Abstract
We investigated the carbon dioxide metabolism of Streptococcus thermophilus, evaluating the phenotype of a phosphoenolpyruvate carboxylase-negative mutant obtained by replacement of a functional ppc gene with a deleted and inactive version, Δppc. The growth of the mutant was compared to that of the parent strain in a chemically defined medium and in milk, supplemented or not with l-aspartic acid, the final product of the metabolic pathway governed by phosphoenolpyruvate carboxylase. It was concluded that aspartate present in milk is not sufficient for the growth of S. thermophilus. As a consequence, phosphoenolpyruvate carboxylase activity was considered fundamental for the biosynthesis of l-aspartic acid in S. thermophilus metabolism. This enzymatic activity is therefore essential for growth of S. thermophilus in milk even if S. thermophilus was cultured in association with proteinase-positive Lactobacillus delbrueckii subsp. bulgaricus. It was furthermore observed that the supplementation of milk with aspartate significantly affected the level of urease activity. Further experiments, carried out with a pureI-gusA recombinant strain, revealed that expression of the urease operon was sensitive to the aspartate concentration in milk and to the cell availability of glutamate, glutamine, and ammonium ions.
Streptococcus thermophilus is a major component of dairy starter used for the manufacture of yogurt and cheeses. One of the main roles of S. thermophilus growing in milk is to provide rapid acidification as a consequence of the production of lactic acid. Previous studies have shown that the availability of peptides and amino acids is critical for the optimal growth of this bacterial species in milk (8, 11). While for other species, such as Lactococcus lactis and Lactobacillus delbrueckii subsp. bulgaricus, growth in milk depends largely on the activity of cell-wall-anchored proteinases, for S. thermophilus the amino acid requirement is satisfied by the efficiency of its biosynthetic capacities and by the cooperation with other bacterial species growing in association in the dairy environment, such as Lactobacillus delbrueckii subsp. bulgaricus. Cell-wall-anchored proteinases are reported to be present only in a minority of S. thermophilus strains studied until now (9), and their physiological and technological roles have been investigated (8, 11). The capacity of S. thermophilus to satisfy its amino acid requirement during growth in milk is different from that of other lactic acid bacterial species. Some S. thermophilus strains appear auxotrophic for at least four amino acids, Glu, Cys, His, and Met, while other lactic acid bacteria are known to be more exigent (16, 19). The relevance of the amino acid biosynthetic pathways for the metabolism of S. thermophilus have been investigated for branched-chain amino acids, proline, and glutamine (13, 20, 24). S. thermophilus genome analysis has revealed a relatively high conservation of functional amino acid biosynthetic genes, which might reflect the importance of amino acid synthesis for the growth of this species in milk (2, 16). In this work, we investigated the aspartic acid biosynthesis of S. thermophilus, focusing our attention on phosphoenolpyruvate (PEP) carboxylase activity and its relationship with urease, an ammonia and carbon dioxide-generating enzyme. Ammonia produced from urea hydrolysis was recently reported to be involved in glutamine biosynthesis (24). An overview of the potential metabolic connections among the aspartate biosynthetic pathway, glutamine synthesis, and urease activity is shown in Fig. 1.
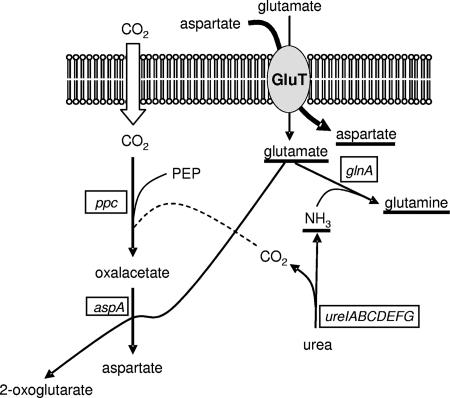
FIG. 1.
Schematic representation of the enzymatic activities involved in aspartate biosynthesis and urea metabolism in Streptococcus thermophilus. The molecules underlined act on transcription of ure operon. ppc, gene coding for phosphoenolpyruvate carboxylase; aspA, gene coding for aspartate aminotransferase; glnA, gene coding for glutamine synthetase, ureIABCDEFG, operon coding for urease; GluT, hypothetical glutamate membrane transporter. The wide arrow crossing the GluT represents the competitive effect of aspartic acid on the uptake of glutamate.
In a previous study on carbon dioxide fixation by S. thermophilus, PEP carboxylase was found to be mainly responsible for bicarbonate fixation in this species (21). This enzymatic activity (EC 4.1.1.31) catalyzes the reaction between carbon dioxide and PEP to obtain oxalacetic acid. Oxalacetic acid is then converted to aspartic acid by an aspartate aminotransferase (EC 2.6.1.1). In the present work, the aspartic acid biosynthesis was studied by analyzing the phenotype of a PEP carboxylase-negative mutant obtained by replacement of a functional ppc gene with a deleted and inactive version, Δppc, demonstrating the following: (i) this enzymatic activity is fundamental for the growth of S. thermophilus in milk; (ii) the aspartic acid concentration in milk is not sufficient for S. thermophilus growth; (iii) aspartate, glutamate, glutamine, and ammonia availability modulate the level of urease activity through a transcriptional regulation mechanism sensitive to a nitrogen-limited condition. Furthermore, culture association between the Δppc mutant and the proteinase-positive/proteinase-negative Lactobacillus delbrueckii subsp. bulgaricus strains was also analyzed with the aim of contributing to a more complete understanding of the protocooperation between these two lactic acid bacterial species.
MATERIALS AND METHODS
Bacterial strains, growth conditions, and reagents.
