Abstract
The genetic analysis of essential genes has been generally restricted to the use of conditional mutations, or inactivating chromosomal mutations, which require a complementing plasmid that must either be counterselected or lost to measure a phenotype. These approaches are limited because they do not permit the analysis of mutations suspected to affect a specific function of a protein, nor do they take advantage of the increasing abundance of structural and bioinformatics data for proteins. Using the dnaC gene as an example, we developed a genetic method that should permit the mutational analysis of other essential genes of Escherichia coli and related enterobacteria. The method consists of using a strain carrying a large deletion of the dnaC gene, which is complemented by a wild-type copy expressed from a plasmid that requires isopropyl-β-d-thiogalactopyranoside for maintenance. Under conditions in which this resident plasmid is lost, the method measures the function of a dnaC mutation encoded by a second plasmid. This methodology should be widely applicable to the genetic analysis of other essential genes.
The genetic study of cellular pathways that are essential for viability has utilized conditionally defective alleles, or chromosomally encoded null mutations that are complemented by a functional copy carried in a plasmid. These approaches can demonstrate the requirement of a specific gene for viability, but the penetrance of a mutation can vary depending on the mutation's effect on the activity of the respective gene product. For example, conditionally defective alleles often give rise to misfolded or partially unstable proteins, which retain limited function to cause a weak phenotype. The accumulation of extragenic suppressors may also affect the phenotype. The alternate approach of studying a phenotype caused by a large deletion relies on a complementing plasmid encoding the gene of interest. If the gene is expressed from a regulatable promoter, the plasmid may be inadequate if the level of expression under noninduced or repressed conditions continues to maintain viability. Mutants carrying complementing plasmids that can be counterselected or that are temperature sensitive for maintenance are useful but limited because the approach cannot address the importance of an individual activity of a multifunctional protein.
For essential proteins with several biochemical functions, site-directed mutagenesis followed by the in vitro characterization of the corresponding mutant protein can reveal the importance of a protein's individual activities if either structural information for a protein is available or conserved amino acids have been identified. However, a genetic method to correlate the biochemical defect to a phenotype is generally not available.
Almost all DNA replication genes are essential, including dnaC, which acts during the initiation of DNA replication (reviewed in reference 16) and the reassembly of collapsed replication forks (7, 19). Like DnaA and subunits of the DnaX or clamp loader complex, as well as their functional counterparts in eukaryotic cells, DnaC is a member of the AAA+ family of ATPases (reviewed in references 10 and 15). Koonin noted a significant similarity in amino acid sequence between Escherichia coli DnaA and DnaC and suggested that they share a common ancestry (17). As DnaC functions at a different step in initiation than DnaA, it appears that each protein has evolved to perform separate functions. However, in contrast to the measurable activities of DnaA protein during initiation, DnaC action at oriC requires the formation of the DnaB-DnaC complex, in which six DnaC monomers interact on one face of the DnaB hexamer (2, 24). Interestingly, despite the strict requirement for the DnaB-DnaC complex in the entry of DnaB at oriC, the complex is not catalytically active (9, 28, 29). To reveal the helicase and ATPase activities of DnaB, DnaC must first dissociate from DnaB in a process requiring the hydrolysis of ATP bound by DnaC. By itself, DnaC binds, albeit weakly, to ATP (Kd [dissociation constant] of 8 μM) (3, 9, 13, 28). Its interaction with single-stranded DNA (3, 9, 18) suggests that DnaC binds to the unwound region of oriC opened by DnaA, which leads to the binding of DnaB. Inasmuch as DnaB's single-stranded DNA binding activity may suffice, this activity of DnaC may not be involved.
Because of our interest in the role of DnaC protein during initiation in bacteria, we sought to obtain novel mutations in the dnaC gene in order to gain insight into DnaC function from the biochemical study of corresponding mutant proteins. In this report, we describe a genetic method for identifying inactivating missense mutations in dnaC. The approach can be used for more-extensive mutational analysis of dnaC and for any other essential gene of E. coli or another closely related bacterium. Using the dnaC gene as an example, the method relies on complementing a null dnaC mutant with a dnaC-carrying plasmid whose maintenance depends on isopropyl-β-d-thiogalactopyranoside (IPTG) in the culture medium. With a separate plasmid that does not depend on IPTG for its maintenance, the host strain cannot survive without IPTG if this plasmid encodes a nonfunctional dnaC allele but remains viable if it carries an active dnaC gene.
MATERIALS AND METHODS
Bacteriological methods, strains, and plasmids.
Bacteriological methods were performed essentially as described previously (22). Bacterial transformation was done either by the calcium chloride method or by electroporation using a Bio-Rad Gene Pulser according to the manufacturer's recommendations. Bacterial strains and their plasmid-containing derivatives (Table 1) were routinely grown in Luria-Bertani (LB) medium supplemented with ampicillin (100 μg/ml), kanamycin (40 μg/ml), tetracycline (10 μg/ml), and/or chloramphenicol (35 μg/ml) as appropriate. To construct one set of dnaC plasmids, the fragment carrying the dnaC gene from pdnaC113 (20) was isolated after cleavage with BamHI nuclease and ligated into the BamHI cleavage site of pAM34 to construct pAM34dnaCL1-4 and pAM34dnaCL1-2, which, respectively, contain the inserted DNAs in the clockwise and counterclockwise orientations relative to the physical map of pBR322. Because the orientation of the dnaC gene in pAM34 did not affect complementation of the ΔdnaC::cat mutant (see Results), pAM34dnaCL1-2 was used in the experiments described unless otherwise stated and is noted as pAM34dnaC for simplicity. A derivative of pBR322, pAM34 initiates DNA replication by extension of the primer transcript from the lac promoter, which replaces the natural primer promoter of pBR322 (14). Maintenance of pAM34 requires IPTG in the culture medium. To construct pACYC184dnaC, the dnaC gene carried in pINCSSD (1) was PCR amplified with oligonucleotide primers AATATTTGCCCATGGATACCGCCAGAAAC and AATGCTCATCCGGAATTCTGTGCCATAAGC; cleavage sites for NcoI and EcoRI endonuclease are underlined. The amplified DNA was digested with the respective endonucleases, and the DNA was ligated into the EcoRI and NcoI sites of pACYC184, disrupting the chloramphenicol resistance gene. DNA sequence analysis of the dnaC gene confirmed the presence of the wild-type allele in the plasmids constructed (Table 1).
TABLE 1.
