Abstract
The Alvord Basin in southeast Oregon contains a variety of hydrothermal features which have never been microbiologically characterized. A sampling of Murky Pot (61°C; pH 7.1) led to the isolation of a novel arsenic-metabolizing organism (YeAs) which produces an arsenic sulfide mineral known as β-realgar, a mineral that has not previously been observed as a product of bacterial arsenic metabolism. YeAs was grown on a freshwater medium and utilized a variety of organic substrates, particularly carbohydrates and organic acids. The temperature range for growth was 37 to 75°C (optimum, 55°C), and the pH range for growth was 6.0 to 8.0 (optimum, pH 7.0 to 7.5). No growth was observed when YeAs was grown under aerobic conditions. The doubling time when the organism was grown with yeast extract and As(V) was 0.71 h. Microscopic examination revealed Gram stain-indeterminate, non-spore-forming, nonmotile, rod-shaped cells, with dimensions ranging from 0.1 to 0.2 μm wide by 3 to 10 μm long. Arsenic sulfide mineralization of cell walls and extracellular arsenic sulfide particulate deposition were observed with electron microscopy and elemental analysis. 16S rRNA gene analysis placed YeAs in the family Clostridiaceae and indicated that the organism is most closely related to the Caloramator and Thermobrachium species. The G+C content was 35%. YeAs showed no detectable respiratory arsenate reductase but did display significant detoxification arsenate reductase activity. The phylogenetic, physiological, and morphological characteristics of YeAs demonstrate that it is an anaerobic, moderately thermophilic, arsenic-reducing bacterium. This organism and its associated metabolism could have major implications in the search for innovative methods for arsenic waste management and in the search for novel biogenic mineral signatures.
Microorganisms play an important role in the biogeochemical cycling of many elements (33) including arsenic. Arsenic is relatively abundant in soils and natural waters, and although it is primarily a consequence of anthropogenic sources, it also occurs naturally in many minerals (18, 25) and in groundwater in contact with geologic formations containing high levels of arsenic. In arsenic-laden environments, the compound can exist in three different oxidation states, including As(V) (arsenate), As(III) (arsenite), and As(−III) (arsine) (10). Microorganisms are capable of transforming the various forms of arsenic by using mechanisms for oxidizing As(III) or reducing As(V) (18). These mechanisms, although commonly used as a mode of detoxification, can also be a source of energy generation through the use of As(V) as a terminal electron acceptor (18) or the use of As(III) as an electron donor. Depending on the type of mechanism utilized, these processes require distinct sets of genes/enzymes to perform the redox reaction (32). A total of 24 prokaryotes, including members of the phyla Crenarchaeota and Aquificae, Chrysiogenes spp., phylum Deferribacteres, low-G+C gram-positive bacteria, Halanaerobacter spp., and the Gamma-, Delta-, and Epsilonproteobacteria, have been observed to utilize arsenic as a terminal electron acceptor (27, 34). Irrespective of the exact method of metabolism, the number and variety of arsenic-utilizing organisms isolated from the environment clearly show that they play an important role in the transformation and geochemical cycling of arsenic.
Murky Pot, located in the Alvord Basin and part of the Mickey hydrothermal system, was initially examined for determination of the bacterial diversity, by using contemporary molecular methods. These studies revealed the presence of a variety of 16S rRNA sequences, the most abundant of which were related to the Deltaproteobacteria and the phylum Acidobacteria (17). This paper describes the isolation, cultivation, and characterization of an arsenic-metabolizing organism (YeAs) from Murky Pot hot spring. Although arsenic metabolism is common among microorganisms, YeAs seems to be distinct in that it produces a novel biogenic mineral, β-realgar. Neither the mineral nor the associated metabolism has been observed among the organisms most closely related phylogenetically to YeAs.
MATERIALS AND METHODS
Field site description.
At the time of sampling, the temperature of Murky Pot (Fig. 1) was 61°C and the pH was 7.1. The aqueous geochemistry of Murky Pot revealed significant levels of arsenic (1.0 mg/liter or about 15 μM) as well as substantial levels of sodium, borate, and carbonate.
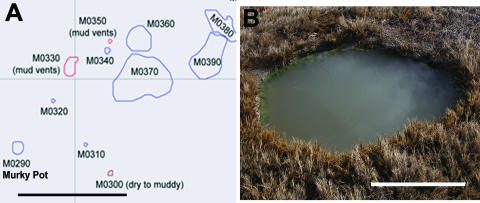
FIG. 1.
(A) Map of a portion of the Mickey hydrothermal area. Murky Pot (M0290) is indicated and lies among several other mud vents and pools. Scale bar represents 10 m. (B) Murky Pot hot spring. Pool is turbid with flocculent materials. Scale bar represents 0.5 m.
Enrichment, isolation, and cultivation.
A growth medium prepared anoxically from 0.22-μm-filtered Murky Pot hot spring water was inoculated in the field with suspensions of sediment/water from Murky Pot and supplemented with 0.5% (wt/vol) yeast extract and 2.5 mM sodium arsenate (Na2HAsO4). Tubes were transported back to the laboratory in thermos bottles filled with hot spring water at 55°C and then transferred to incubators at the desired temperature of 55°C. When growth was evident, as judged by microscopy and the appearance of a yellow precipitate, cultures were transferred to a synthetic freshwater medium, Orex-4 (25), with 0.5% (wt/vol) yeast extract and 2.5 mM As(V). Orex-4 medium contained the following salts (g/liter): 0.14 g of KH2PO4, 0.25 g of NH4Cl, 0.5 g of KCl, 1.0 g of NaCl, and 0.62 g of MgCl2. The base medium (per liter) was amended with 1.0 ml of Wolfe's mineral solution (37), 1.0 ml of Wolfe's vitamin stock solution (37), 20 ml of 2.5% (wt/vol) cysteine solution, 20 ml of 9.5% (wt/vol) bicarbonate solution, and 20 ml of 0.75% (wt/vol) CaCl2 solution. The Hungate technique was used throughout, and culture headspaces contained an atmosphere of N2/CO2 (80:20) (21). One subculture, designated YeAs, was propagated for further study. In order to obtain single colonies of YeAs, samples of enrichment cultures were used as inoculum for Orex-4 solid medium containing 1.5% high-melting-point agarose. Solid medium was prepared in anaerobic glass bottles, and the Hungate technique was employed to create an anoxic atmosphere. Following sterilization, the anaerobic bottles were inoculated and allowed to solidify. Yellow colonies appeared after 5 days of incubation, and these colonies were picked and transferred to fresh solid medium. This procedure was repeated two more times, after which the YeAs culture was maintained in liquid freshwater medium with 0.5% (wt/vol) yeast extract and 2.5 mM As(V) and incubated at 55°C, unless otherwise stated.