Wild-type PpC+ S. thermophilus DSM 20617T and its PpC− derivative, A18(Δppc), were maintained in M17 broth (Difco Laboratories, Detroit, MI) at 37°C, Lactobacillus delbrueckii subsp. bulgaricus ATCC 11842, CNRZ 397, and 1159 (CNRZ 397 PrtB negative) were maintained in MRS broth at 37°C. Plasmid-containing S. thermophilus strains were maintained in M17 broth (10 g liter−1 lactose) supplemented with 5 μg of erythromycin per ml at 28°C, while strains containing the pG+host9-derived vector integrated into the chromosome were maintained in the same medium supplemented with 2 μg of erythromycin per ml at 42°C. Escherichia coli strains were routinely maintained in Luria broth at 37°C with aeration supplemented with 10 μg of kanamycin per ml and when necessary with 200 μg of erythromycin per ml. The auxotrophy for aspartic acid and the growth behavior of S. thermophilus wild type and A18(Δppc) in the presence of urea, l-aspartate, or NaHCO3 was evaluated in a chemically defined medium (CDM) derived from the medium described by Reiter and Oram (30). It contained lactose (10 g liter−1), sodium acetate (1 g liter−1), ascorbic acid (0.5 g liter−1), NH4Cl (0.5 g liter−1), adenine (5 mg liter−1), guanine (5 mg liter−1), xanthine (5 mg liter−1), uracil (5 mg liter−1), pyridoxal hydrochloride (2 mg liter−1), niacin (1 mg liter−1), thiamine hydrochloride (1 mg liter−1), riboflavin (1 mg liter−1), calcium panthotenate (1 mg liter−1), para-aminobenzoic acid (10 μg liter−1), biotin (10 μg liter−1), folic acid (1 μg liter−1), vitamin B12 (1 μg liter−1), MgCl2·6H2O (200 mg liter−1), CaCl2 (50 mg liter−1), FeCl3·6H2O (5 mg liter−1), ZnSO4·7H2O (5 mg liter−1), CoCl2·6H2O (2.5 mg liter−1), CuSO4·5H2O (2.5 mg liter−1), NiSO4·7H2O (10 mg liter−1), MnSO4·1H2O (10 mg liter−1), l-cysteine hydrochloride (200 mg liter−1), l-leucine (200 mg liter−1), l-alanine (150 mg liter−1), l-lysine hydrochloride (800 mg liter−1), l-arginine (450 mg liter−1), l-proline (540 mg liter−1), l-isoleucine (100 mg liter−1), l-methionine (70 mg liter−1), l-phenylalanine (100 mg liter−1), l-serine (200 mg liter−1), l-threonine (100 mg liter−1), l-tryptophan (100 mg liter−1), l-valine (150 mg liter−1), glycine (200 mg liter−1), l-hystidine hydrochloride·1H2O (550 mg liter−1), l-glutamate (3.2 g liter−1), l-glutamine (3.2 g liter−1), and l-tyrosine (100 mg liter−1). The medium was reconstituted using concentrated stock solutions containing the nutrients. A mixture containing lactose, salts, and vitamins was prepared at a concentration double that in the CDM. After its pH was adjusted to 7 using NaOH, the mixture was autoclaved for 15 min at 110°C. The amino acids were prepared as a five-times-concentrated solution filter sterilized (0.22 μm) after adjustment of its pH to 7. When appropriate, this medium was supplemented with l-Asp, urea, and NaHCO3 using filter-sterilized concentrated solution. For growth measurements, S. thermophilus strains were grown in reconstituted (10% [wt/vol]) skimmed milk (SM) (Difco Laboratories, Detroit, MI) sterilized for 10 min at 110°C.
PCR protocols and DNA sequencing.
As previously described, total bacterial DNA was extracted (25) starting from 100 μl of M17 broth culture. A PCR approach for the amplification of the ppc gene was developed on the basis of the genome sequence of S. thermophilus LMG13811 and CNRZ1066 (accession numbers CP000023 and CP000024) (2). The amplification of a DNA region of about 3,200 bp encompassing all the ppc gene was performed as recommended by the supplier using 0.5 μmol liter−1 of primers PpCF (5′-GGAAAATCGGTGAGAAAGCT-3′) and PpCR (5′-CTGCTCAGCTTTTCAATCGT-3′) and 2 U of ExTaq DNA polymerase (Takara Bio, Inc., Shida, Japan). The PCR conditions were as follows: 35 cycles at 94°C for 1 min, 60°C for 35 s, and 72°C for 2 min and a single final extension at 72°C for 7 min. All amplification reactions were performed in a Mastercycler (Eppendorf Italia s.r.l., Milan, Italy). The PCR product was purified (NucleoSpin extract; Machery-Nagel GmbH and Co, Düren, Germany) and sequenced using the PpCF and PpCR primers followed by primer walking. The sequence reactions were analyzed in a 310 automatic DNA sequencer (Applera, Monza, Italy) with fluorescent dideoxy chain terminators (Big Dye Terminator cycle sequencing kit version 2.0; Applera, Monza, Italy). The sequence obtained was analyzed with ORF Finder and BLAST services at the National Center for Biotechnology Information and subsequently manually aligned with the homologous ppc gene of S. thermophilus LMG13811 and CNRZ1066.
Phylogenetic analysis.
Sequences of ppc genes of Streptococcus agalactiae, Streptococcus pneumoniae, Streptococcus pyogenes, Streptococcus mutans, Lactobacillus delbrueckii subsp. bulgaricus, Lactobacillus acidophilus, and Bifidobacterium longum were obtained from genome sequence data (GenBank/EMBL accession numbers AE009948, AE007317, AF004092, AE014133, CR954253, CP000033, and AE014295), while the ppc sequence of Lactobacillus gasseri was obtained from genome sequence data of strain ATCC 33323 produced by the Joint Genome Institute (http://genome.jgi-psf.org/mic_home.html). Sequences were phylogenetically aligned using ClustalX v.1.83 (32). Alignments were checked manually, and poorly aligned or divergent regions were eliminated using Gblocks software v.0.91b (5) with a minimum block of five and allowed gap positions equal to half. Tree construction was done with Treecon v.1.3b, using neighbor-joining methods and a bootstrap analysis with 1,000 replicates.
Replacement of ppc gene with a deleted version, Δppc.
DNA manipulation of the pG+host9 vector and derivatives was carried out using Escherichia coli VE7108. Plasmid isolation was performed using the Nucleospin plasmid kit according to the manufacture's instructions (Machery-Nagel GmbH and Co, Düren, Germany).