E. coli K12 strains and plasmids
| Strain or plasmid | Genotype and/or properties (reference); source |
|---|---|
| Strains | |
| PC2 | leuB6 thyA47 dnaC2 dnaT12 deoC3 rpsL153 λ− (5); laboratory stock |
| SS1020 | λ−rph-1 zjj202::Tn10 dnaC2; Steven Sandler |
| HME5 | F− λ− IN (rrnD-rrnE) ΔlacU169 λcI857 Δ(cro-bioA) (32); Donald Court |
| MC1061 | araD139 Δ(ara, leu)7697 ΔlacX74 galU galK rpsL hsdR2 (rK− mK+) mcrB1; laboratory stock |
| KH1061 | araD139 Δ(ara, leu)7697 ΔlacX74 galU galK rpsL hsdR2 (rK− mK+) mcrB1 ΔdnaC::cat; this work |
| MF1061 | araD139 Δ(ara, leu)7697 ΔlacX74 galU galK rpsL hsdR2 (rK− mK+) mcrB1 ΔdnaC::cat recA::Kanr; Magdalena M. Felczak of this laboratory by P1 transduction from BW26355 (relevant genotype, ΔrecA635::Kanr) (8) into MC1061, followed by selection for kanamycin resistance; the transductant and BW26355 were confirmed to be UV sensitive |
| Plasmids | |
| pACYC184 | cat, Tetr (6) |
| pACYC184dnaC | Tetr, dnaC (this work) |
| pAM34 | Ampr, IPTG-dependent maintenance (32); Kaymeuang Cam |
| pAM34dnaCL1-2 | Ampr, IPTG-dependent maintenance, dnaC (this work) |
| pAM34dnaCL1-4 | Ampr, IPTG-dependent maintenance, dnaC (this work) |
| pINCSSD | Kanr, pBR322 derivative, dnaC under araBAD promoter control and downstream from the ribosome binding site for gene 10 of T7 phage (1) |
| pdnaC113 | Ampr, pBR322 derivative, dnaC under araBAD promoter control (20) |
| pdnaC97 (Pro108Ser) | Kanr, pINCSSD derivative, missense mutation encoding Pro108Ser substitution (20) |
| pdnaC116 (Leu44Pro) | Kanr, pINCSSD derivative, missense mutation encoding Leu44Pro substitution (20) |
| pdnaC250 (Leu11Gln) | Kanr, pINCSSD derivative, missense mutation encoding Leu11Gln substitution (20) |
| pHK-dnaC | Ampr, pET11a derivative (Novagen), dnaC joined at the 5′ end to sequence encoding protein kinase A phosphorylation site and six histidine residues (9); Mike O'Donnell |
| pHK-dnaCK112R | Ampr, derivative of pHK-dnaC encoding a Lys112Arg substitution (9); Mike O'Donnell |
Construction of ΔdnaC::cat mutants.
A null dnaC mutant was constructed using the E. coli λ Red targeted-mutagenesis system (8, 32). Briefly, a chloramphenicol resistance cassette was PCR amplified from pACYC184 DNA with oligonucleotide primers (GACAGCCAAATTCCACCAGGATTCAGAGGGTAACGAGCGGCTATTTAACGACCCTG and CATCATCATTACTCAAGGTGGAATTGTGTCGCAGTATACCTGTGACGGAAGATCAC) that are complementary to the dnaC coding region, as indicated by underlined sequences, and the gene encoding chloramphenicol acetyltransferase (cat) carried in pACYC184. Following transformation with 50 ng of the PCR product into E. coli HME5 carrying the dnaC plasmid, pINCSSD, or without a plasmid as a negative control, selection for chloramphenicol-resistant colonies led to the substitution of the dnaC gene with the cat gene by homologous recombination. The mutation was then transduced via bacteriophage P1 into MC1061 carrying pdnaC113. This strain was transformed by pAM34dnaC, followed by growth in medium lacking ampicillin but supplemented with 0.5 mM IPTG to obtain the strain carrying only the latter plasmid. In these strains, the presence of the chromosomal ΔdnaC::cat mutation was confirmed by PCR analysis with 25 pmol of oligonucleotide primers (AGCCAAATTCCACCAGGATTC and CATCATCATTACTCAAGGTGG) that are complementary to yjjA and dnaT bordering the chromosomal dnaC gene and 25 ng of bacterial DNA, which was isolated using a DNeasy tissue kit (QIAGEN). To construct the asnA::cat strain, oligonucleotides AGCGGGCGATAGCGAAAG and CGGTCAGCTTAAACGTGG, which are complementary to asnA, were used to amplify a DNA fragment from an oriC minichromosome carrying an insertion of the cat gene in asnA (25). After transforming the amplified DNA into HME5, the ΔasnA::cat mutation was verified by PCR analysis with 25 ng of bacterial DNA and 25 pmol of the oligonucleotide primers as described above.
Mutagenesis and screening.
Mutagenesis of the dnaC gene was carried out by error-prone PCR amplification with Taq DNA polymerase (Promega), pINCSSD as the DNA template, and the primers used for the construction of pACYC184dnaC. The reaction conditions were essentially as described by the manufacturer, but assays were supplemented with 0.1 to 0.5 mM Mn2+. The amplified DNA was electrophoresed on an agarose gel, purified using a QIAGEN gel extraction kit, and then digested with EcoRI and NcoI endonuclease. After gel purification, the DNA carrying the mutagenized dnaC gene was ligated into the EcoRI and NcoI endonuclease sites of pACYC184 with T4 DNA ligase (New England Biolabs). The ligation mixture was then transformed into KH1061 (ΔdnaC::cat) carrying pAM34dnaC and plated on LB medium supplemented with 0.5 mM IPTG, 10 μg/ml tetracycline, and 100 μg/ml ampicillin. Transformants obtained after overnight incubation at 37°C were picked by hand or with a GeneMachine Mantis colony picker and transferred to microtiter plates containing the medium described above. In the absence of IPTG to maintain pAM34dnaC, cells carrying pACYC184, which carries an inactivating dnaC mutation, will fail to grow. To identify such transformants, the cultures after overnight growth at 37°C were diluted with LB medium about 104-fold into microtiter plates, and about 2.0 μl was transferred with a pronged replica plater onto the antibiotic-supplemented medium with or without IPTG. From transformants that showed a growth dependence for IPTG, plasmid DNA was isolated by using an Autogen 850 DNA purification robot, followed by DNA sequence analysis with a high-throughput Applied Biosystems 3730XL genetic analyzer and oligonucleotide primers (AATGCTCATCCGGAATTCTGTGCCATAAGC and ATCACAGACGGCATGATGAAC) that are complementary to pACYC184 and border the dnaC gene in pACYC184dnaC.
