Abstract
Retrospective molecular and phenotypic characterization of a vaccine-derived poliovirus (VDPV) type 1 isolate (7/b/97) isolated from sewage in Athens, Greece, in 1997 is reported. VP1 sequencing of this isolate revealed 1.87% divergence from the VP1 region of reference strain Sabin 1, while further genomic characterization of isolate 7/b/97 revealed a recombination event in the nonstructural part of the genome between a vaccine strain and a nonvaccine strain probably belonging to Enterovirus species C. Amino acid substitutions commonly found in previous studies were identified in the capsid coding region of the isolate, while most of the attenuation and temperature sensitivity determinants were reverted. The ultimate source of isolate 7/b/97 is unknown. The recovery of such a highly divergent derivative of a vaccine strain emphasizes the need for urgent implementation of environmental surveillance as a supportive procedure in the polio surveillance system even in countries with high rates of OPV coverage in order to prevent cases or even outbreaks of poliomyelitis that otherwise would be inevitable.
Poliovirus is a positive single-stranded RNA virus whose genome is approximately 7,500 bases long. It contains four regions: a 5′ untranslated region (5′ UTR), a coding region, a 3′ UTR, and a poly(A) tail. After translation of the coding region, a polyprotein is produced, which is then enzymatically cleaved, yielding all the viral proteins (VP4, VP2, VP3, VP1, 2A, 2B, 2C, 3A, 3B, 3C, 3D).
Poliovirus is the pathogen responsible for paralytic poliomyelitis and is classified into three serotypes (PV1, PV2, and PV3) based on the pattern of neutralization by monoclonal antibodies specific for each serotype (56).
The initiative for eradication of poliomyelitis worldwide was launched by the World Health Organization (WHO) in 1988. By the end of 2005, the reported polio-associated cases had declined from 350,000 in 1988 to less than 2,000. The two basic cornerstones of the initiative for poliomyelitis eradication are massive OPV immunization and sensitive poliovirus surveillance (31). Surveillance for poliovirus consists of investigation of acute flaccid paralysis (AFP) cases and virologic studies of isolates originating either from clinical specimens or from the environment (environmental surveillance) (33).
Environmental surveillance is a powerful and highly sensitive tool for detection of poliovirus circulation in high-risk areas in which complete and effective AFP surveillance cannot be accomplished. Environmental poliovirus surveillance is implemented mainly through investigation of wastewater samples. A previous study proved that environmental poliovirus surveillance is capable of detecting a poliovirus strain even if it is excreted by only 1 of 10,000 people (30). Furthermore, there are examples of detection of poliovirus strains in the environment even in the absence of reported AFP cases in the population (5, 41, 53).
The genomes of most RNA viruses are characterized by high evolution rates, which are many orders of magnitude greater than those of DNA viruses (18). Poliovirus is considered one of the most rapidly evolving RNA viruses (22, 39, 42). The mechanisms underlying rapid poliovirus evolution are mutation and recombination events.
OPV consists of live attenuated poliovirus strains (Sabin strains) of the three poliovirus serotypes. OPV proved to be a very effective vaccine for interrupting wild poliovirus transmission. However, due to its genetic instability there are two major disadvantages of its use. The most commonly recognized adverse effect is vaccine-associated paralytic poliomyelitis, whose clinical symptoms are identical to those of paralytic poliomyelitis caused by wild poliovirus (33). During multiplication of Sabin strains in the human intestine, mutations at specific sites of the genome are associated with the attenuated phenotype of Sabin strains and recombination between Sabin strains may result in the loss of the attenuating phenotype of OPV strains and the acquisition of traits characteristic of wild poliovirus, such as increased neurovirulence and loss of temperature sensitivity (1, 10, 21).The rate of vaccine-associated paralytic poliomyelitis worldwide is 1 case per 750,000 primary vaccinees (13).
In addition, OPV is the potential source of vaccine-derived polioviruses (VDPVs). An OPV-related isolate is considered to be a VDPV if the VP1 nucleotide sequence has diverged more than 1% from the VP1 nucleotide sequence of the reference OPV strain (57, 59). The ≥1% threshold implies that replication and/or circulation of the strain has occurred for at least 1 year after administration of the OPV dose, which is substantially longer than the 4 to 6 weeks which is the normal period of virus replication in vaccinees.
