem. 38:514-515. [Google Scholar]
- 41.Mo, C., J. Douek, and B. Rinkevich. 2002. Development of a PCR strategy for thraustochytrid identification based on 18S rDNA sequence. Mar. Biol. 140:883-889. [Google Scholar]
- 42.Nakahara, T., T. Yokochi, T. Higashihara, S. Tanaka, T. Yaguchi, and D. Honda. 1996. Production of docosahexaenoic and docosapentaenoic acids by Schizochytrium sp. isolated from Yap Islands. J. Assoc. Oil Chem. Soc. 73:1421-1426. [Google Scholar]
- 43.Nyyssölä, A., and M. Leisola. 2001. Actinopolyspora halophila has two separate pathways for betaine synthesis. Arch. Microbiol. 176:294-300. [DOI] [PubMed] [Google Scholar]
- 44.Ogawa, S., and M. Kanto. 2007. Synthesis of valiolamine and some precursors for bioactive carbaglycosylamines from (−)-vibo-quercitol produced by biogenesis of myo-inositol. J. Nat. Prod. 70:493-497. [DOI] [PubMed] [Google Scholar]
- 45.Orthen, B., and M. Popp. 2000. Cyclitols as cryoprotectants for spinach and chickpea thylakoids. Environ. Exp. Bot. 44:125-132. [DOI] [PubMed] [Google Scholar]
- 46.Pachaly, P., and H. Khosravian. 1988. Tilitriandrine, ein neues Bisbenzylisochinolin-Alkaloid aus Tiliacora triandra. Planta Med. 6:516-519. [DOI] [PubMed] [Google Scholar]
- 47.Passarinho, J. A. P., P. Lamosa, J. P. Baeta, H. Santos, and C. P. P. Ricardo. 2006. Annual changes in the concentration of minerals and organic compounds of Quercus suber leaves. Physiol. Plant 127:100-110. [Google Scholar]
- 48.Raghukumar, S. 2002. Ecology of the marine protists, the Labyrinthulomycetes (Thraustochytrids and Labyrinthulids). Eur. J. Protistol. 38:127-145. [Google Scholar]
- 49.Rontein, D., G. Basset, and A. D. Hanson. 2002. Metabolic engineering of osmoprotectant accumulation in plants. Metab. Eng. 4:49-56. [DOI] [PubMed] [Google Scholar]
- 50.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 51.Salamci, E., H. Secen, Y. Sütbeyaz, and M. Balci. 1997. A concise and convenient synthesis of dl-proto-quercitol and dl-gala-quercitol via ene reaction of singlet oxygen combined with [2+4] cycloaddition to cyclohexadiene. J. Org. Chem. 62:2453-2457. [DOI] [PubMed] [Google Scholar]
- 52.Sleator, R. D., and C. Hill. 2001. Bacterial osmoadaptation: the role of osmolytes in bacterial stress and virulence. FEMS Microbiol. Rev. 26:49-71. [DOI] [PubMed] [Google Scholar]
- 53.Street, T. O., D. W. Bolen, and G. D. Rose. 2006. A molecular mechanism for osmolyte induced protein stability. Proc. Natl. Acad. Sci. USA 103:13997-14002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Takahashi, A., K. Kanbe, T. Tamamura, and K. Sato. 1999. Bioconversion of myo-inositol to rare cyclic sugar alchohols. Anticancer Res. 19:3807. [Google Scholar]
- 55.Takao, Y., K. Nagasaki, K. Mise, T. Okuno, and D. Honda. 2005. Isolation and characterization of a novel single-stranded RNA virus infectious to a marine fungoid protist, Schizochytrium sp. (Thraustochytriaceae, Labyrinthulea). Appl. Environ. Microbiol. 71:4516-4522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wegmann, K. 1986. Osmorgulation in eukaryotic algae. FEMS Microbiol. Rev. 39:37-43. [Google Scholar]
- 58.Welsh, D. T. 2000. Ecological significance of compatible solute accumulation by micro-organisms: from single cells to global climate. FEMS Microbiol. Rev. 24:263-290. [DOI] [PubMed] [Google Scholar]
- 59.Welsh, D. T., R. H. Reed, and R. A. Herbert. 1991. The role of trehalose in the osmoadaptation of Escherichia coli NCIB 9484: interaction of trehalose, K+ and glutamate during osmoadaptation in continuous culture. J. Gen. Microbiol. 137:745-750. [DOI] [PubMed] [Google Scholar]
- 60.Wethered, J. M., and D. H. Jennings. 1985. Major solutes contributing to solute potential of Thraustochytrium aureum and T. roseum after growth in media of different salinities. Trans. Br. Mycol. Soc. 85:439-446. [Google Scholar]
- 61.Wolf, A. V., M. G. Brown, and P. G. Prentiss. 1976-1977. Concentrative properties of aqueous solutions, p. D218-D267. In R. C. Weast (ed.), CRC handbook of chemistry and physics, 57th ed. CRC Press, Cleveland, OH.
- 62.Yaguchi, T., S. Tanaka, T. Yokochi, T. Nakahara, and T. Higashihara. 1997. Production of high yields of docosahexaenoic acid by Schizochytrium sp. strain SR21. J. Assoc. Oil Chem. Soc. 74:1431-1434. [Google Scholar]
- 63.Yancey, P. H., M. E. Clark, S. C. Hand, R. D. Bowlus, and G. N. Somero. 1982. Living with water stress: evolution of osmolyte systems. Science 217:1214-1222. [DOI] [PubMed] [Google Scholar]
- 64.Yokochi, T., D. Honda, T. Higashihara, and T. Nakahara. 1998. Optimization of docosahexaenoic acid production by Schizochytrium limacinum SR21. Appl. Microbiol. Biotechnol. 49:72-76. [Google Scholar]