For some growth experiments, cysteine was eliminated from the medium to prevent interference of the arsenic sulfide mineral precipitation. All other components and concentrations [including the yeast extract and As(V)] remained the same. Even with the removal of cysteine, trace amounts of sulfur were still present in the medium from the addition of the vitamin and mineral solutions; however, mineral production was significantly reduced.
Determination of growth parameters.
Cell growth was measured by using direct cell counts. YeAs was grown in Orex-4 and in Orex-4 (−Cys) supplemented with yeast extract and As(V) or in Orex-4 with yeast extract only. Subsamples were taken at time 0 and every 3 hours following until the cells were in stationary phase. At each time point, 1 μl of each sample was stained with 400 μl of 4′,6′-diamidino-2-phenylindole (DAPI) stock solution (2.5 μg/ml) for 25 min and then vacuum filtrated onto 0.22-μm black polycarbonate membranes (Millipore, Billerica, MA). The membranes were then mounted onto glass slides with a drop of immersion oil. Four fields were counted for each sample under a total magnification of ×1,000, using a Leica DMRB microscope fitted with a DAPI filter.
To determine the temperature range for growth, YeAs was inoculated into Orex-4 (−Cys) containing yeast extract and As(V) and incubated at temperatures ranging from 37°C to 75°C at pH25°C 6.8. The pH range was tested from pH55°C 4 to 9. All tubes were incubated at 55°C. The pH was tested prior to inoculation and also during late log phase to ensure proper buffering. YeAs was also tested for its ability to grow aerobically on Orex-4 and on Orex-4 (−Cys) supplemented with yeast extract and As(V). While organisms were growing in the presence of oxygen, all tubes were incubated in a tissue culture rotator (Lab-Line Cel-Gro; Fisher Scientific, Pittsburgh, PA) at 30 rpm. For temperature, pH, and oxygen tolerance experiments, culture turbidity was determined by measuring the optical density (at 600 nm) until the cells were in stationary phase. All growth parameter experiments were done in duplicate.
Substrate utilization.
The substrates utilized by YeAs were assessed using Biolog Anaerobic MicroPlates (Hayward, CA) (4, 5). The MicroPlates test the ability of a microorganism to utilize or oxidize an array of carbon sources under anaerobic conditions using an artificial tetrazolium colorimetric electron acceptor. The plates were inoculated with YeAs cells in triplicate, and wells exhibiting a strong color change due to the reduction of the tetrazolium dye compared to that of the negative control were selected as positive for substrate utilization.
Microscopy and morphological characterization.
The Gram type was tested using the Gram stain (12), the KOH test (7), and the peptidoglycan isolation test for cell wall analysis. The Schaeffer-Fulton endospore stain (30) was used to determine if spores were present in a YeAs culture grown for 24 h in Orex-4 with yeast extract and As(V) and in a culture incubated for 2 weeks under the same conditions. Motility was determined by using 0.4% and 0.7% semisolid Orex-4 and Orex-4 (−Cys) deeps supplemented with yeast extract and As(V). Deeps were inoculated with YeAs cells and incubated for 24 h at 55°C.
Two drops of the 24-h liquid culture of YeAs grown in Orex-4 with yeast extract and As(V) were placed on a microscope slide with a coverslip and then observed using Nomarski differential interference contrast microscopy (1). Specimens were examined using a Leica DMRB microscope equipped with SPOT imaging software (Diagnostic Instruments, Sterling Heights, MI).
Transmission and scanning electron microscopy (TEM; SEM) were performed at the Environmental Molecular Sciences Laboratory (EMSL) at the Pacific Northwest National Laboratory. YeAs cells grown for 24 h in Orex-4 with yeast extract and As(V) were used for the microscopy, and EM samples were prepared anaerobically in a glove bag. Copper grids coated with a Formvar support film and sputtered with carbon (Electron Microscopy Sciences, Hatfield, PA) were used for the whole-mount TEM imaging. A drop of cellular suspension was applied to a grid and gently blotted on a filter paper. Imaging and analyses were performed with a JEOL 2010 high-resolution TEM operating at 200 kV. Elemental analysis was performed by energy dispersive spectroscopy (ISIS, Oxford University, Oxford, United Kingdom) coupled to the TEM. Images were collected digitally and analyzed using Gatan digital micrograph software (Gatan, Pleasanton, CA). Samples for SEM were fixed in 2.5% glutaraldehyde, gradually dehydrated in an ethanol series, and applied on a carbon tape. They were processed by critical point drying, sputter-coated with carbon, and imaged with a LEO 942 SEM at 2 kV.
Raman spectroscopy.
Precipitate samples for Raman spectroscopy were prepared under a 95% N2:5% H2 atmosphere and, where necessary, under a 785-nm illumination (to prevent photo alteration of the sample). Precipitates were removed from YeAs culture tubes with a syringe and needle and collected by centrifugation at 5,000 × g for 15 min. The collected precipitates were immediately transferred to a nuclear magnetic resonance tube, and the tube was sealed with a plastic cap under anaerobic conditions before being transferred to the spectrometer. To prevent exposure to light, all tubes were transferred wrapped in aluminum foil and placed in the spectrometer in a darkened laboratory. Spectra were obtained using a FT-Raman 960 spectrometer (Nicolet; Thermo Electron Corp.) employing 1,064 nm of excitation from a Nd:YVO4 laser and a liquid-N2-cooled Ge detector. Spectra were collected at room temperature with 800 mW of laser power at the sample and at a spectral resolution of 1 cm−1. For phototransformation studies, samples were illuminated for 12 h using a 900-mW ultraviolet light source (R-52 Mineralight; Ultra-violet Products Inc., San Gabriel, CA) or were placed in direct sunlight for 2 weeks.