VDPVs are subdivided into three categories: immunodeficient VDPVs (iVDPVs), circulating VDPVs (cVDPVs), and ambiguous VDPVs (aVDPVs). iVDPVs have been isolated from patients with primary B-cell immunodeficiencies and occasionally have caused paralytic disease in vaccinees (9, 29, 34, 42). cVDPVs emerge in communities with gaps in OPV coverage, where circulation of wild poliovirus has already ceased. The most important properties of cVDPVs are sustained person-to-person transmission and an increased capacity to cause poliomyelitis (14). Furthermore, the vast majority of cVDPV isolates have been found to recombine with Enterovirus group C in the nonstructural region of the genome. In recent years, polio outbreaks in which the causal agent was a cVDPV strain have been documented. The polio outbreak in Hispaniola in 2000 and 2001 was caused by a type 1 cVDPV (32). Similarly, in 2001 in The Philippines a type 1 cVDPV was implicated in three AFP cases (52), and a type 2 cVDPV was the underlying cause of five AFP cases in Madagascar in 2002 (51), while three AFP cases were connected to a type 1 cVDPV in China in 2004 (36). aVDPVs are a more recent category which contains isolates excreted from vaccinees with no known immunodeficiencies and environmental isolates whose ultimate source has not been identified. aVDPV strains with highly divergent genomes have been isolated from sewage in Israel (53) and Estonia (5) in the absence of reported AFP cases, revealing the importance of environmental surveillance even in adequately immunized populations.
In this paper we report in retrospect the isolation of a type 1 VDPV isolated from sewage in Athens, Greece, in 1997. Almost complete genomic sequencing revealed the recombinant vaccine/nonvaccine nature of this isolate. The ultimate source of the isolate was not known. The results of this study emphasize the need for enhanced environmental surveillance in order to identify highly divergent isolates which could be potential sources of epidemics, even in regions where the rates of OPV coverage are high.
MATERIALS AND METHODS
Environmental sampling.
Environmental samples were collected from the sewage treatment plant of Athens from April to October 1997. Fourteen 1-liter samples were collected and stored at −80°C. The samples were stored at the Hellenic Pasteur Institute. Environmental samples were transported in dry ice to the Biochemistry & Biotechnology Department in Larissa, Greece, in 2004, where processing of the samples took place.
Processing of samples: virus isolation.
Processing of the environmental samples was carried out using the method proposed by the WHO (58). A concentrate of every environmental sample was added to 10× minimal essential medium (MEM) (9 ml of concentrate and 1 ml of 10× MEM) and inoculated into 96-well plates. Fifty microliters of the concentrate was inoculated into the 96-well plates containing confluent HEp-2 cells in 100 μl of Eagle MEM supplemented with 3% fetal calf serum (FCS). Following absorption of the virus for 2 h at 37°C, the medium in each well was replaced by 100 μl of Eagle MEM supplemented with 1% FCS, and the 96-well plates were incubated at 37°C for 4 to 5 days. Then the plates were frozen and thawed three times before a second passage in cell culture. This procedure was repeated. The second passage lasted for approximately 5 to 6 days. The wells in which cytopathic effects (CPE) were observed were marked, and their contents were inoculated into 25-cm2 flasks containing HEp-2 cells in 3 ml of Eagle MEM and 2% FCS in order to confirm the CPE. RNA extraction (12), reverse transcription, and PCR with primer pair UG52/UC53 (24) were performed in each flask in which CPE were observed in order to confirm the enterovirus CPE. Following confirmation of enterovirus CPE, serial 10-fold dilutions were prepared in order to avoid viral mixtures. The last dilution in which CPE was observed was inoculated into a 25-cm2 flask containing HEp-2 cells, and the presence of enterovirus was retested as described above.
Preliminary characterization of the isolates.
For preliminary characterization of the isolates, RNA extraction (12), reverse transcription, and PCR with primer pair 292/222 (48) were performed. PCR amplicons were sequenced at Macrogen Inc. (Seoul, Korea). After sequencing the identified poliovirus isolates were subjected to further characterization.
Complete VP1 sequencing of poliovirus isolates.
The primer pairs used for complete VP1 gene sequencing were UG1/222 (2, 47) for the first half of the VP1 gene and serotype-specific primer pairs S137/S1688, S230/S2688, and S326/S3651 for Sabin 1, Sabin 2, and Sabin 3 isolates, respectively, for the second half of the VP1 gene (17). CLUSTALW (www.ebi.ac.uk/clustalw) was used for alignment of VP1 sequences of the isolated poliovirus Sabin strains with sequences of the reference poliovirus Sabin strains. The accession numbers of the reference Sabin strains that were used in this study were AY184219, AY184220, and AY184221 for Sabin 1, Sabin 2, and Sabin 3 strains, respectively. Isolate 7/b/97 exhibited high sequence divergence from the reference Sabin 1 strain and was studied further.