RESULTS
An efficient genetic screen to isolate mutations in dnaC, an essential gene of E. coli.
To summarize our general approach, the strategy was to construct a null dnaC mutant whose deficiency is complemented by a dnaC plasmid that is conditionally maintained. If we introduce a separate plasmid carrying a mutagenized dnaC gene into the null mutant, the strain should remain viable after the loss of the resident plasmid if the incoming plasmid encodes a functional dnaC gene. If the incoming plasmid bears a nonfunctional gene, the strain cannot survive when it lacks the resident plasmid, thus providing a method to identify inactivating dnaC mutations. Our long-term objective is to study mutant DnaC proteins at the biochemical level to gain new insight into the role of DnaC during initiation or replication fork restart.
Construction of a null dnaC mutant.
A plasmid that encodes the wild-type dnaC gene should sustain a null dnaC mutant. As a control experiment to identify suitable plasmids, we measured the complementation of dnaC2(Ts) mutants at a nonpermissive temperature by dnaC plasmids named pdnaC113 and pINCSSD. Both DNAs carry dnaC downstream from the araBAD promoter and were examined under noninduced conditions, but pINCSSD also bears the ribosome binding site of T7 bacteriophage gene 10 ahead of the dnaC gene (Table 1). These plasmids, as well as pAM34dnaC, are derivatives of pBR322. However, maintenance of pAM34dnaC requires IPTG in the culture medium for reasons described in detail below. We also constructed a derivative of pACYC184 encoding the dnaC gene, named pACYC184dnaC. As shown in Table 2, all of the dnaC plasmids complemented the temperature sensitivity of the dnaC2 mutants, although the basal level of dnaC expression by plasmids that carry dnaC downstream from the araBAD promoter was only partially effective.
TABLE 2.
Plasmids carrying dnaC complement the temperature-sensitive phenotype of dnaC mutants
| Strain | Plasmid | Relative frequency of colony formationa |
|---|---|---|
| SS1020 (dnaC2) | None | <10−8 |
| SS1020 (dnaC2) | pBR322 | 1.2 × 10−7 |
| SS1020 (dnaC2) | pdnaC113 | 7 × 10−2 |
| SS1020 (dnaC2) | pINCSSD | 2 × 10−2 |
| PC2 (dnaC2) | None | <10−8 |
| PC2 (dnaC2) | pACYC184dnaC | ∼1 |
| SS1020 (dnaC2) | None | <10−8 |
| SS1020 (dnaC2) | pAM34 | <10−8 |
| SS1020 (dnaC2) | pAM34dnaCL1-2 | ∼1b |
| SS1020 (dnaC2) | pAM34dnaCL1-4 | ∼1b |
Cultures of E. coli PC2 (dnaC2) or SS1020 (dnaC2) carrying the indicated plasmids were grown at 30°C in LB media supplemented with 10 μg/ml tetracycline, 100 μg/ml ampicillin or 50 μg/ml kanamycin to select for the respective plasmid. After serial dilution, the samples were plated on antibiotic-supplemented LB media followed by incubation at 30°C or 42°C for about 14 h. Transformants carrying pAM34 and its derivatives were plated on medium with or without 0.5 mM IPTG. The relative frequency of colony formation is the ratio of the number of colonies detected at 42°C to that at 30°C.
Colony formation, which was observed at 42°C for the transformants carrying pAM34dnaCL1-2 and pAM34dnaCL1-4, required IPTG in the culture medium.
We then constructed a null dnaC strain by replacing the dnaC coding region with a chloramphenicol resistance cassette via the λ recombination system (8, 32) (see Materials and Methods). In the construction, we tested if we could construct the dnaC mutant with a strain carrying the dnaC plasmid, pINCSSD, despite its incomplete complementation of the dnaC2(Ts) mutants (Table 2), because we plan to examine later if a limiting level of DnaC compared to the natural level can exacerbate the bias in the loading of DnaB that moves leftward from oriC relative to DnaB that moves to the right (4). We obtained chloramphenicol-resistant colonies only when the strain (HME5) carried the dnaC plasmid, suggesting the substitution of the chromosomal dnaC locus with the chloramphenicol resistance cassette. As a control, we obtained antibiotic-resistant recombinants when we used a chloramphenicol resistance cassette that should replace the chromosomal asnA gene regardless of the presence of the dnaC plasmid. The asnA gene is not essential for growth in rich medium. Although we obtained fivefold more recombinants of ΔasnA::cat than ΔdnaC::cat for the plasmid-bearing strain, we do not know if variability in the efficiency of electroporation or another factor, such as a limiting level of DnaC, causes the difference.
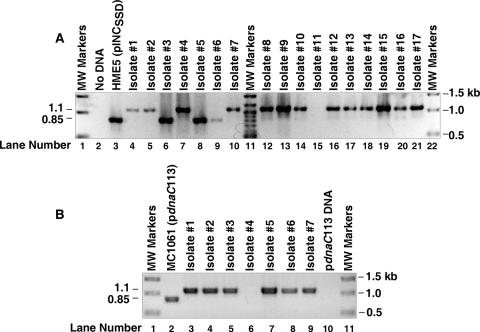
To obtain direct evidence that we were able to replace the chromosomal dnaC gene with the chloramphenicol resistance cassette, we isolated bacterial DNA from 17 randomly chosen recombinants of HME5 that also carried the dnaC plasmid, pINCSSD, as described in Materials and Methods. We then performed PCR analysis using oligonucleotide primers that are complementary to sequences that flank the chromosomal dnaC locus (Fig. 1A) and are known not to anneal to any of the dnaC plasmids described in this work (data not shown; see below). If the dnaC gene remains intact, PCR amplification should produce a DNA fragment of about 0.85 kb, which we observed with bacterial DNA isolated from HME5 carrying pINCSSD (Fig. 1A, lane 3). In comparison, the failure to detect a PCR product for the reaction lacking a bacterial DNA template (lane 2) shows that PCR amplification of any DNA depends on the addition of a DNA template. If the recombinants contain the ΔdnaC::cat mutation, the PCR product should be about 1.1 kb in size. Of the isolates analyzed, the expected DNA fragment was observed in 13 recombinants. PCR analysis of three isolates revealed a DNA fragment of about 0.85 kb (lanes 6, 8, and 9), indicating that the chloramphenicol resistance cassette had not replaced the chromosomal dnaC gene and may have inserted elsewhere in the chromosome. No PCR product was detected in one isolate (lane 15), suggesting that PCR amplification failed. Because no other PCR products except those described above were detected in any of the reactions, the results confirm our expectation that the oligonucleotide primers are specific for the chromosomal dnaC locus. Hence, Fig. 1 shows only the relevant portion of the gel. Analogous PCR analysis confirmed the construction of the ΔasnA::cat strain (data not shown; see Materials and Methods).