Strain A18 contains a deletion of 1,106 bp in the ppc gene, referred to as Δppc. The Δppc gene was obtained by PCR as previously described (26; also data not shown). Briefly, DNA fragments located upstream and downstream of the 1,106-bp deletion were independently amplified using the PpC1-PpC2 and PpC3-PpC4 primer pairs (PpC1, CAGGAAATCATCTCTGAAGA; PpC2, ACCAATCCACATTCCCATTG; PpC3, ATAATCCAACACCTATTACAATGGGAATGTGGATTGGTTCAGAACGAGTTGACCAAGA; PpC4, CAAGACGTTGAAGTATGGCA). Primer PpC3 has a 38-bp 5′ region complementary to the 5′ region of amplified product obtained using the PpC1-PpC2 primer set. These two PCR fragments were diluted to a final concentration of 100 fmol and mixed with 5 μl of 10× PCR buffer, 200 μM of each deoxynucleoside triphosphate, and 1.5 U of Taq DNA polymerase in 50 μl (Amersham-Pharmacia Biotech, Milan, Italy) with the aim of generating a new template DNA containing a deleted version of the ppc gene using the following thermal protocol: denaturation at 94°C for 2 min, reassociation at 40°C for 5 min, and extension at 72°C for 10 min. Following this step, primers PpC5 and PpC6 (PpC5, TTATTACTGCAGCTGCAGATGATGCAACACGAGGCATA; PpC6, TTATTACTGCAGCTGCAG AGCACCATATCCACGTTAGA) carrying a PstI site at their 5′ end were added to the reaction mixture at a final concentration of 0.5 μM and subjected to the following amplification protocol: 40 cycles consisting of 94°C for 45 s, 58°C for 35 s, and 72°C for 50 s, followed by a final extension at 72°C for 10 min. The resulting PCR fragment, Δppc, was ligated into the dephosphorylated PstI site of pG+host9, generating pMI49. pMI49 was introduced into S. thermophilus DSM20617T using a previously described protocol (26). The procedure of gene replacement described by Biswas and colleagues (1) was then applied to the ppc gene. The resulting PEP carboxylase-negative mutant was named A18(Δppc).
Evaluation of PEP carboxylase activity.
At the end of the exponential growth phase, S. thermophilus cells were harvested by centrifugation of 250 ml of culture for 15 min at 14,000 × g and 4°C. They were then washed with 250 ml of water, resuspended in 1.5 ml of Tris-HCl buffer (100 mM, pH 7.8), and placed in a 2-ml tube containing 600 mg of 0.1-mm-diameter glass beads (PolyLabo, Strasbourg, France). They were then disrupted in a cell disruptor (FP120 FastPrep; Savant Instruments, Inc.) for 30 s at 4°C and at speed 6.5. The cell crude extract was recovered after centrifugation for 5 min at 21,000 × g and 4°C. The protein content of the total cell crude extract was evaluated using the Bradford method (4). PEP carboxylase activity was evaluated (17) by coupling the malate dehydrogenase reaction to the spectrophotometric determination of NADH at 340 nm. Briefly, the enzymatic reaction was assayed at 37°C with Tris-HCl buffer (pH 7.2; 100 mM), MnSO4 (5 mM), PEP (5 mM), NADH (0.6 mM), malate dehydrogenase (5 U ml−1), and total cell crude extract (about 0.2 mg protein). The reaction was started with the addition of KHCO3 (10 mM). Specific activity was expressed as nanomoles of reacted PEP per minute per milligram of protein. The values were averages of three independent determinations.
Evaluation of casein hydrolysis by cell-wall-anchored proteinases.
Streptococcus thermophilus and Lactobacillus delbrueckii strains were subjected to evaluation of casein hydrolysis by PrtS and PrtB cell-wall-anchored proteinases. Briefly, 10 ml of an overnight culture was harvested by centrifugation, washed with 0.1 M Tris-HCl, pH 8, and resuspended in 500 μl of the same buffer with or without the addition of 2 to 10 mM of CaCl2. One hundred microliters of cell suspension containing 100 μg of caseins (VWR, Milan, Italy) per ml was incubated at 37°C for 3 h. After incubation, the cell suspension was centrifuged at maximum speed and the supernatant recovered and analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis using a 12% acrylamide gel in a Mini-Protean III apparatus (Bio-Rad, Milan, Italy). A solution containing 100 μg of caseins per ml in Tris-HCl buffer, incubated at 37°C for 3 h, was used as a negative control (undigested caseins).
Growth curves and measurement of acidifying activity.
Growth curves of DSM 20167T and the A18 derivative mutant were evaluated in triplicate in CDM and in SM at 37°C. Briefly, cells from of an overnight M17 culture were harvested by centrifugation, washed twice, resuspended in sterile NaCl (9 g liter−1) and inoculated at a concentration equivalent to 0.7 absorbance unit (575 nm) in CDM and reconstituted SM. Sterile 8-ml tubes were filled with 7.8 ml of inoculated CDM or milk and hermetically sealed. Growth in CDM was measured spectrophotometrically. Growth in SM was measured with the M680 microplate reader (Bio-Rad Laboratories, Hercules, CA). A clarification procedure was necessary for the evaluation of growth in milk (10). Some cultures were done in the presence of different combination of the following chemicals: aspartic acid, urea, NaHCO3, and fluorofamide, a potent inhibitor of urease. For the evaluation of milk acidifying activity, the pH was continuously measured during 24 h using a CINAC apparatus (Ysebaert, Frépillon, France).
Mixed cultures of S. thermophilus and L. delbrueckii.
Mixed cultures of S. thermophilus and L. delbrueckii and their derivative mutants were performed at 37°C by inoculating SM with approximately 5 × 106 CFU per ml of stock cultures of each strain. For each strain, cells from stock cultures were washed three time in sterile NaCl (9 g liter−1) before inoculation to avoid peptides or amino acids being supplied by the mixed cultures. The bacterial population was estimated by the plating method on specific medium, M17 for S. thermophilus and MRS (pH 5.5) for Lactobacillus delbrueckii. The l-Asp content of uncultured milk and l-Asp changes during growth of mixed cultures were determined by ion exchange chromatography using liquid chromatography equipment (Biochrom 30; Biochrom, Cambridge, United Kingdom) operating with ninhydrin (Flucka, Buchs, Switzerland) postcolumn derivatization and detection at 530 nm. The chromatographic conditions described by Resmini et al. (31) and a multipoint calibration were adopted.
GusA assay.
A recombinant S. thermophilus strain harboring the pMI200 vector was grown in SM or in M17 broth supplemented with 0.75 to 2 mM l-Asp and/or 0.75 mM NH4Cl, 1 mM l-Glu, or 0.75 mM l-Glu. Vector pMI200 (26) is a pNZ273 (29) derivative containing a transcriptional fusion between a 219-bp region located upstream of the ureI gene and containing pureI and the promoterless gusA reporter gene coding for the β-glucuronidase (GusA) enzyme. Cells from milk cultures were harvested at the end of the exponential growth phase after a clarification procedure (10), washed once with GusA buffer (10 mM β-mercaptoethanol, 1 mM EDTA, 0.1% Triton X-100 in 50 mM sodium phosphate buffer at pH 7), and then resuspended in 200 μl of the same buffer. The cell density was measured spectrophotometrically. Cell suspensions were subjected to mechanical disruption in the presence of 100 μl of glass beads (0.1 mm diameter) by homogenization for 5 min at 4°C. The amount of total proteins of each lysate was measured using the Bradford method (4) with bovine serum albumin as the standard. For the determination of GusA activity, 50 μl of protein extract was added to 950 μl of GusA buffer containing 1.25 mM para-nitro-β-d-glucoronic acid (VWR International, Milano, Italy). Measurement of GusA activity was performed at 37°C by monitoring the optical density at 420 nm (OD420) with the microplate reader M680 (Bio-Rad Laboratories, Hercules, CA) programmed for a reading set of 60 repetitions with intervals of 30 s. The GusA activity was expressed in milli-OD420 units per mg of protein for four independent determinations.