β-Realgar production by YeAs and phylogenetically related bacteria.
Thermobrachium celere and Caloramator coolhaasii were tested for their ability to produce β-realgar while cultured under YeAs standard conditions [Orex-4 with yeast extract and As(V)]. They were also tested using their published growth media supplemented with 0.05% cysteine, 0.5% yeast extract, and 2.5 mM As(V) (11, 29).
G+C content and 16S rRNA gene analysis.
The G+C content of YeAs was determined by high-performance liquid chromatography at Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH (DSMZ) (19). Genomic DNA was isolated from YeAs by using a QIAGEN DNeasy tissue kit (Valencia, CA), and the 16S rRNA gene was amplified by PCR with eubacterial primers 8F (5′-AGA GTT TGA TCM TGG CTC AG-3′) and 1492R (5′-GGY TAC CTT GTT ACG ACT T-3′). PCR products were purified using a QIAquick PCR purification kit (QIAGEN, Valencia, CA). Sequencing was performed using an ABI Prism 377 DNA sequencer (Applied Biosystems, Foster City, CA) with ABI Prism BigDye terminator cycle sequencing reagent and the following primers: 8F, 1492 R, and 338F (5′-ACI CCT ACG GGI GGC WGC-3′). Sequences of each type strain within the Clostridiaceae family, based on the designations found in Bergey's Manual Taxonomic Outline of the Prokaryotes, release 5.0 (11a), and sequences of species within the Caloramator/Thermobrachium clade were aligned and masked in ARB (http://www.arb-home.de/) (17a). Only unambiguously alignable nucleotide positions were kept for subsequent phylogenetic analysis as described by Lane (16a). Phylogenetic analysis was carried out with the program PAUP* 4.0 beta 10 (34a) and included 1,195 nucleotide positions. The tree topology was inferred by neighbor joining using the Jukes and Cantor model to estimate evolutionary distances. Bootstrap proportions were determined from 1,000 resamplings. Three Bacillus species—B. subtillis (X60646), B. megaterium (D16273), and B. thuringiensis (D16281)—were used as outgroups.
Enzyme assays for respiratory and detoxification arsenate reductases.
Cell extracts of YeAs were prepared by homogenization of cells harvested from a culture grown on Orex-4 (−Cys) with yeast extract and As(V) and were used for the following experiments. In order to identify the enzyme responsible for arsenate reduction in YeAs, three approaches were used. These included a spectrophotometric assay for respiratory arsenate reductase (Arr), zymography for the detection of Arr, and a spectrophotometric assay for detoxification arsenate reductase (Ars). Arr was assayed spectrophotometrically by using the reduced viologen method (23). An in-gel zymography approach was also used to identify Arr activity. The Thermus spp. strain AO3C was used as a positive control in this experiment as it has been shown to respire on arsenate (S. A. Connon et al., submitted). Nondenaturing polyacrylamide gel electrophoresis was employed to resolve the respiratory arsenate reductase enzyme complex in an active state. To visualize activity, the respiratory arsenate reductase activity stain was used and adopted from the respiratory nitrate reductase enzyme activity stain as published by Vallejos (36). The only change in the procedure was the addition of 1 g of sodium arsenate in place of 1 g of potassium nitrate. Negative control gels (no arsenate added) were run to ensure visualized bands were due to arsenate reductase activity. Ars was assayed using a NADPH/glutathione assay (22). All spectrophotometric assays and activity stains were performed at 55°C.
Nucleotide sequence accession number.
The 16S rRNA gene sequence of the YeAs isolate has been submitted to the DDBJ/EMBL/GenBank databases under accession number EU051651.
RESULTS
Culture characteristics and morphology.
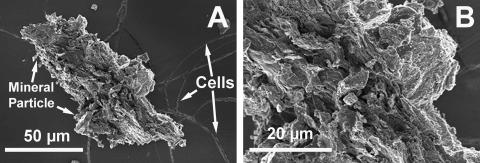
Copious amounts of yellow precipitate formed within 24 h, when the cells were grown on Orex-4 with yeast extract and As(V) (Fig. 2A), and microscopy examination revealed that the cells were straight to slightly curved rods (Fig. 2B). The cells occurred primarily singly, and no endospores were observed by microscopy in a 24-h or 2-week-old culture. Cell wall analyses yielded no conclusive results as to the cell wall type; thus, YeAs was considered to be a gram-indeterminate organism. No motility was observed when cultures were grown anaerobically in Orex-4 and in Orex-4 (−Cys) deeps (0.4% and 0.7% agar, respectively) supplemented with yeast extract and As(V). Additionally, no flagella were seen in the microscopy examinations. The average cell length was 6 μm, with a range of 3 μm to 10 μm, while the average width was 100 nm and up to 200 nm (Fig. 3 to 5). Mineral deposition on the outer cell wall contributed to cell thickness. Electron microscopy analysis revealed unusual rod-shaped cell morphology and branching at the cell terminus (Fig. 3) when organisms were grown on Orex-4 with yeast extract and As(V). These branching structures did not appear when the cells were grown with yeast extract only. Considerable mineral deposition was observed on cell surfaces, and energy-dispersive X-ray spectroscopy (EDS) showed that the elemental composition of the mineral deposition was primarily arsenic and sulfur (Fig. 4 and 5), consistent with Raman spectroscopy data of mineral precipitates in cultures.
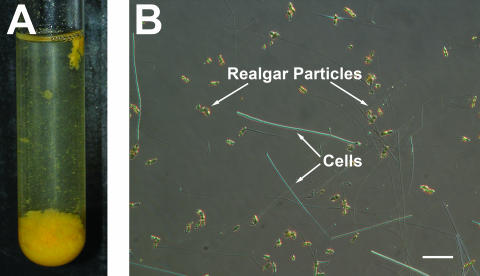
FIG. 2.
(A) Culture of YeAs after 24 h, showing β-realgar production when grown with 0.5% yeast extract and 2.5 mM arsenic at 55°C. (B) Nomarski differential interference microscopy of YeAs, showing long thin rods. Cells were in late log phase when the image was taken. Scale bar represents 2 μm.