Genomic sequencing of isolate 7/b/97.
Reverse transcription and PCRs for isolate 7/b/97 were performed in an Eppendorf Mastercycler personal. In brief, 5 μl of extracted RNA and 2 μl of random d(N6) primers (New England Biolabs, United States) were incubated for 5 min at 70°C. The reaction mixture contained 5 μl of 5× Moloney murine leukemia virus (M-MLV) reaction buffer, 200 U M-MLV reverse transcriptase (Promega Corporation, Madison, WI), 5 μl deoxynucleoside triphosphates (1 mM), 40 U RNase inhibitor (Promega Corporation, Madison, WI), and enough RNase-free distilled water to bring the final volume to 18 μl. The 18-μl reaction mixture was added to the annealed template-RNA solution and incubated for 1 h at 37°C for cDNA synthesis and then at 94°C for 5 min for M-MLV reverse transcriptase inactivation.
The primer pairs used in this study are shown on Table 1. Primer pairs S123/S1976, S182/S1542, S1350/S1693, S1476/S1154, and S1013/S1669 were designed using the reference Sabin 1 strain (accession number AY184219), while primer pair CVD1/CVD2 was designed using the sequence of the 2C region of isolate 7/b/97 in order to cover a 50-base gap. For design of all other primer pairs the program Primer 3 (www.genome.wi.-mit.edu/genomesoftware/other/) was used.
TABLE 1.
Primers used for complete genomic sequencing of isolate 7/b/97
| Primer | Polarity | Sequence (5′→3′)a | Reference |
|---|---|---|---|
| 72437 | Sense | 001-TTAAAACAGCTCTGGGGTTG-020 | 47 |
| 216616 | Antisense | 545-GAAACACGGACACCCAAAGTA-565 | 6 |
| S123 | Sense | 323-GAGTCTGGACATCCCTCAC-342 | This study |
| S1976 | Antisense | 957-TACCCGCAAGCCTCTATGTT-976 | This study |
| S182 | Sense | 882-CTCAAGACCCTTCCAAGTTCA-902 | This study |
| S1542 | Antisense | 1523-ATTATCTGGTGCGGGAACAC-1542 | This study |
| S1350 | Sense | 1350-GGGATAGCAACACCACTACCA-1370 | This study |
| S1693 | Antisense | 1675-TGGAATCTCTGGGGAGGAC-1693 | This study |
| S1476 | Sense | 1476-CGGTGGATTACCTCTTTGGA-1495 | This study |
| S1154 | Antisense | 2136-AGTTTGCCAGTTGCCATCA-2154 | This study |
| S1013 | Sense | 2013-TCTGCCTGTCACTCTCTCCA-2032 | This study |
| S1669 | Antisense | 2648-TGGTTTGCACTGTATCAGAAGG-2669 | This study |
| UG1 | Sense | 2402-TTTGTGTCAGCGTGTTAATG-2421 | 2 |
| 222 | Antisense | 2982-CICCIGGIGGIAYRWACA-2999 | 48 |
| S137 | Sense | 2837-AGGAAATTGGAGTTCTTCACC-2857 | 17 |
| S1688 | Antisense | 3468-ACATGACGTTCACTGCGTTTT-3488 | 17 |
| 71935 | Sense | 3206-GTCAATGATCACAACCC-3222 | 46 |
| EUC2 | Antisense | 4454-TTTGCACTTGAACTGTATGTA-4474 | 11 |
| UG23 | Sense | 4169-AAGGGATTGGAGTGGGTGTC-4188 | 25 |
| UC15 | Antisense | 4948-CATCTCTTGAAGTTTGCT-4965 | 25 |
| CVD1 | Sense | ACGACCTGAACCAAAATCCA | This study |
| CVD2 | Antisense | CAATCCACCCCTTCTTCTCAC | This study |
| S142 | Sense | 4962-GATGCTGTCCTTTAGTGTGTGG-4984 | 49 |
| S1952 | Antisense | 5853-GATGCTGTCCTTTAGTGTGTGG-5873 | 49 |
| S170 | Sense | 5769-TATGTATGTTCCTGTCGGTGCT-5791 | 49 |
| S1299 | Antisense | 5977-ACTGGATTTCACCTTGACTCTG-5999 | 49 |
| S1450 | Antisense | 6851-ATTGAAGTGCCTGAGCAACC-6871 | 49 |
| S1198 | Sense | 6728-GCACTAAAGATGGTGCTTGAGAAA-6752 | 49 |
| S1885 | Antisense | 7391-CTACAACAGTATGACCCAATCCAA-7415 | 49 |
Primer positions are based on those of the Sabin 1 reference strain (accession number AY184219).