Urease activity.
Urease activity was determined on cell biomass obtained from 40 ml of SM culture after a clarification procedure as previously described. Cells were suspended in 1 ml of distilled water, and the cell density (OD600) was evaluated. Cells were recovered by centrifugation and suspended in 1 ml of urease assay solution containing 0.3 M urea, 85 mM NaCl, 0.002% phenol red, 6% (vol/vol) ethanol, 13 mM potassium buffer, pH 6.5, for the evaluation of urease activity. Measurement of the urease activity was performed at 37°C. The generation of ammonium ions due to urea hydrolysis led to a pH change and a development of a red-violet color detected at 580 nm. Urease activity was measured as the increment of OD580 units after 15 min of incubation at 37°C, as the mean of four independent determinations. The OD580 measurement was performed using the microplate reader M680.
RESULTS AND DISCUSSION
Optimal growth of S. thermophilus in milk requires hydrolysis of caseins, internalization and degradation of the resulting peptides, and de novo amino acid biosynthesis. Genomic analysis of S. thermophilus has revealed that it has maintained the capacity to synthesize several amino acids throughout evolution (16). Despite the relevance of amino acid biosynthetic pathways in milk adaptation of S. thermophilus, only the functionality of genes involved in branched-chain-amino-acid, proline, and glutamine biosynthesis was experimentally investigated in this species (13, 20, 24).
In this study the aspartate biosynthesis of S. thermophilus was studied, focusing attention on the first step of the pathway, the fixation of CO2 by a phosphoenolpyruvate carboxylase. The ppc gene coding for PEP carboxylase of strain DSM 20617T was sequenced, and the mutant A18(Δppc) was obtained by using a gene replacement approach.
Sequence and phylogenetic analysis of ppc gene.
Streptococcus thermophilus DSM 20617T ppc gene sequence analysis (AM167938) revealed a high sequence similarity (99%) with ppc gene sequences of the genomes of strains LMG13811 and CNRZ1066. The ppc gene consisted of 2,833 bp and encoded a 940-amino-acid PpC protein with a predicted molecular mass of 107 kDa. The average percent G+C of the ppc gene was 39.0%, as expected for the S. thermophilus species (2, 16). In the available genome of streptococci and lactic acid bacterial species, ppc genes were identified in pathogenic S. pneumoniae, S. mutans, S. pyogenes, and S. agalactiae and in Lactobacillus delbrueckii subsp. bulgaricus, L. acidophilus, and L. gasseri. Lactococcus lactis, Lactobacillus plantarum, and Lactobacillus salivarius were characterized by the presence of pyruvate carboxylase (E.C. 6.4.1.1) and/or PEP carboxykinase (E.C. 4.1.1.49). Other lactic acid bacterial species did not show genes coding for these enzymatic activities, which are involved in the anaplerotic assimilation of carbon dioxide. Phylogenetic analysis revealed that the ppc gene of S. thermophilus clustered with the orthologs of pathogenic streptococci but represented an independent lineage. The ppc gene of streptococci appeared well separated from those of Lactobacillus and Bifidobacterium species, with the L. delbrueckii subsp. bulgaricus ppc gene clustering in a phylogenetic branch with Bifidobacterium longum, according to the high G+C content of the L. delbrueckii genome and the evolutionary scenario recently depicted by van de Guchte et al. (33).
PEP carboxylase-negative mutant is auxotroph for l-Asp.
The wild type and the A18(Δppc) mutant were subjected to evaluation of PEP carboxylase activity. The wild type displayed similar enzymatic activities whether the cells were grown in M17 (97 ± 25 U), in CDM (137 ± 20 U), or in CDM supplemented with l-Asp (0.75 mM) (126 ± 20 U). As expected, no PEP carboxylase activity could be detected in strain A18(Δppc).
Evaluation of the growth of the wild type and the PEP carboxylase-negative mutant in CDM revealed that strain A18(Δppc) is auxotrophic for l-Asp (Fig. 2). The addition of aspartate to the CDM medium was found to meet only partially the nutritional requirement of A18(Δppc). The cell density of mutant A18(Δppc) did not reach a level similar to that of the parental strain even when aspartate was added to the medium at a final concentration of 2.25 mM (Fig. 2). The supplementation of CDM with aspartate in the range of concentration between 0.075 and 2.25 mM did not show any effect on the growth of the wild type. Addition of urea to CDM (urea hydrolyzed by urease activity yields ammonia and carbon dioxide, a substrate for PEP carboxylase activity) did not have any effect on the growth of either the parental or the mutant strain, indicating that carbon dioxide dissolved in the medium was not present in a limiting concentration. Similar data were obtained when bicarbonate was added to CDM (data not shown).
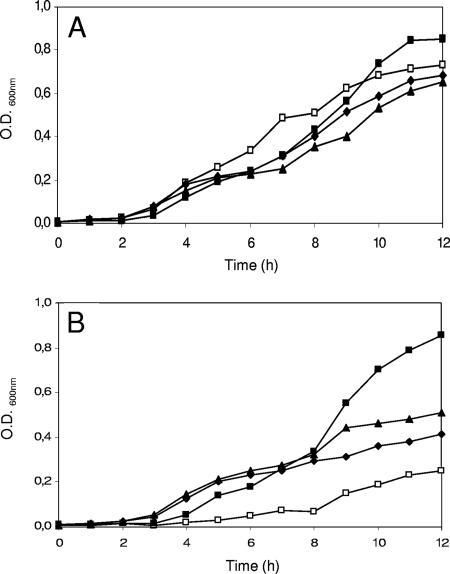
FIG. 2.