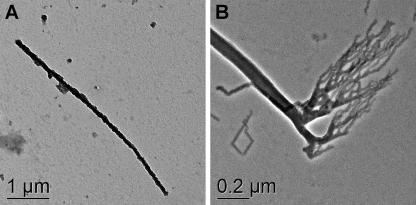
FIG. 3.
Transmission electron microscopy of YeAs cells grown under standard conditions with yeast extract and As(V). (A) Mineral (realgar)-encrusted cell. (B) Unusual structures commonly found at cell termini.
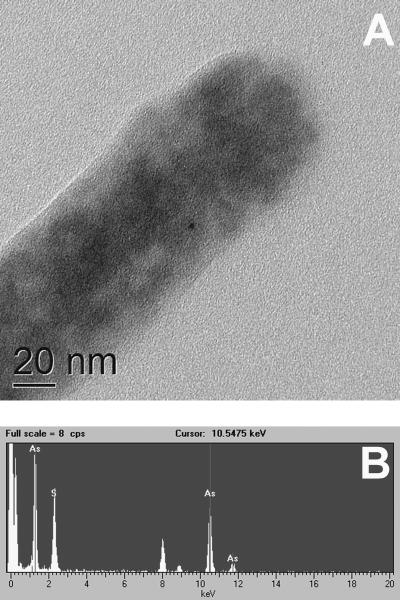
FIG. 5.
Energy-dispersive spectroscopy analysis of mineral formed by YeAs at the cell surface. (A) The tip of the cell was examined by EDS for elemental composition. (B) Results of EDS analysis showing significant arsenic and sulfide distribution on the cell surface.
FIG. 4.
(A) Scanning electron microscopy of YeAs, showing long thin cells and large β-realgar particle formed during growth. (B) Enlarged view of panel A. Note evidence of layers running diagonally across the specimen.
Mineral precipitate characterization and production.
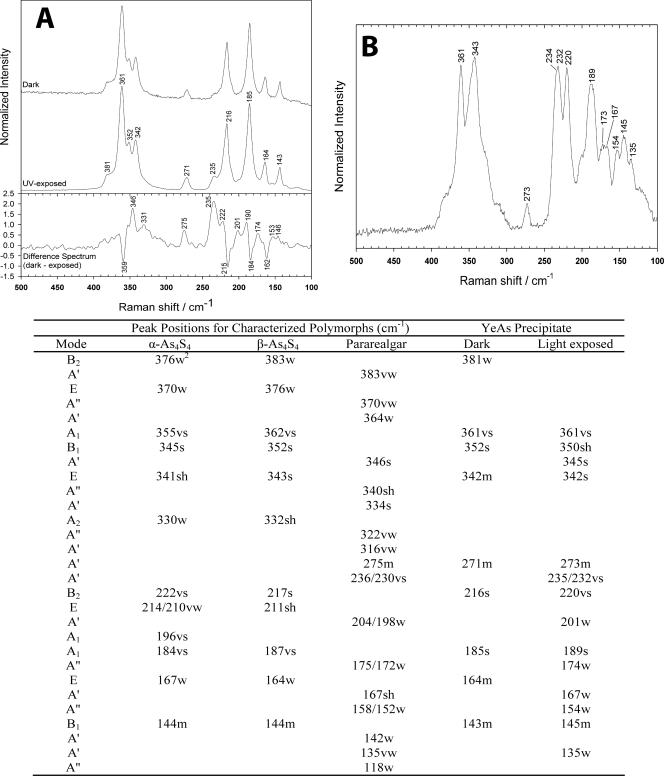
Raman spectra collected from the precipitate produced by YeAs in culture that had not been exposed to light exhibited a set of prominent peaks in the 100 to 450 cm−1 region (Fig. 6A). Consideration of the likely products of arsenic reduction in the culture medium, combined with a comparison of the experimentally obtained spectra with previously published spectra, suggested that the peaks were derived from an arsenic chalcogenide structure of the general form As4S4 (24). The lack of a principal peak at 220 cm−1 and a peak at 472 cm−1 indicated that the observed spectrum was not due to the presence of either α or β polymorphs of sulfur (16). The absence of strong spectral features at 294 and 309 cm−1 also precluded the presence of arsenic sulfides of the orpiment (As2S3) type (8). Tetra-arsenic tetrasulfide exists in a number of well-studied polymorphs, including realgar (α-As4S4), pararealgar (light-induced polymorph of realgar), and a high-temperature-phase β-realgar (β-As4S4). Most instructive in structural discrimination are the A1, B1, and E modes of α- and β-realgar and the A′ mode of pararealgar, which gives rise to strong peaks between 340 and 365 cm−1 (24). These modes are dominated by νAs-S vibrations, with smaller contributions from δAs-S-As, and δS-As-S. The observed peak positions, 361 cm−1 (A1), 352 cm−1 (B1), and 342 cm−1 (E), all indicated the presence of the β-realgar structure, the modes being shifted to lower wave numbers in α-realgar, and the pararealgar with a single peak at 346 cm−1 in this region (Fig. 6A). The presence of a peak at 216 cm−1 was, again, more consistent with the B2 mode (principally δS-As-S vibrations) of the β-realgar polymorph than that of either realgar or pararealgar. The peak at 271 cm−1 could not be ascribed to β-realgar but was consistent with an A′ mode of pararealgar dominated by δS-As-S vibrations; however, there was no evidence for the stronger pararealgar doublet at 236/230 cm−1, which would have definitively indicated the presence of this polymorph. The remaining peaks were consistent with β-realgar assignment, although they are less instructive when determining structure. Further confirmation of the β-realgar assignment could be gained from following the photo-induced transformation of the putative structure to pararealgar. Exposure to wavelengths shorter than 550 nm typically results in a rapid transformation of both α and β polymorphs to pararealgar. When cultures were exposed to ultraviolet light, the YeAs precipitate proved uncharacteristically photo stable. Figure 6A also shows the FT-Raman spectrum of the β-realgar precipitate following 12 h of UV exposure. Ostensibly, there was little difference between the dark and UV light-exposed spectra. However, a difference spectrum (Fig. 6A) suggests that limited transformation occurred during the exposure period. In this representation, peaks with negative intensity are reduced due to UV light exposure, while positive peaks increase in intensity. The four negative peaks were all consistent with β-realgar vibrations, demonstrating that some structure had been lost. Simultaneously, a number of positive peaks were observed, suggesting the formation of a new structure within the sample. The strongest peaks at 235 and 346 cm−1 were consistent with A′ modes of pararealgar, due to νAs-As and νAs-S vibrations, respectively (24). Other A′ modes were evident at 275, 331, 201 and 146 cm−1. A″ modes were also evident at 190, 174, and 153 cm−1. Thus, there is evidence to suggest that exposure of the precipitate to UV light induces limited transformation to pararealgar. More extensive transformation was observed for cultures exposed to sunlight for 2 weeks. The spectrum collected from the precipitates after extended light exposure is shown in Fig. 6B and essentially represents a mixture of β-realgar and pararealgar. Strong contributions from β-realgar were again evident at 361 cm−1 (A1 mode), 220 cm−1 (B2 mode), and 189 cm−1 (A1 mode). A weaker B1 mode was also observed at 145 cm−1. The relative intensity of these peaks suggests that a significant amount of β structure remains in the precipitate. However, a significant amount of transformation occurred since strong peaks were observed for pararealgar at 345 cm−1 (A′ mode), a characteristic doublet at 234/232 cm−1 (A′ mode), and a weaker contribution at 273 cm−1 (A′ mode).