FIG. 1.
Construction of a ΔdnaC::cat mutant. E. coli HME5 carrying the dnaC plasmid, pINCSSD, was transformed as described in Materials and Methods with a DNA fragment encoding the cat gene, which is flanked by DNA that is homologous to the 5′ and 3′ ends of the dnaC gene. PCR analysis of genomic DNA isolated from individual chloramphenicol-resistant colonies was performed with oligonucleotide primers that are complementary to sequences in yjjA and dnaT that border the chromosomal dnaC locus. In panel A, the lanes at the extreme left and right contain DNA fragments whose sizes are indicated at the right of the figure. Lane 11 contains DNA fragments of 1.5, 1.2, 1.0, 0.9, 0.8, 0.7, 0.6, and 0.5 kb. A PCR lacking bacterial DNA corresponds to lane 2. Lanes 3 to 10 and 12 to 21 contain bacterial DNA from HME5 carrying pINCSSD or from the indicated chloramphenicol-resistant recombinants that also contain the dnaC plasmid. The estimated sizes of the amplified DNA are indicated at the left of the figure. In panel B, the ΔdnaC::cat mutation was transferred into E. coli MC1061(pdnaC113) by P1 transduction, using isolate 4 in panel A as the donor strain, followed by selection for chloramphenicol-resistant transductants. PCR analysis was performed as described for panel A either with bacterial DNA isolated from seven independent transductants, with bacterial DNA isolated from MC1061 carrying pdnaC113 (lane 2), or with pdnaC113 DNA (25 ng; lane 10). Lanes 1 and 10 contain DNA fragments whose sizes are indicated at the right of the figure. The estimated sizes of the amplified DNA are noted at the left.
We have moved the ΔdnaC::cat mutation into various strains by P1 transduction with reasonable transduction frequencies but only when a dnaC plasmid was present. As a representative example, we constructed the strain named KH1061 by transferring the ΔdnaC::cat mutation by P1 transduction from isolate 4 of Fig. 1A into E. coli MC1061 carrying pdnaC113. As a control, we used the strain lacking the dnaC plasmid and obtained chloramphenicol-resistant transductants only when the strain carried the dnaC plasmid. When we analyzed the bacterial DNA from seven randomly chosen transductants, we found that six contained the ΔdnaC::cat mutation, as indicated by the presence of a PCR product of about 1.1 kb (Fig. 1B, lanes 3 to 5 and 7 to 9). We did not detect a PCR product in lane 6; the reason for this negative result was not determined. By comparison, we observed a 0.85-kb DNA when we analyzed the genomic DNA isolated from MC1061 carrying pdnaC113 (lane 2) to correlate with the strain's dnaC+ genotype. No amplified DNA was detected in the reaction containing the dnaC plasmid as the template for PCR analysis (lane 10), indicating that the oligonucleotide primers are specific for the chromosomal dnaC locus.
Isolation of novel dnaC mutations.
To obtain mutations in dnaC, we used a derivative of pBR322 that requires IPTG for plasmid DNA replication. To explain the role of this inducer of the lac operon for plasmid maintenance, we first summarize the mechanism of pBR322 replication. After synthesis of the pBR322 primer transcript (RNA II), RNase H processes RNA II in the absence of a small antisense RNA (RNA I). DNA polymerase I then extends the primer transcript during the initial steps of plasmid DNA replication. Gil and Bouché have described pAM34, a pBR322 derivative that has its primer transcript regulated by the lac promoter region. The plasmid also encodes lacIQ, which causes repression of the lac promoter in the absence of IPTG (14). Repression of the primer transcript leads to rapid loss of the plasmid such that only 0.1% of cells in culture contained the plasmid after 14 generations (less than an overnight incubation).
To confirm the properties of pAM34, we observed that the plasmid carrying the dnaC+ gene (pAM34dnaC) complemented the ΔdnaC strain (KH1061) also bearing pACYC184 but only when the growth medium contained IPTG (Table 3). The transformation frequency was more than 105-fold lower in the absence of IPTG than in its presence. Thus, viability of the ΔdnaC strain depended on IPTG. With the strain originally bearing pAM34dnaC, we showed that it remained viable on medium lacking IPTG after selection for antibiotic-resistant transformants carrying pACYC184 encoding dnaC+ but without selection for antibiotic resistance conferred by the resident pAM34dnaC plasmid. Apparently, the incoming plasmid complements the ΔdnaC strain when the resident plasmid is lost.
TABLE 3.
In absence of IPTG required to maintain the resident dnaC plasmid, pAM34dnaC, viability of E. coli KH1061 (ΔdnaC::cat) depends on separate dnaC plasmid
| Plasmid | Relative plating efficiencya |
|---|---|
| pACYC184 | <2 × 10−6 |
| pACYC184dnaC | 0.95 |
| pINCSSD | 2.4 × 10−2 |
| pdnaC97 (Pro108Ser) | 5 × 10−2 |
| pdnaC116 (Leu44Pro) | 1.5 × 10−2 |
| pdnaC250 (Leu11Gln) | 2.3 × 10−2 |
The indicated plasmids were transformed into KH1061 (ΔdnaC::cat) carrying the plasmid pAM34dnaC. Various dilutions of the transformation mixture were plated on LB medium lacking or supplemented with 0.5 mM IPTG and either tetracycline or kanamycin to select for the respective plasmids. Ampicillin was included in the medium containing IPTG to select for colonies carrying pAM34dnaC. Incubation was carried out at 37°C overnight. Relative plating efficiency is the ratio of the number of colonies detected in the absence of IPTG to that in its presence.