Growth of S. thermophilus wild type (A) and PEP carboxylase-negative mutant A18(Δppc) (B) in CDM (□) or CDM supplemented with l-Asp at a 0.075 mM (⧫), 0.23 mM (▴), 0.75 mM (▪), or 2.25 mM (•) concentration. All CDM cultures were repeated three times, and all standard deviations were <0.05 OD600 units.
PEP carboxylase activity is essential for growth of S. thermophilus in milk.
The A18(Δppc) mutant showed only limited growth in milk and was unable to coagulate milk within 24 h of incubation (Fig. 3). Supplementation of SM with 0.75 mM aspartic acid resulted in a growth rate similar to that obtained with the parental strain. The use of a higher aspartate concentration, 2 mM, was found to inhibit the growth of the parental strain. The growth-inhibiting effect due to a high aspartate concentration in milk was not observed when aspartate supplementation was counterbalanced by the addition of glutamic acid (data not shown), thus confirming the existing competition in the uptake system of these two amino acids, as reported by Bracquart et al. (3). The inhibitory effect of a high aspartate concentration on S. thermophilus growth was not observed in CDM, probably because in this medium glutamate was present in a higher concentration (20 mM) than in milk (0.30 mM). The extremely low growth rate of mutant A18(Δppc) was also reflected in limited milk acidification, less than 0.6 pH units in 12 h of incubation (Fig. 4). An acidification rate similar to that of the parental strain was not observed for the mutant A18(Δppc), even when aspartate was supplemented at the highest concentration. After 12 h of incubation, the parental strain acidified the medium to pH 5.1 while strain A18(Δppc) grown in aspartate-supplemented milk reached the pH value of 5.6.
FIG. 3.
Growth of S. thermophilus wild type (A) and PEP carboxylase negative mutant A18(Δppc) (B) in reconstituted SM (□) or SM supplemented with l-Asp at 0.075 mM (⧫), 0.23 mM (▴), or 0.75 mM (▪). All milk cultures were repeated three times, and standard deviations were always <0.08 OD600 units.
FIG. 4.
Acidification rate of S. thermophilus wild type (A) and PEP carboxylase-negative mutant A18(Δppc) (B) in reconstituted SM (□), 0.75 mM l-Asp (▪), or 0.75 mM l-Asp and flurofamide (10 μM) (▵). The decrease in the acidification rate due to urease activity is indicated by arrows. All milk cultures were repeated three times, and the standard deviations were always <0.08 pH units.
Inhibition of growth by a high aspartate concentration was reported by Bracquart et al. (3) and was explained by the competitive effect of aspartic acid on the uptake of glutamic acid. The authors reported that S. thermophilus harbors a glutamate transport system characterized by a marked specificity in the uptake of glutamic acid with aspartic acid being a strong competitive inhibitor. During growth in milk, glutamate is required in large amounts for the optimal growth of S. thermophilus, auxotrophic for this amino acid, and the presence of an excess of aspartic acid saturates the entry sites inhibiting glutamate uptake and therefore bacterial growth. Unfortunately, experimental data on S. thermophilus amino acid transport systems are very limited, and in silico analysis performed on the available genomes has revealed the presence of several symporters, a branched-chain amino acid ABC transporter, and several glutamine ABC transporters, but a specific system for the uptake of glutamate has not been identified (16).
The availability of aspartic acid affects the rate of milk acidification of S. thermophilus.
It is interesting to note that in aspartate-supplemented milk, the typical reduction of the acidification rate due to the urease activity detectable when the culture pH is near 6 (Fig. 4A) is remarkably different from that observed without the addition of aspartate. The same phenomenon was observed with mutant A18(Δppc) (Fig. 4B). The addition of fluorofamide, a potent urease inhibitor, to the aspartate-supplemented milk confirmed that the reduction of the acidification rate detected near pH 6 was related to urea hydrolysis as previously demonstrated (22, 26, 28). Moreover, the addition of fluorofamide in aspartate-supplemented milk allowed mutant A18(Δppc) to reach a pH value similar to that of the parental strain.
Aspartate-dependent glutamate uptake deficiency revealed that urease is transcriptionally regulated by a mechanism sensitive to a nitrogen-limited condition.
The data obtained in this work and in a previous study (3, 24) and the information deduced by analysis of the S. thermophilus genome highlight close relationships among the aspartic acid biosynthetic pathway, glutamate/glutamine cellular equilibrium, and urease activity, as shown in Fig. 1. Carbon dioxide, generated by urea hydrolysis, might be used as a substrate in several anaplerotic reactions mainly involved in amino acid and nucleic acid biosynthesis, while ammonia has been previously reported to be involved in glutamine biosynthesis (24). In light of these considerations, the effect of aspartate availability on the levels of urease activity was investigated.
With the aim of verifying our hypothesis, the recombinant strain MIM200 was grown in milk in the presence of different combinations of aspartate, glutamate, and glutamine or aspartate and ammonium chloride for the evaluation of GusA as reporter activity. In the recombinant strain MIM200, GusA reporter activity was under the control of the urease operon promoter pureI (27). With the aim of highlighting the influence of aspartate, glutamate, glutamine, and ammonia concentrations on the expression levels of urease and to avoid the induction of urease biogenesis due to the culture acidification (27), S. thermophilus MIM200 was cultured in forced glutamate starvation obtained by adding to milk a growth-inhibiting concentration of aspartate (2 mM). The experiments were performed using an early induction approach in which recombinant S. thermophilus MIM200 was cultured for 2 h in milk in the presence of aspartate in a growth-inhibiting concentration (2 mM), followed by an additional 10 min of incubation without or with the addition to the culture of glutamate, glutamine, or ammonium chloride. The results obtained and shown in Table 1 underlined that urease expression was sensitive to aspartate, glutamate, glutamine, and ammonia availability. Here the addition of glutamate, glutamine, and ammonia to S. thermophilus cells cultured in the presence of a growth-inhibiting concentration of aspartate resulted in a significant reduction (25 to 33%) of urease expression. The decrease in the expression level was confirmed by the reduction of urease activity (26 to 53%) in the same experimental condition (Table 1). In conclusion, the data obtained suggest that apart from the primary transcriptional regulation by environmental pH, reported in a previous study (27), urease biogenesis also appears to be regulated through a transcriptional regulation system sensitive to a nitrogen-limited condition and specifically sensitive to glutamate, glutamine, and ammonia concentrations. The induction of ure genes under a nitrogen-limited condition has been previously demonstrated for Bacillus subtilis (12) but was not investigated with Streptococcus salivarius, the closest urease-positive neighbor of S. thermophilus (6, 7). Glutamic acid and glutamine have a central role in the nitrogen metabolism of many bacteria (23). Previous studies carried out on the glutamine synthetase activity of S. thermophilus (24) supported the idea that one of the physiological functions of urease was to supply ammonia for the synthesis of glutamine. In this context, the transcriptional regulation of the ure operon by glutamate availability corroborates this hypothesis, highlighting a key role of urease activity in the nitrogen metabolism of S. thermophilus (Fig. 1). Despite the relevance of nitrogen sources for the growth of lactic acid bacteria in milk, relatively little is known about central nitrogen regulation of this group of bacteria (18). Only recently it was demonstrated that glutamine synthetase, the enzyme that converts glutamate and ammonium into glutamine, is a key aspect of the metabolic adaptation of Lactococcus lactis to milk (15) and is essential for the growth in milk of Streptococcus thermophilus (24). In this context, the modulation of urease activity in aspartate-supplemented milk might be interpreted as a cellular response to a nitrogen-limited condition due to glutamate and glutamine starvation.