FIG. 6.
(A) Raman spectra of mineral precipitates kept in the dark versus spectra of those exposed to UV light for 12 h. (B) Spectrum of mineral precipitate following exposure to sunlight for 2 weeks, representing a mixture of β-realgar and pararealgar. In the accompanying table, peak positions for the polymorphs (α-As4S4, β-As4S4, and pararealgar; taken from reference 24) and for this study can be found. vs, very strong; s, strong; m, medium; w, weak; vw, very weak; sh, shoulder.
No β-realgar was produced by either T. celere or C. coolhaasii. In fact, very little to no growth was seen with these organisms when grown with As(V), suggesting that the arsenic is significantly more toxic to these strains.
Growth parameters and electron donor utilization.
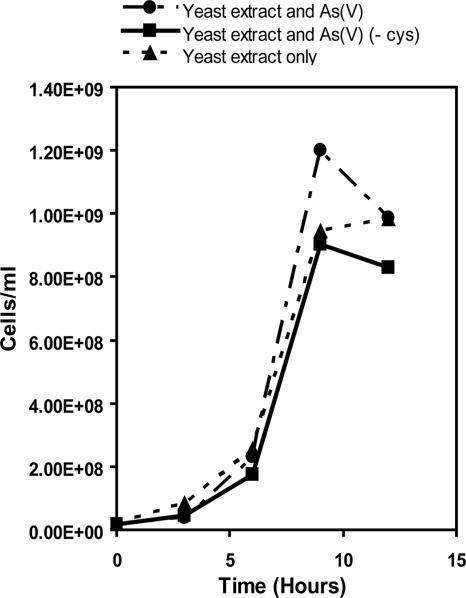
The doubling time for YeAs was 0.71 h. During late log phase, there were 1.20 × 109 cells/ml for YeAs grown on Orex-4 with yeast extract and As(V), 9.04 × 108 cells/ml when grown on Orex-4 (−Cys) with yeast extract and As(V), and 9.84 × 108 cells/ml on yeast extract only (Fig. 7). These data suggest that YeAs is a fermentative microorganism and does not rely on As(V) as a respiratory electron acceptor.
FIG. 7.
YeAs direct cell count data plotted against time. Note the similarity in growth rates between cells grown with and without As(V), suggesting that As(V) is not used as an electron acceptor.
The temperature range at pH25°C 6.8 for YeAs was 37°C to 75°C, with an optimal growth temperature of 55°C. YeAs grew at a pH55°C of 6.0 to 8.0 and displayed optimal growth between pH55°C 7.0 and 7.5. Growth was observed under anaerobic conditions only.
Based on Biolog plate results, YeAs was able to utilize the following substrates: fructose, fucose, galactose, galacturonic acid, d-glucose-6-phosphate, palatinose, rhamnose, turanose, and pyruvic acid. YeAs was not able to utilize the following substrates: acetate, formate, lactate, succinate, malate, alanine, and valine.
G+C content and 16S rRNA gene analysis.
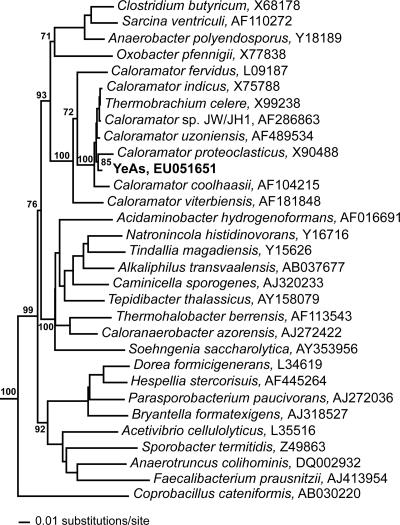
The G+C content of YeAs was 35.0 mol% G+C. Based on16S rRNA gene analysis, the sequence data revealed that YeAs branches within the family Clostridiaceae. The phylogenetic tree indicates that the organisms most closely related to YeAs include Thermobrachium celere (11), Caloramator coolhaasii (29), Caloramator proteoclasticus (35), Caloramator viterbiensis (31), Caloramator indicus (9), Caloramator fervidus (28), and Caloramator uzoniensis (Fig. 8).
FIG. 8.
Phylogenetic tree of 16S rRNA genes illustrating affiliations of the YeAs isolate with members of the bacterial family Clostridiaceae. Scale bar corresponds to 0.01 substitution per nucleotide position. Only bootstrap values for the major clades within the Clostridiaceae are shown along with positions in the Caloramator/Thermobrachium clade relevant to placing YeAs within this clade.
Enzyme detection for respiratory and detoxification arsenate reductases.