Interestingly, under the conditions that fail to maintain the resident pAM34dnaC plasmid, derivatives of pINCSSD (pdnaC97, pdnaC116, and pdnaC250; Table 1) encoding mutant DnaCs with defects in binding to ATP or forming a complex with DnaB (20) complemented the ΔdnaC strain as effectively as the wild-type dnaC plasmid (pINCSSD) (Table 3). These observations suggest that the primer transcript of these plasmids can act in trans by annealing to the homologous region of pAM34dnaC to prime plasmid DNA replication. Because of these results, we chose pACYC184 as the incoming plasmid to carry the DNA fragment encoding dnaC. To introduce mutations in the dnaC gene, we PCR amplified the DNA under error-prone conditions. Transformants obtained on medium supplemented with IPTG and antibiotics for both resident (pAM34dnaC) and incoming (pACYC184dnaC) plasmids were picked manually or with a robotic instrument and grown in microtiter plates. We then screened the isolates on solid medium supplemented with or lacking 0.5 mM IPTG, as described in Materials and Methods, but with selection for antibiotic resistance conferred by pACYC184 to identify those that either did not grow or grew poorly. At this step, we diluted the cultures 104-fold. One reason for the dilution is that we had found that the frequency of colony formation of KH1061 (ΔdnaC::cat) carrying pAM34dnaC was reduced by only about twofold at 0.05 mM IPTG and by (1 to 2) × 102-fold at 0.005 mM IPTG. Thus, an undiluted culture or a lesser dilution contains an amount of IPTG that is almost sufficient to maintain the resident pAM34dnaC plasmid. A second reason for diluting the cultures was to increase the sensitivity of the assay. By reducing the number of viable bacterial cells of each culture ([0.5 to 1] × 109/ml) to approximately 100 to 200 per screened sample of about 2 μl, we should be able to identify dnaC mutations that are partially active.
Characterization of the dnaC mutations by DNA sequence analysis and by relative plating efficiencies of plasmid-bearing strains.
DNA sequence analysis of the pACYC184 derivatives from isolates identified in the genetic screen was then performed (Table 4). The results confirm the presence of dnaC mutations, substantiating the genetic method.
TABLE 4.
Novel dnaC mutations and their phenotypes
| Mutant plasmid | Nucleotide substitution(s) | Amino acid substitution(s) | Relative plating efficiencya
|
|
|---|---|---|---|---|
| KH1061 (recA+ ΔdnaC::cat) | MF1061 (recA::Kanr ΔdnaC::cat | |||
| pACYC184dnaC | None | None | 1.2 | 1.3 |
| pACYC184 | Not relevant | Not relevant | 1.8 × 10−3 | 1.2 × 10−5 |
| 31-f3 | G289A, T318A (silent) | Gly97Ser | 1.0 × 10−3 | 2.6 × 10−5 |
| 157-c2 | A428G | Lys143Arg | 1.4 × 10−3 | 1.3 × 10−5 |
| 36-c4 | G289A | Gly97Ser | 2.1 × 10−4 | 1.4 × 10−5 |
| 29-d3 | A534G (silent), G716T | Arg239Leu | 1.6 × 10−4 | 2.6 × 10−5 |
| 55-c6 | T425C, A521G | Met142Thr, Gln174Arg | 7.7 × 10−5 | 9.3 × 10−6 |
| 97-h9 | T129C (silent), A218G | Asn73Ser | 2.0 × 10−4 | 2.0 × 10−5 |
| 107-g12 | A40G, T178C | Met14Val, Ser60Pro | 3.3 × 10−4 | 2.8 × 10−5 |
| 125-c1 | G359A | Cys120Tyr | 2.1 × 10−4 | 7.9 × 10−6 |
| 145-f1 | T89C, A237G (silent), A394T, G565A | Leu30Pro, Ile132Phe, Asp189Asn | 4.1 × 10−4 | 2.0 × 10−5 |
| 151-a2 | T86C, A248G | Leu29Pro, Asn83Ser | 2.2 × 10−4 | 1.7 × 10−5 |
| 239-e10 | T178A, T213C (silent), A336G (silent), C504A (silent), A632G | Ser60Thr, Lys211Arg | 3.1 × 10−4 | 1.2 × 10−5 |
| 164-g2 | G82A, T207C (silent), A676T | Glu28Lys, Ser226Cys | 6.0 × 10−4 | 1.9 × 10−5 |
| 40-g4 | A71G, A107G, T189C (silent), T254A, A354T (silent), A400G, C483T (silent) | Lys24Arg, Gln36Arg, Leu85Stop, Thr134Ala | 1.6 × 10−5 | 1.1 × 10−5 |
| 4-g1 | A712T | Ser238Cys | 1.2b | 1.2b |
| 38-e4 | T68C, T207C (silent) | Phe23Ser | 1.7b | 1.2b |
| 54-b6 | A84T, T706A | Glu28Val, Tyr236Asn | 1.0b | 0.7b |
| 234-h9 | T358C, T686A | Cys120Arg, Val229Glu | 1.4b | 0.7b |
| 35-b4 | T650A | Val217Glu | 6.1 × 10−6 | 1.8 × 10−5 |
| 246 | T131C | Leu44Pro | 4.6 × 10−6 | 2.3 × 10−5 |
| 256-f12 | A451C, G659A | Thr151Pro, Arg220His | 1.0 × 10−5 | 2.1 × 10−5 |
Cultures of the indicated strains carrying pAM34dnaC and the pACYC184 derivative encoding the respective dnaC mutation(s) were serially diluted and plated as described in footnote a of Table 3, but the plating medium was additionally supplemented with chloramphenicol. Relative plating efficiency is defined in footnote a of Table 3.
On medium lacking IPTG, the colonies were either heterogeneous in size or uniformly smaller (for Phe23Ser).
To characterize the plasmid-borne mutations genetically, we measured the relative plating efficiencies of strains initially carrying both pAM34dnaC and the respective pACYC184 derivatives on media with and without IPTG (Table 4). The relative plating efficiency is the ratio of the number of colonies observed on medium lacking IPTG to the number of colonies observed on IPTG-supplemented media. This experiment utilizing strains with coresident plasmids differs from that for Table 3, which measures the relative plating efficiency after transformation to reflect the establishment of the incoming plasmid. We also compared isogenic recA+ and recA::Kanr strains to determine the effect of homologous recombination on the genetic method. Via DNA recombination, substitution of the mutant sequence carried in the multicopy pACYC184 derivative by the wild-type dnaC sequence from pAM34dnaC produces a mixed population of the pACYC184 derivative. If the mutation is recessive and insensitive to gene dosage, the bacterial cell encoding both the wild-type gene and the dnaC mutation should be phenotypically dnaC+ in the absence of IPTG to select against pAM34dnaC. Alternatively, because DnaC function requires the assembly of the DnaB-DnaC complex with a stoichiometry of six DnaC monomers per DnaB hexamer, the formation of an active DnaB-DnaC complex should depend on the relative ratio of wild-type to mutant plasmid encoding a recessive mutation. Either circumstance may lead to a relative increase in colony formation depending on the frequency of DNA recombination with the recA+ strain compared with the recA mutant when the pAM34dnaC plasmid has been lost. As for the behavior of dominant-negative mutations, we describe them below and in the Discussion.