TABLE 1.
Influence of aspartate, glutamate, and glutamine availability on urease operon transcriptional level of S. thermophilus DSM 20617, measured as GusA reporter activity
| Culture conditions | GusA activitya | Urease activity (%)b |
|---|---|---|
| Milk, aspartate (2 mM) | 862 ± 28 | 100 |
| Milk, aspartate (2 mM), glutamate (0.75 mM) | 579 ± 18 | 53 |
| Milk, aspartate (2 mM), glutamine (0.75 mM) | 649 ± 22 | 33 |
| Milk, aspartate (2 mM), NH4Cl (0.75 mM) | 634 ± 24 | 26 |
GusA activity was expressed as milli-OD420 units per mg of protein as the mean of four independent determinations ± the standard deviation. Experiments were performed by incubating recombinant S. thermophilus MIM200 for 2 h in milk in the presence of aspartate at a growth-limiting concentration (2 mM), followed by 10 min in the presence of glutamate, glutamine, or ammonium chloride or without any other addition.
Urease activity was evaluated with the same conditions used for GusA activity experiments and was expressed as a percentage of urease activity measured in aspartate-supplemented milk as a mean of three independent determinations; standard deviations were always less than 12%. The pH of the cultures at the end of the incubation time was 6.6.
Behavior of wild type and mutant A18(Δppc) in mixed culture with proteinase-positive Lactobacillus delbrueckii subsp. bulgaricus.
With the aim of testing whether the aspartate requested by mutant A18(Δppc) may be provided by the proteolytic activity of other lactic acid bacterial species traditionally used in association with S. thermophilus in dairy preparations, mixed cultures with L. delbrueckii subsp. bulgaricus were performed. All the strains used in mixed cultures were preventively checked for the presence and activity of cell-wall-anchored proteinases. S. thermophilus DSM 20617T (wild type) did not show any activity on casein solution and gave a negative response to a prtS-specific PCR assay and was therefore designated proteinase negative/PrtS negative. On the contrary, L. delbrueckii subsp. bulgaricus ATCC 11842 and CNRZ 397 showed a CaCl2-dependent proteolytic activity against casein, as expected for a PrtB cell wall proteinase. Mutant CNRZ 1159 (CNRZ 397 PrtB negative) (14) did not show any activity against casein solution (data not shown). L. delbrueckii subsp. bulgaricus ATCC 11842 and CNRZ 397 were characterized by a different growth rate in milk, and despite both being proteinase positive, the behavior of strain ATCC 11842 in milk was similar to that observed for mutant CNRZ 1159 (CNRZ 397 PrtB negative) (data not shown).
Mutant A18(Δppc) was grown in association with strains L. delbrueckii subsp. bulgaricus ATCC 11842 and CNRZ 397. As shown in Fig. 5, the aspartate requirement of A18(Δppc) was not satisfied by the proteolytic activity of L. delbrueckii strains. Despite the weakly increased growth rate of A18(Δppc) during association with CNRZ 397, cell density reached at the end of growth was low. The growth of parental strain DSM 20617T was not incremented during association with CNRZ 397 but was significantly reduced in association with mutant CNRZ 1159 (CNRZ 397 PrtB negative) (Fig. 5).
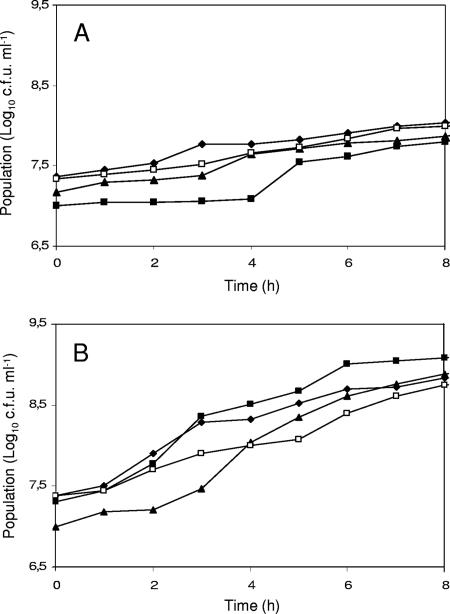
FIG. 5.
Growth of mixed cultures of S. thermophilus mutant A18(Δppc) and DSM 20617 wild type with Lactobacillus delbrueckii subsp. bulgaricus ATCC 11842, CNRZ 397, or CNRZ 1159 (CNRZ 397 PrtB negative) in reconstituted SM. (A) Population of A18(Δppc) cultured alone (⧫) or A18(Δppc) in mixed culture of A18(Δppc)/ATCC 11842 (▴), A18(Δppc)/CNRZ 397 (▪), or A18(Δppc)/CNRZ 1159 (□). (B) Population of DSM 20617 cultured alone (⧫) or DSM 20617 in mixed culture of DSM 20617/ATCC 11842 (▴), DSM 20617/CNRZ 397 (▪), or DSM 20617/CNRZ 1159 (□). All milk cultures were repeated three times, and standard deviations were always <0.2 log10 CFU ml−1.
Monitoring of the l-Asp concentration during growth of mixed cultures (Table 2) revealed that the concentration of this amino acid decrease rapidly in the first hours of growth up to the beginning of the exponential phase. After 8 h of incubation, the aspartate concentration was most likely increased by the activity of the L. delbrueckii proteinase. The increase of the aspartate concentration was not observed in the A18(Δppc)/ATCC 11842 culture or in the mixed cultures performed with mutant CNRZ 1159. In a comparison of the overall changes in the aspartate concentration, it was evident that the highest consumption of aspartate was detected, as expected, in mixed cultures containing mutant A18(Δppc) and/or CNRZ 1159. Nevertheless, the free aspartate was not totally consumed and remained detectable at values ranging between 11 and 18 μM.