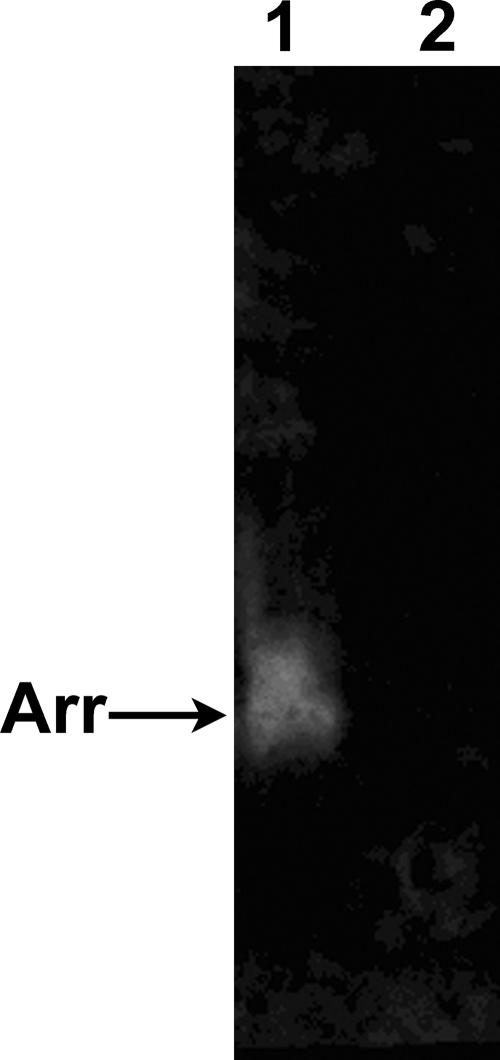
Strain YeAs had no detectable respiratory arsenate reductase, as judged by both spectrophotometric and in-gel assays (Fig. 9). YeAs demonstrated significant detoxification arsenate reductase activity with a specific activity of 3.70 μmol NADPH/min/mg protein at 55°C. This activity was glutathione dependent, consistent with other reports of bacterial arsenic detoxification reductases (20).
FIG. 9.
Respiratory arsenate reductase activity stain showing the absence of activity in YeAs. Forty micrograms of protein was loaded per lane. Lane 1, Thermus strain AO3C; lane 2, YeAs.
DISCUSSION
YeAs is an anaerobic, moderately thermophilic, arsenic-metabolizing bacterium that was suspected to be novel based on its ability to reduce arsenic and to biologically produce copious amounts of the mineral β-realgar. Phylogenetic analysis revealed that YeAs is most closely related to a group of organisms in the family Clostridiaceae, most specifically Thermobrachium celere and six Caloramator sp. Table 1 shows the general characteristics of six of the most closely related organisms (detailed data for Caloramator uzoniensis are unpublished).
TABLE 1.
General characteristics of YeAs and the most closely related organisms
| Strain | Characteristic
|
Reference | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Habitat | Geographic location | Cell width (μm) | Cell length (μm) | Gram stain | Spore formation | G+C content (mol%) | Growth temp (°C)
|
pH
|
Oxygen tolerance | β-Realgar production | ||||
| Optimal | Range | Optimal | Range | |||||||||||
| YeAs | Hot spring | Alvord Basin, Oregon, United States | 0.1-0.2 | 3-10 | Indeterminate | − | 35 | 55 | 37-75 | 7.0-7.5 | 6.0-8.0 | Strict anaerobe | + | This study |
| T. celere | Several locations | Several locations | 0.5-0.8 | 1.5-13 | + | − | 31 | 66 | 43-75 | 8.2 | 5.4-9.5 | Strict anaerobe | − | 11 |
| C. coolhaasii | Thermophilic granular sludge | Wageningen, The Netherlands | 0.5-0.7 | 2-40 | + | − | 31.7 | 50-55 | 37-65 | 7.0-7.5 | 6.0-8.5 | Strict anaerobe | − | 29 |
| C. proteoclasticus | Mesophilic granular sludge | Montevideo, Uruguay | 0.4 | 2.4-4 | − | + | 31 | 55 | 30-68 | 7.0-7.5 | 5.5-8.7 | Strict anaerobe | NDa | 35 |
| C. viterbensis | Hot spring | Viterbo, Italy | 0.4-0.6 | 2-3 | + | − | 32 | 58 | 33-64 | 6.0-6.5 | 5.0-7.8 | Strict anaerobe | ND | 31 |
| C. indicus | Thermophilic aquifer | Gujarat, India | 0.6-0.8 | 10-100 | − | − | 25 | 60-65 | 37-75 | 7.5-8.1 | 6.2-9.2 | Strict anaerobe | ND | 9 |
| C. fervidus | Hot spring | New Zealand | 0.65-0.75 | 2-3 | − | + | 39 | 68 | 37-80 | 7.0-7.5 | 5.5-9.0 | Strict anaerobe | ND | 28 |
ND, not determined.
Although YeAs does not use As(V) as a terminal electron acceptor, as indicated by growth experiments, the reduction of As(V) by this organism is confirmed by two lines of evidence: the detection of arsenate reductase activity in cell extracts and the production of copious amounts of β-realgar, a mineral composed of As(III) and sulfide (3). Our observation is the first for bacterially mediated β-realgar production. Huber et al. (14, 15) reported a hyperthermophilic archaeon capable of realgar production and characterized the mineral using X-ray diffraction, SEM, and EDS. Additional reports have also mentioned realgar formation in the presence of arsenate-reducing bacteria (13, 26). Although realgar formation has been cited several times, in no case was the polymorph of the realgar determined. β-Realgar is normally produced abiotically at high temperatures, above 250°C (6). Additionally, arsenic sulfide minerals are typically extremely photosensitive; however, the β-realgar produced by YeAs undergoes limited transformation when exposed to UV light.
One striking finding of these studies is that none of the phylogenetically related Thermobrachium and Caloramator isolates tested was able to metabolize arsenate and form β-realgar. In this case, phylogenetic relatedness makes a poor predictor of metabolic metal-transforming capability. Another unusual feature of this bacterium is the presence of structures at cell termini, resembling attachment structures or perhaps budding cells (2). This feature is not commonly observed among the bacteria and has not been reported for related genera of bacteria. It is suspected that these structures are not flagella or pili as they appear branched along the length of the structure, whereas flagella and pili tend to be single-stranded structures with an origin along the cell surface and with no branching along the length. Because these interesting structures only appear on cells grown with As(V) and are not seen on cells grown with yeast extract only, it is also speculated that they could be inorganic mineral remnants. Further work is proceeding to determine the origin, composition, and potential function of these structures.