With pACYCdnaC as a control, we observed essentially no difference in the relative plating efficiencies between the recA+ and recA::Kanr strains (Table 4). With pACYC184, the ratios of 1.8 × 10−3 and 1.2 × 10−5 for the recA+ strain and the recA mutant, respectively, confirm the requirement of the IPTG-dependent plasmid for viability, but we were expecting comparable ratios for the two strains. We observed ratios within a twofold range (the range of error for the assay) with a separate isolate of the recA mutant originally bearing pACYC184 and pAM34dnaC, and with two other independent isolates of the recA+ strain carrying pACYC184 and the dnaC plasmid, indicating that the results are reproducible (data not shown). For the recA+ strain, we presume that the difference in the relative plating efficiency compared with the ratio given in Table 3 relates to the different methods. To attempt to explain the results with pACYC184, we considered the possibility that a single crossover event between pACYC184 and pAM34dnaC joins the two plasmids. Based on comparative DNA sequence analysis of pACYC184 and pAM34dnaC, the plasmids share short regions of homology in the replication origins and a longer region of 219 bp that corresponds to a DNA sequence downstream from the tetracycline resistance gene in pACYC184 (23, 30). The sequence remained in pAM34dnaC after the tetracycline resistance gene in the parental plasmid was replaced with a DNA fragment containing the lacIQ gene and the lac promoter (14). If formed, the cointegrant would maintain the dnaC+ gene by virtue of DNA replication from the pACYC184 replication origin to preserve viability on medium lacking IPTG, explaining the higher ratio for the recA+ strain than for the recA::Kanr mutant. Whereas the values for the recA+ strain carrying plasmids 31-f3 and 157-c2 were within a twofold range of the ratio for the strain carrying pACYC184, supporting this interpretation, the recA+ strain carrying most of the remaining dnaC mutant plasmids showed more greatly reduced ratios of plating efficiency compared with pACYC184 (Table 4, rows 5 to 15 and 20 to 22). These last results undermine the suggestion of cointegrate formation. Other exceptions are described below. We do not have an alternate explanation for the results obtained with pACYC184, but this lack of understanding does not weaken the method.
Comparing the relative plating efficiencies, the ratios for the recA+ strain carrying most of the mutant dnaC plasmids are roughly one to two orders of magnitude greater than the ratios for the recA mutant bearing the respective plasmids (Table 4). Since homologous recombination should lead to a population of the pACYC184 derivative carrying both the wild-type dnaC sequence and the dnaC mutation, their proportion in individual cells and the associated phenotype depend on when recombination occurred in an earlier cell cycle or the present cycle and on the random probability of replicating the plasmid carrying wild-type dnaC compared to the mutant dnaC plasmid. In support, dissimilar ratios were observed for the recA+ strain but not the recA mutant with two plasmids encoding a Gly97Ser substitution. The greater relative plating efficiencies in the recA+ strain suggest the RecA-dependent formation of cointegrates and/or the substitution of the dnaC mutation by the wild-type dnaC sequence in these pACYC184 derivatives. However, the latter possibility appears to be infrequent, because DNA sequence analysis did not indicate the presence of both wild-type and mutant sequences (data not shown). In summary and despite the exceptions discussed below, the results in Table 4 indicate that homologous recombination does not interfere with the method.
Exceptions were observed with two groups of plasmids. One group encodes the Ser238Cys and Phe23Ser substitutions and the Glu28Val-Tyr236Asn and Cys120-Val229Glu double substitutions where the ratios are near 1 regardless of the host strain (Table 4). Relative to the colonies of the recA+ strain carrying pACYCdnaC, which were similar in size on medium with or without IPTG, colony sizes for the other plasmid-bearing strains were comparable on medium supplemented with IPTG. On medium lacking IPTG, the colonies were either heterogeneous in size or uniformly smaller (for Phe23Ser), which suggests that these dnaC alleles encode partially active proteins that ineffectively support DNA replication, affecting colony size. Because we diluted the cultures prior to screening to improve the sensitivity of the assay, these observations support the conclusion that the genetic method can identify dnaC mutations that encode proteins having partial activity.
The second group encodes Val217Glu, Leu44Pro, and Thr151Pro Arg220His substitutions. The ratios for these plasmids in the recA+ strain were lower by 34-fold, 20-fold, and 2-fold, respectively, than for the recA mutant (Table 4). For the latter plasmid, the difference is within the experimental error. The reduced plating efficiency caused by plasmids encoding the Val217Glu and Leu44Pro substitutions in the recA+ strain relative to that of the recA mutant when pAM34dnaC has been lost suggests that these dnaC mutations are weakly dominant-negative compared with the wild-type dnaC gene in a mixture of plasmid molecules in a bacterial cell. Mutations that are strongly dominant-negative to dnaC+ are not expected because they may be lethal.
DISCUSSION
Method for analyzing functions of essential genes.
In this work, we describe the construction of a null dnaC mutant and a method to identify dnaC mutations that disrupt or partially inactivate DnaC function. Our intention is to obtain a large collection of missense mutations from which we can identify those that are potentially interesting. Further study may reveal activities of DnaC that are poorly understood, such as the mechanism of the cooperative binding of DnaC to DnaB (12, 31), the significance of its interaction with single-stranded DNA (3, 9, 18), and its role in the entry of DnaB at the unwound region of oriC (9, 21).
The method can be adapted to the phenotypic analysis of site-directed mutations in other essential genes, but it is restricted to bacteria that can maintain pAM34 and its derivatives. Because of the abundance of structural and bioinformatics data on a variety of essential proteins, the approach permits the genetic analysis of specific amino acids to complement structure-function studies. It also can correlate in vivo effects with biochemical results.
Mutational analysis of dnaC.