TABLE 2.
Changes in level of aspartic acid during growth of S. thermophilus wild type and mutant A18(Δppc) in association with Lactobacillus delbrueckii subsp. bulgaricus strains
| Mixed culture |
l-Asp concn (μM)a
|
||
|---|---|---|---|
| 3 h | 5 h | 8 h | |
| DSM 20617/ATCC 11842 | 12.3 ± 0.6 | 12.8 ± 0.7 | 20.7 ± 1.0 |
| DSM 20617/CNRZ 397 | 16.0 ± 0.9 | 24.5 ± 1.4 | 67.1 ± 3.4 |
| DSM 20617/CNRZ 1159 | 20.5 ± 1.4 | 24.4 ± 1.0 | 17.7 ± 1.1 |
| A18(Δppc)/ATCC 11842 | 14.6 ± 0.8 | 10.6 ± 0.6 | 11.2 ± 0.5 |
| A18(Δppc)/CNRZ 397 | 17.0 ± 0.9 | 16.8 ± 0.9 | 37.9 ± 2.2 |
| A18(Δppc)/CNRZ 1159 | 27.6 ± 1.4 | 21.2 ± 1.2 | 18.0 ± 0.8 |
| DSM 20617 | 15.0 ± 1.2 | 14.3 ± 1.0 | 12.2 ± 0.8 |
| A18(Δppc) | 14.1 ± 1.1 | 11.5 ± 0.7 | 9.0 ± 0.5 |
| ATCC 11842 | 25.0 ± 1.4 | 17.9 ± 1.0 | 24.4 ± 1.3 |
| CNRZ 397 | 21.7 ± 1.2 | 31.5 ± 1.8 | 103.1 ± 5.0 |
| CNRZ 1159 | 23.6 ± 1.3 | 25.4 ± 1.2 | 31.4 ± 1.6 |
The l-Asp concentration in uncultured SM is 28.2 ± 1.5 μM.
The overall data obtained showed that the amounts of aspartic acid present in milk or generated as a consequence of the proteolytic activity of L. delbrueckii strains were always lower than those required for the optimal growth of the A18(Δppc) mutant. Moreover, additional experiments showed that the growth rate of mutant A18(Δppc) was significantly increased in amino acid-enriched milk and not in casein peptone-enriched milk, underlining the presence in S. thermophilus of an oligopeptide uptake system unable to compete with L. delbrueckii to satisfy its amino acid requirements (data not shown).
It was therefore concluded that peptides and amino acids generated by L. delbrueckii proteolysis were not sufficient to restore the growth of an S. thermophilus phosphoenolpyruvate carboxylase-negative mutant. If we consider that S. thermophilus during its adaptation to the milk environment retained its ability to synthesize almost of all the amino acids (2), it could be hypothesized that the supplementation of oligopeptides and amino acids by L. delbrueckii should not be considered the main aspect of the protocooperation with S. thermophilus. Nevertheless, a previous study (8) has demonstrated that the activity of L. delbrueckii PrtB was necessary for the optimal growth and acidification of S. thermophilus. In light of these considerations, it could be deduced that the number of factors that play a relevant role in the protocooperation between these two species is strain dependent. It is possible to say that perhaps some strains of S. thermophilus adapted to the milk environment independently, while others coevolved with protease-producing L. bulgaricus as already hypothesized by van de Guchte et al. (33).
Acknowledgments
The financial support of Ministero dell'Istruzione, dell'Università e della Ricerca (MIUR) (Prin 2006) is gratefully acknowledged. The project was also supported by the Protocol for Scientific and Technological Collaboration between Italy and the Republic of India (Bilateral Research Project no. BS4).
We thank Lorenzo Brusetti for phylogenetic analysis and Francoise Rul for providing L. delbrueckii strains. A special thanks to Paolo Mora for the critical review of the manuscript.
Footnotes
Published ahead of print on 27 July 2007.
REFERENCES
- 1.Biswas, I., A. Gruss, D. Ehrlich, and E. Maguin. 1993. High-efficiency gene inactivation and replacement system for gram-positive bacteria. J. Bacteriol. 175:3628-3635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bolotin, A., B. Quinquis, P. Renault, A. Sorokin, S. D. Ehrlich, S. Kulakauskas, A. Lapidus, E. Goltsman, M. Mazur, G. D. Pusch, M. Fonstein, R. Overbeek, N. Kyprides, B. Purnelle, B. Prozzi, K. Ngui, D. Masuy, F. Hancy, S. Burteau, M. Boutry, J. Delcour, A. Goffeau, and P. Hols. 2004. Complete sequence and comparative genome analysis of the dairy bacterium Streptococcus thermophilus. Nat. Biotechnol. 22:1554-1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bracquart, P., J.-Y. Le Deaut, and G. Linden. 1989. Uptake of glutamic acid by Streptococcus salivarius subsp. thermophilus CNRZ 302. J. Dairy Res. 56:107-116. [DOI] [PubMed] [Google Scholar]
- 4.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 5.Castresana, J. 2000. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 17:540-552. [DOI] [PubMed] [Google Scholar]
- 6.Chen, Y. Y., and R. A. Burne. 1996. Analysis of Streptococcus salivarius urease expression using continuous chemostat culture. FEMS Microbiol. Lett. 135:223-229. [DOI] [PubMed] [Google Scholar]
- 7.Chen, Y. Y., C. A. Weaver, D. R. Mendelsohn, and R. A. Burne. 1998. Transcriptional regulation of the Streptococcus salivarius 57. I urease operon. J. Bacteriol. 180:5769-5775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Courtin, P., V. Monnet, and F. Rull. 2002. Cell-wall proteinases PrtS and PrtB have a different role in Streptococcus thermophilus/Lactobacillus bulgaricus mixed cultures in milk. Microbiology 148:3413-3421. [DOI] [PubMed] [Google Scholar]
- 9.Derzelle, S., A. Bolotin, M. Y. Mistou, and F. Rul. 2005. Proteome analysis of Streptococcus thermophilus grown in milk reveals pyruvate formate-lyase as the major upregulated protein. Appl. Environ. Microbiol. 71:8597-8605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dudley, E. G., and J. L. Steele. 2001. Lactococcus lactis LM0230 contains a single aminotransferase involved in aspartate biosynthesis, which is essential for growth in milk. Microbiology 147:215-224. [DOI] [PubMed] [Google Scholar]