The ability of YeAs to produce such a novel mineral has implications in several scientific fields, including nanotechnology (bionano materials), bioremediation, and exobiology. YeAs could be considered a solution for immobilizing arsenic in contaminated waters. The rapid and efficient precipitation of arsenic by YeAs is a noteworthy mechanism that has the potential for widespread use in these contaminated water systems. Additionally, β-realgar has the potential to be utilized as an indicator of life in both characterized and uncharacterized extreme environments. Because the abiotic formation of the high-temperature-phase mineral is thermodynamically unfavorable (6), YeAs arsenic metabolism is unique, and the mineral it forms could be a significant “life signature” in the search for life on Mars and other worlds.
Acknowledgments
We thank Amanda Weaver for critical review of the manuscript. We also thank the Alvord Ranch and the U.S. Bureau of Land Management for granting permissions to the field site.
This research was funded by a Research Infrastructure Improvement grant (award no. 0132626) from the National Science Foundation-Experimental Program to Stimulate Competitive Research (J. Shreeve, principal investigator, University of Idaho) and by NASA-EPSCoR. R.N.L. was supported by a NASA-Idaho Space Grant Consortium Fellowship. Our work was facilitated by the Molecular Research Core facility (E. O'Leary-Jepson and M. Andrews) and the Center for Biological Imaging (W. Siebold and C. Anderson).
Footnotes
Published ahead of print on 13 July 2007.
REFERENCES
- 1.Allen, R. D., G. B. David, and G. Nomarski. 1969. The Zeiss-Nomarski differential interference equipment for transmitted-light microscopy. Z. Wiss. Mikrosk. 69:193-221. [PubMed] [Google Scholar]
- 2.Angert, E. R. 2005. Alternatives to binary fission in bacteria. Nat. Rev. Microbiol. 3:214-224. [DOI] [PubMed] [Google Scholar]
- 3.Benning, L. G., and B. W. Mountain. 1996. Metal distribution in modern arsenic mineralization associated with a hot spring environment: Uzon Caldera, Kamchatka, Russia, p. 32. In Geochemistry of crustal fluids. ERC, Seefeld, Austria.
- 4.Bochner, B. R. 1989. “Breathprints” at the microbial level. ASM News 55:536-539. [Google Scholar]
- 5.Bochner, B. R. 1989. Sleuthing out bacterial identities. Nature 339:157-158. [DOI] [PubMed] [Google Scholar]
- 6.Bonazzi, P., S. Menchetti, G. Pratesi, M. Muniz-Miranda, and G. Sbrana. 1996. Light-induced variations in realgar and β-As4S4: X-ray diffraction and Raman studies. Am. Mineral. 81:874-880. [Google Scholar]
- 7.Buck, J. D. 1982. Nonstaining (KOH) method for determination of Gram reactions of marine bacteria. Appl. Environ. Microbiol. 44:992-993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Èernošek, Z., E. Èernošeková, and L. Beneš. 1999. Crystalline arsenic trisulfide: preparation, differential scanning calorimetry and Raman scattering measurements. Mat. Lett. 38:336-340. [Google Scholar]
- 9.Chrisostomos, S., B. K. C. Patel, P. P. Dwivedi, and S. E. Denman. 1996. Caloramator indicus sp. nov., a new thermophilic anaerobic bacterium isolated from the deep-seated nonvolcanically heated waters of an Indian artesian aquifer. Int. J. Syst. Bacteriol. 46:479-501. [DOI] [PubMed] [Google Scholar]
- 10.Dowdle, P. R., A. M. Laverman, and R. S. Oremland. 1996. Bacterial dissimilatory reduction of arsenic(V) to arsenic(III) in anoxic sediments. Appl. Environ. Microbiol. 62:1664-1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Engle, M., Y. Li, F. Rainey, S. DeBlois, V. Mai, A. Reichert, F. Mayer, P. Messner, and J. Wiegel. 1996. Thermobrachium celere gen. nov., sp. nov., a rapidly growing thermophilic, alkalitolerant, and proteolytic obligate anaerobe. Int. J. Syst. Bacteriol. 46:1025-1033. [DOI] [PubMed] [Google Scholar]
- 11a.Garrity, G. M., J. A. Bell, and T. G. Lilburn. 2004. Taxonomic outline of the prokaryotes. Bergey's manual of systematic bacteriology, release 5.0, 2nd ed. Springer, New York, NY. http://141.150.157.80/bergeysoutline/main.htm. Accessed 15 March 2007.
- 12.Gram, H. C. 1884. Über die isolierte Färbung der Schizomyceten in Schnitt- und Trockenpräparaten. Fortschr. Med. 2:185-189. [Google Scholar]
- 13.Hollibaugh, J. T., C. Budinoff, R. A. Hollibaugh, B. Ransom, and N. Bano. 2006. Sulfide oxidation coupled to arsenate reduction by a diverse microbial community in a soda lake. Appl. Environ. Microbiol. 72:2043-2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huber, R., H. Huber, and K. O. Stetter. 2000. Towards the ecology of hyperthermophiles: biotopes, new isolation strategies and novel metabolic properties. FEMS Microbiol. Rev. 24:615-623. [DOI] [PubMed] [Google Scholar]
- 15.Huber, R., M. Sacher, A. Vollmann, H. Huber, and D. Rose. 2000. Respiration of arsenate and selenate by hyperthermophilic archaea. Syst. Appl. Microbiol. 23:305-314. [DOI] [PubMed] [Google Scholar]
- 16.Kalampounias, A. G., D. T. Kastrissios, and S. N. Yannopoulos. 2003. Structure and vibrational modes of sulfur around the λ-transition and the glass-transition. J. Non-Crystalline Solids 326/327:115-119. [Google Scholar]
- 16a.Lane, D. J. 1991. 16S/23S rRNA sequencing, p. 115-147. In E. Stackebrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. John Wiley & Sons, New York, NY.