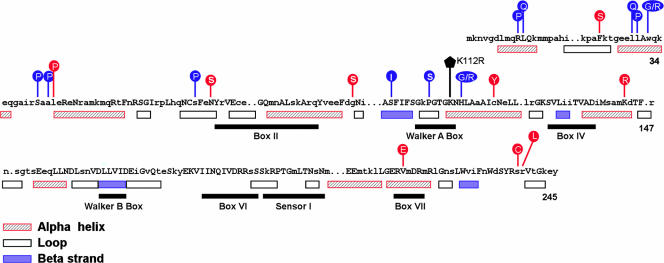
Lacking a three-dimensional structure for DnaC to provide a framework for correlating its biochemical activities, current structural information relies on cryoelectron microscopy (2, 24) and on sequence alignment of DnaC proteins from various bacteria (20). The latter analysis reveals motifs shared by AAA+ proteins (Fig. 2). As with other oligomeric AAA+ proteins that form a bipartite nucleotide binding pocket between adjacent monomers (reviewed in references 10 and 15), the Walker A box and other AAA+ motifs of DnaC are known or presumed to function in ATP binding or in coupling of the relative movement of domains with ATP hydrolysis, respectively. As direct evidence on the functional importance of the Walker A box motif, Davey et al. showed that a Lys112Arg substitution inhibited ATP binding (9). Based on this information, the mutant DnaC proteins in Table 4, with amino acid substitutions such as G97S, C120Y, and K143R near the Walker A box (Fig. 2), may be defective in ATP binding. The proviso on interpreting the mutant DnaCs is that these and other mutant proteins are as stable to proteases as wild-type DnaC in vivo. The Val217Glu substitution is in the box VII motif of AAA+ proteins. Because conserved residues in this element in other proteins are suggested to differentiate ATP from ADP (10, 15) or to form an oligomer (11, 26), we speculate that the substitution affects one or both activities.
FIG. 2.
Amino acid substitutions in mutant DnaC proteins. The amino acid sequence of E. coli DnaC shows conserved residues found in five of nine homologous DnaC proteins (uppercase letters) and gaps in the alignment (20). The numbers at the ends refer to the coordinates for the E. coli DnaC protein. Its predicted secondary structure is indicated by the open (loop), hatched (alpha-helix), or filled (beta-strand) boxes. Above the consensus sequence, the locations of the unique single amino acid substitutions in mutant DnaCs are also shown. Those in red were identified in the present work; substitutions in blue were previously described (20). The filled pentagon represents the Lys112Arg substitution encoded by pKH-dnaCK112R (9). AAA+ motifs are also shown. (Adapted from reference 20 with permission of the publisher.)
Of the other single missense mutations in Table 4, those encoding the Phe23Ser and Leu44Pro substitutions were isolated previously using genetic selection to obtain mutant proteins that fail to interact with DnaB (20). Because the Leu44Pro substitution and others that map near the N terminus were shown biochemically to impair the binding of DnaC to DnaB, we speculate that Phe23Ser shares this defect. The Asn73Ser substitution alters a highly conserved residue, whereas the Ser238Cys and Arg239Leu substitutions near the carboxyl terminus (residue 245) change nonconserved residues. Because the function of these regions is unknown, the biochemical characterization of these mutant proteins may provide new insight.
Dominant-negative dnaC mutations.
With two plasmids coresident in a null dnaC mutant, we were initially concerned that the assembly of mutant and wild-type DnaC to form an inactive DnaB6-DnaC6 complex might lead to a failure to duplicate the bacterial chromosome and inviability. If so, the genetic method might select against nonfunctional dnaC mutations. Fostering this concern, Davey et al. reported that combining the wild-type DnaC protein with increasing amounts of the mutant DnaC with the Lys112Arg substitution proportionally inhibited oriC plasmid replication (9). At an equivalent ratio of mutant DnaC and DnaC+, the DnaB-DnaC complex was inactive. To address this issue, we compared a plasmid encoding the Lys112Arg substitution (pHK-dnaCK112R) with the corresponding dnaC+ plasmid (pHK-dnaC) (9) by introducing the DNAs into isogenic strains [MC1061 (dnaC+) and KH1061 (ΔdnaC)]. Both strains carried pdnaC113, which encodes dnaC downstream from the araBAD promoter, and were examined under noninduced conditions. With both strains, we observed less than a twofold reduction in transformation frequency for the mutant dnaC plasmid compared with the dnaC+ plasmid (K. Hupert-Kocurek and J. Kaguni, unpublished results). These findings indicate that the nonfunctional dnaC allele marginally affects viability. Thus, the genetic method should not yield a biased collection of dnaC mutations.
As a control, we showed that the dnaC+ plasmid (pHK-dnaC) complemented a dnaC2(Ts) mutant (SS1020) at the nonpermissive temperature whereas the empty vector was inactive (Hupert-Kocurek and Kaguni, unpublished). Because the dnaC alleles of these plasmids are downstream from a T7 RNA polymerase promoter and the strains in the above experiments lack T7 RNA polymerase, read-through transcription from an upstream promoter apparently adequately complements the dnaC2 mutant. If the cellular abundance of DnaC is comparable to the 10 to 20 DnaB monomers estimated per cell (27), an extremely low expression level appears to suffice.
Acknowledgments
This research was supported by grant GM33992 from the National Institutes of Health and by the Michigan Agricultural Experiment Station.
We thank Steven Sandler, Donald Court, Kaymeuang Cam, and Mike O'Donnell for generously providing some of the E. coli strains and plasmids used in this study. In the Kaguni lab, we thank Lyle Simmons for his helpful suggestions during the development of the work and Magdalena M. Felczak for E. coli MF1061.
Footnotes
Published ahead of print on 14 September 2007.