- 11.Fernandez-Espla, M. D., M. Dolores, P. Garault, V. Monnet, and F. Rul. 2000. Streptococcus thermophilus cell wall-anchored proteinase: release, purification, and biochemical and genetic characterization. Appl. Environ. Microbiol. 66:4772-4778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fisher, S. H. 1999. Regulation of nitrogen metabolism in Bacillus subtilis: vive la difference! Mol. Microbiol. 32:223-232. [DOI] [PubMed] [Google Scholar]
- 13.Garault, P., C. Letort, V. Juillard, and V. Monnet. 2000. Branched-chain amino acid biosynthesis is essential for optimal growth of Streptococcus thermophilus in milk. Appl. Environ. Microbiol. 66:5128-5133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gilbert, C., B. Blanc, J. Frot-Coutaz, R. Portalier, and D. Atlan. 1997. Comparison of cell surface proteinase activities within the Lactobacillus genus. J. Dairy Res. 64:561-571. [Google Scholar]
- 15.Gitton C., M. Meyrand, J. Wang, C. Caron, A. Trubuil, A. Guillot, and M.-Y. Mistou. 2005. Proteomic signature of Lactococcus lactis NCDO763 cultivated in milk. Appl. Environ. Microbiol. 71:7152-7163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hols, P., F. Hancy, L. Fontaine, B. Grossiord, D. Prozzi, N. Leblond-Bourget, B. Decaris, A. Bolotin, C. Delorme, S. D. Ehrlich, E. Guédon, V. Monnet, P. Renault, and M. Kleerebezem. 2005. New insights in the molecular biology and physiology of Streptococcus thermophilus revealed by comparative genomics. FEMS Microbiol. Rev. 29:435-463. [DOI] [PubMed] [Google Scholar]
- 17.Lapujade, P., M. Cocaign-Bosquet, and P. Loubiere. 1998. Glutamate biosynthesis in Lactococcus lactis subsp. lactis NCDO 2118. Appl. Environ. Microbiol. 64:2485-2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Larsen, R., T. G. Kloosterman, J. Kok, and O. P. Kuipers. 2006. GlnR-mediated regulation of nitrogen metabolism in Lactococcus lactis. J. Bacteriol. 188:4978-4982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Letort, C., and V. Jauillard. 2001. Development of a minimal chemically-defined medium for the exponential growth of Streptococcus thermophilus. J. Appl. Microbiol. 91:1023-1029. [DOI] [PubMed] [Google Scholar]
- 20.Limauro, D., A. Falciatore, A. L. Basso, G. Forlani, and M. De Felice. 1996. Proline biosynthesis in Streptococcus thermophilus: characterization of the proBA operon and its products. Microbiology 142:3275-3282. [DOI] [PubMed] [Google Scholar]
- 21.Louailéche, H., P. Bracquart, C. Grimont, and G. Linden. 1996. Carbon dioxide fixation by cells and cell-free extracts of Streptococcus thermophilus. J. Dairy Res. 63:321-325. [Google Scholar]
- 22.Martin, B., J. B. Coulon, J. F. Chamba, and C. Bugaud. 1997. Effect of milk urea content on characteristics of matured Reblochon cheeses. Lait 77:505-514. [Google Scholar]
- 23.Merrick, M. J., and R. A. Edwards. 1995. Nitrogen control in bacteria. Microbiol. Rev. 59:604-622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Monnet, C., D. Mora, and G. Corrieu. 2005. Glutamine synthesis is essential for growth of Streptococcus thermophilus in milk and is linked to urea catabolism. Appl. Environ. Microbiol. 71:3376-3378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mora, D., G. Ricci, S. Guglielmetti, D. Daffonchio, and M. G. Fortina. 2003. 16S-23S rRNA intergenic spacer region sequence variation in Streptococcus thermophilus and related dairy streptococci and development of a multiplex ITS-SSCP analysis for their identification. Microbiology 149:807-813. [DOI] [PubMed] [Google Scholar]
- 26.Mora, D., E. Maguin, M. Masiero, C. Parini, G. Ricci, P. L. Manachini, and D. Daffonchio. 2004. Characterization of urease genes cluster of Streptococcus thermophilus, J. Appl. Microbiol. 96:209-219. [DOI] [PubMed] [Google Scholar]
- 27.Mora, D., C. Monnet, C. Parini, S. Guglielmetti, A. Mariani, P. Pintus, F. Molinari, D. Daffonchio, and P. L. Manachini. 2005. Urease biogenesis in Streptococcus thermophilus. Res. Microbiol. 156:897-903. [DOI] [PubMed] [Google Scholar]
- 28.Pernoud, S., C. Fremaux, A. Sepulchre, G. Corrieu, and C. Monnet. 2004. Effect of the metabolism of urea on the acidifying activity of Streptococcus thermophilus. J. Dairy Sci. 87:550-555. [DOI] [PubMed] [Google Scholar]
- 29.Platteeuw, C., G. S. Simons, and W. M. de Vos. 1994. Use of the Escherichia coli β-glucuronidase (gusA) gene as a reporter gene for analyzing promoters in lactic acid bacteria. Appl. Environ. Microbiol. 60:587-593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reiter, B., and J. D. Oran. 1962. Nutritional studies on cheese starters. I. Vitamin and amino acid requirements of single strain starters. J. Dairy Res. 29:63-77. [Google Scholar]
- 31.Resmini, P., J. A. Hogenboom, C. Pazzaglia, and L. Pellegrino. 1993. Gli amminoacidi liberi nella caratterizzazione analitica del formaggio Grana Padano. Sci. Tecnica Lattiero-Casearia 44:7-19. [Google Scholar]
- 32.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.van de Guchte, M., S. Pernaud, C. Grimaldi, V. Barbe, K. Bryson, P. Nicolas, C. Robert, S. Oztas, S. Mangenot, A. Coulox, V. Loux, R. Dervyn, R. Bossy, A. Bolotin, J.-M. Batto, T. Walunas, J.-F. Gibrat, P. Bessières, J. Weissenbach, S. D. Ehrlich, and E. Maguin. 2006. The complete genome sequence of Lactobacillus bulgaricus reveals extensive and ongoing reductive evolution. Proc. Natl. Acad. Sci. USA 103:9274-9279. [DOI] [PMC free article] [PubMed] [Google Scholar]