- 17.Lee, M. H., J. L. Keams, D. W. Helzer, O. P. Leiser, M. A. Ochoa, S. A. Connon, T. S. Magnuson, and M. E. Watwood. 2007. Evaluation of viral and prokaryotic community dynamics in Alvord Desert hot springs, Oregon. Aquat. Microb. Ecol. 48:19-26. [Google Scholar]
- 17a.Ludwig, W., O. Strunk, R. Westram, L. Richter, H. Meier, Yadhukumar, A. Buchner, T. Lai, S. Steppi, G. Jobb, W. Förster, I. Brettske, S. Gerber, A. W. Ginhart, O. Gross, S. Grumann, S. Hermann, R. Jost, A. König, T. Liss, R. Lüßmann, M. May, B. Nonhoff, B. Reichel, R. Strehlow, A. Stamatakis, N. Stuckmann, A. Vilbig, M. Lenke, T. Ludwig, A. Bode, and K.-H. Schleifer. 2004. ARB: a software environment for sequence data. Nucleic Acids Res. 32:1363-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Macur, R. E., C. R. Jackson, L. M. Botero, T. R. McDermott, and W. P. Inskeep. 2004. Bacterial populations associated with the oxidation and reduction of arsenic in an unsaturated soil. Environ. Sci. Technol. 38:104-111. [DOI] [PubMed] [Google Scholar]
- 19.Mesbah, M., U. Premachandran, and W. B. Whitman. 1989. Precise measurement of the G+C content of deoxyribonucleic acid by high-performance liquid chromatography. Int. J. Syst. Bacteriol. 39:159-167. [Google Scholar]
- 20.Messens, J., and S. Silver. 2006. Arsenate reduction: thiol cascade chemistry with convergent evolution. J. Mol. Biol. 362:1-17. [DOI] [PubMed] [Google Scholar]
- 21.Miller, T. L., and M. J. Wolin. 1974. A serum bottle modification of the Hungate technique for cultivating obligate anaerobes. Appl. Microbiol. 27:985-987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mukhopadhyay, R., and B. P. Rosen. 2002. Arsenate reductases in prokaryotes and eukaryotes. Environ. Health Perspect. 110(Suppl. 5):745-748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mukhopadhyay, R., J. Shi, and B. P. Rosen. 2000. Purification and characterization of ACR2p, the Saccharomyces cerevisiae arsenate reductase. J. Biol. Chem. 275:21149-21157. [DOI] [PubMed] [Google Scholar]
- 24.Muniz-Miranda, M., G. Sbrana, P. Bonazzi, S. Menchetti, and G. Pratesi. 1996. Spectroscopic investigation and normal mode analysis of As4S4 polymorphs. Spectrochim. Acta A 52:1391-1401. [Google Scholar]
- 25.Newman, D. K., E. K. Kennedy, J. D. Coates, D. Ahmann, D. J. Ellis, D. R. Lovley, and F. M. Morel. 1997. Dissimilatory arsenate and sulfate reduction in Desulfotomaculum auripigmentum sp. nov. Arch. Microbiol. 168:380-388. [DOI] [PubMed] [Google Scholar]
- 26.Niggemyer, A., S. Spring, E. Stackebrandt, and R. F. Rosenzweig. 2001. Isolation and characterization of a novel As(V)-reducing bacterium: implications for arsenic mobilization and the genus Desulfitobacterium. Appl. Environ. Microbiol. 67:5568-5580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oremland, R. S., and J. F. Stolz. 2005. Arsenic, microbes and contaminated aquifers. Trends Microbiol. 13:45-49. [DOI] [PubMed] [Google Scholar]
- 28.Patel, B. C. K., C. Monk, H. Littleworth, H. W. Morgan, and R. M. Daniel. 1987. Clostridium fervidus sp. nov., a new chemoorganotrophic acetogenic thermophile. Int. J. Syst. Bacteriol. 37:123-126. [Google Scholar]
- 29.Plugge, C. M., E. G. Zoetendal, and A. J. Stams. 2000. Caloramator coolhaasii sp. nov., a glutamate-degrading, moderately thermophilic anaerobe. Int. J. Syst. Evol. Microbiol. 50:1155-1162. [DOI] [PubMed] [Google Scholar]
- 30.Schaeffer, A. B., and M. D. Fulton. 1933. A simplified method of staining endospores. Science 77:194. [DOI] [PubMed] [Google Scholar]
- 31.Seyfried, M., D. Lyon, F. A. Rainey, and J. Wiegel. 2002. Caloramator viterbensis sp. nov., a novel thermophilic, glycerol-fermenting bacterium isolated from a hot spring in Italy. Int. J. Syst. Evol. Microbiol. 52:1177-1184. [DOI] [PubMed] [Google Scholar]
- 32.Silver, S., and L. T. Phung. 2005. Genes and enzymes involved in bacterial oxidation and reduction of inorganic arsenic. Appl. Environ. Microbiol. 71:599-608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stolz, J. F., P. Basu, and R. S. Oremland. 2002. Microbial transformation of elements: the case of arsenic and selenium. Int. Microbiol. 5:201-207. [DOI] [PubMed] [Google Scholar]
- 34.Stolz, J. F., P. Basu, J. M. Santini, and R. S. Oremland. 2006. Arsenic and selenium in microbial metabolism. Annu. Rev. Microbiol. 60:107-130. [DOI] [PubMed] [Google Scholar]
- 34a.Swofford, D. L. 2002. PAUP*: Phylogenetic Analysis Using Parsimony (*and other methods), 4.0, 4.0b10 ed. Sinauer Associates, Sunderland, MA.
- 35.Tarlera, S., L. Muxi, M. Soubes, and A. J. Stams. 1997. Caloramator proteoclasticus sp. nov., a new moderately thermophilic anaerobic proteolytic bacterium. Int. J. Syst. Bacteriol. 47:651-656. [DOI] [PubMed] [Google Scholar]
- 36.Vallejos, C. E. 1983. Enzyme activity staining. p. 469. In S. D. Tanskley and T. J. Orton (ed.), Isozymes in plant genetics and breeding, part A. Elsevier, Amsterdam, The Netherlands.
- 37.Wolin, E. A., M. J. Wolin, and R. S. Wolfe. 1963. Formation of methane by bacterial extracts. J. Biol. Chem. 238:2882-2886. [PubMed] [Google Scholar]