REFERENCES
- 1.Allen, G. J., and A. Kornberg. 1991. Fine balance in the regulation of DnaB helicase by DnaC protein in replication in Escherichia coli. J. Biol. Chem. 266:22096-22101. [PubMed] [Google Scholar]
- 2.Barcena, M., T. Ruiz, L. E. Donate, S. E. Brown, N. E. Dixon, M. Radermacher, and J. M. Carazo. 2001. The DnaB.DnaC complex: a structure based on dimers assembled around an occluded channel. EMBO J. 20:1462-1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Biswas, S. B., S. Flowers, and E. E. Biswas-Fiss. 2004. Quantitative analysis of nucleotide modulation of DNA binding by the DnaC protein of Escherichia coli. Biochem. J. 379:553-562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Breier, A. M., H. U. Weier, and N. R. Cozzarelli. 2005. Independence of replisomes in Escherichia coli chromosomal replication. Proc. Natl. Acad. Sci. USA 102:3942-3947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carl, P. L. 1970. Escherichia coli mutants with temperature-sensitive synthesis of DNA. Mol. Gen. Genet. 109:107-122. [DOI] [PubMed] [Google Scholar]
- 6.Chang, A. C., and S. N. Cohen. 1978. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J. Bacteriol. 134:1141-1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cox, M. M., M. F. Goodman, K. N. Kreuzer, D. J. Sherratt, S. J. Sandler, and K. J. Marians. 2000. The importance of repairing stalled replication forks. Nature 404:37-41. [DOI] [PubMed] [Google Scholar]
- 8.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davey, M. J., L. Fang, P. McInerney, R. E. Georgescu, and M. O'Donnell. 2002. The DnaC helicase loader is a dual ATP/ADP switch protein. EMBO J. 21:3148-3159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Erzberger, J. P., and J. M. Berger. 2006. Evolutionary relationships and structural mechanisms of AAA+ proteins. Annu. Rev. Biophys. Biomol. Struct. 35:93-114. [DOI] [PubMed] [Google Scholar]
- 11.Felczak, M. M., and J. M. Kaguni. 2004. The box VII motif of Escherichia coli DnaA protein is required for DnaA oligomerization at the E. coli replication origin. J. Biol. Chem. 279:51156-51162. [DOI] [PubMed] [Google Scholar]
- 12.Galletto, R., M. J. Jezewska, and W. Bujalowski. 2003. Interactions of the Escherichia coli DnaB helicase hexamer with the replication factor the DnaC protein. Effect of nucleotide cofactors and the ssDNA on protein-protein interactions and the topology of the complex. J. Mol. Biol. 329:441-465. [DOI] [PubMed] [Google Scholar]
- 13.Galletto, R., S. Rajendran, and W. Bujalowski. 2000. Interactions of nucleotide cofactors with the Escherichia coli replication factor DnaC protein. Biochemistry 39:12959-12969. [DOI] [PubMed] [Google Scholar]
- 14.Gil, D., and J. P. Bouche. 1991. ColE1-type vectors with fully repressible replication. Gene 105:17-22. [DOI] [PubMed] [Google Scholar]
- 15.Hanson, P. I., and S. W. Whiteheart. 2005. AAA+ proteins: have engine, will work. Nat. Rev. Mol. Cell Biol. 6:519-529. [DOI] [PubMed] [Google Scholar]
- 16.Kaguni, J. M. 2006. DnaA: controlling the initiation of bacterial DNA replication and more. Annu. Rev. Microbiol. 60:351-375. [DOI] [PubMed] [Google Scholar]
- 17.Koonin, E. V. 1992. DnaC protein contains a modified ATP-binding motif and belongs to a novel family of ATPases including also DnaA. Nucleic Acids Res. 20:1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Learn, B. A., S. J. Um, L. Huang, and R. McMacken. 1997. Cryptic single-stranded-DNA binding activities of the phage lambda P and Escherichia coli DnaC replication initiation proteins facilitate the transfer of E. coli DnaB helicase onto DNA. Proc. Natl. Acad. Sci. USA 94:1154-1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lopper, M., R. Boonsombat, S. J. Sandler, and J. L. Keck. 2007. A hand-off mechanism for primosome assembly in replication restart. Mol. Cell 26:781-793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ludlam, A. V., M. W. McNatt, K. M. Carr, and J. M. Kaguni. 2001. Essential amino acids of Escherichia coli DnaC protein in an N-terminal domain interact with DnaB helicase. J. Biol. Chem. 276:27345-27353. [DOI] [PubMed] [Google Scholar]
- 21.Marszalek, J., and J. M. Kaguni. 1994. DnaA protein directs the binding of DnaB protein in initiation of DNA replication in Escherichia coli. J. Biol. Chem. 269:4883-4890. [PubMed] [Google Scholar]
- 22.Miller, J. H. 1992. A short course in bacterial genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 23.Rose, R. E. 1988. The nucleotide sequence of pACYC184. Nucleic Acids Res. 16:355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.San Martin, C., M. Radermacher, B. Wolpensinger, A. Engel, C. S. Miles, N. E. Dixon, and J. M. Carazo. 1998. Three-dimensional reconstructions from cryoelectron microscopy images reveal an intimate complex between helicase DnaB and its loading partner DnaC. Structure 6:501-509. [DOI] [PubMed] [Google Scholar]
- 25.Simmons, L. A., and J. M. Kaguni. 2003. The dnaAcos allele of Escherichia coli: hyperactive initiation is caused by substitution of A184V and Y271H, resulting in defective ATP binding and aberrant DNA replication control. Mol. Microbiol. 47:755-765. [DOI] [PubMed] [Google Scholar]
- 26.Song, H. K., C. Hartmann, R. Ramachandran, M. Bochtler, R. Behrendt, L. Moroder, and R. Huber. 2000. Mutational studies on HslU and its docking mode with HslV. Proc. Natl. Acad. Sci. USA 97:14103-14108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ueda, K., R. McMacken, and A. Kornberg. 1978. dnaB protein of Escherichia coli. Purification and role in the replication of φX174 DNA. J. Biol. Chem. 253:261-269. [PubMed] [Google Scholar]
- 28.Wahle, E., R. S. Lasken, and A. Kornberg. 1989. The dnaB-dnaC replication protein complex of Escherichia coli. I. Formation and properties. J. Biol. Chem. 264:2463-2468. [PubMed] [Google Scholar]
- 29.Wahle, E., R. S. Lasken, and A. Kornberg. 1989. The dnaB-dnaC replication protein complex of Escherichia coli. II. Role of the complex in mobilizing dnaB functions. J. Biol. Chem. 264:2469-2475. [PubMed] [Google Scholar]
- 30.Watson, N. 1988. A new revision of the sequence of plasmid pBR322. Gene 70:399-403. [DOI] [PubMed] [Google Scholar]
- 31.Watt, S. J., T. Urathamakul, P. M. Schaeffer, N. K. Williams, M. M. Sheil, N. E. Dixon, and J. L. Beck. 2007. Multiple oligomeric forms of Escherichia coli DnaB helicase revealed by electrospray ionisation mass spectrometry. Rapid Commun. Mass Spectrom. 21:132-140. [DOI] [PubMed] [Google Scholar]
- 32.Yu, D., H. M. Ellis, E. C. Lee, N. A. Jenkins, N. G. Copeland, and D. L. Court. 2000. An efficient recombination system for chromosome engineering in Escherichia coli. Proc. Natl. Acad. Sci. USA 97:5978-5983. [DOI] [PMC free article] [PubMed] [Google Scholar]