For a PCR, 3 μl of cDNA of isolate 7/b/97, 2 μl of each primer pair, 5 μl of deoxynucleoside triphosphates (10 mM), 2 μl of MgCl2 (50 mM), 0.5 μl (2.5 U) of Taq DNA polymerase (Biotaq, Moscow, Russia), and enough double-distilled H2O to bring the final volume to 50 μl were added to a tube. For primer pairs S123/S1976, S182/S1542, S1350/S1693, S1476/S1154, S1013/S1669, and CVD1/CVD2 40 cycles were performed. Each cycle consisted of 20 s of denaturation at 95°C, 20 s of annealing at 55°C, and 20 s of extension at 72°C. The final extension step was carried out at 78°C for 15 min. For the other primer pairs the PCR conditions were the same as those described in the original papers (Table 1). Analysis of PCR products was performed by agarose gel electrophoresis (2% agarose, 1 μg/ml ethidium bromide in Tris-boric acid-EDTA buffer), and the HaeIII φX174 ladder (Invitrogen, Life Technologies, Paisley, United Kingdom) was used. Where necessary, PCR products were cleaned up with a PCR gel extraction kit (QIAGEN GmbH, Hilden, Germany). All PCR products were sequenced at Macrogen Inc. (Seoul, Korea).
Computational analysis.
CLUSTALW was used for complete alignment of the genomes of isolate 7/b/97 and the Sabin 1 reference strain (accession number AY184219). Amino acid sequences of isolate 7/b/97 were obtained by using Gene-Runner V 3.05, and alignment of amino acid sequences of isolate 7/b/97 and the Sabin 1 reference strain (accession number AY184219) was also performed by using CLUSTALW. Isolate 7/b/97's amino acid substitutions in the capsid region were depicted with Rasmol V.2.7.1.1 (www.umass.edu/microbio/rasmol). For visualization of amino acid substitutions of isolate 7/b/97, the capsid protomer structure of the poliovirus type 1 Mahoney strain (Protein Data Bank accession number 1ASJ.pdb) was used (55). Finally, the Simplot program (version 2.5) was used for depiction of the recombination event.
RCT marker.
In an attempt to determine the reproductive capacity at different temperatures (RCT assay) the reproductive capacity of viral isolate 7/b/97 was tested at two different temperatures. The titers of the viral stock in HEp-2 cells were calculated and expressed as the 50% tissue culture infective dose TCID50/0.1 ml at 36 and at 39.5°C. Two different incubators were used. The temperature of one incubator was adjusted to 36°C (optimal temperature for virus reproduction), while the temperature of the other incubator was adjusted to 39.5°C (supraoptimal temperature for virus reproduction). Viral isolates with differences between the titers at 36 and 39.5°C greater than 2 logarithms were considered temperature sensitive.
Nucleotide sequence accession number.
The almost complete genomic sequence of isolate 7/b/97 (from nucleotide 48 to nucleotide 7406) has been deposited in the GenBank library under accession number EF456706.
RESULTS
Preliminary characterization of the isolates: complete VP1 sequencing.
Partial sequencing of the VP1 region of the isolates identified five poliovirus isolates. Two of the isolates were of Sabin 2 vaccine origin, two were of Sabin 3 vaccine origin, and one was of Sabin 1 vaccine origin.
We further studied the poliovirus isolates by performing complete sequencing of the VP1 capsid protein. Complete sequencing of the VP1 genomic region revealed that the isolates of Sabin 2 and Sabin 3 vaccine origin were very closely related to the corresponding OPV reference strains (levels of homology, >99.5%). In contrast, isolate 7/b/97, which was found to be of Sabin 1 vaccine origin, exhibited 1.87% divergence in the VP1 region from the corresponding OPV reference strain. Thus, it was characterized as a type 1 VDPV and was studied further by sequencing a large part of the genome.
Genomic sequencing of isolate 7/b/97.
The almost complete genome (from base 48 to base 7406) of isolate 7/b/97 was sequenced and was aligned with the genome of the reference Sabin 1 strain. Several mutations were identified in the 5′ UTR and in the capsid coding region, and a recombination event was identified in the 2A region of the genome.
In the 5′ UTR two mutations were identified, a transition mutation (C→T) at nucleotide 471 and a transversion mutation (C→G) at nucleotide 583 (Table 2). Furthermore, the A→G transition mutation at nucleotide 480, which is a well-known determinant of loss of the attenuated phenotype of Sabin 1 strains, was not identified (7).
TABLE 2.
Nucleotide mutations and amino acid substitutions identified in the 5′ UTR and P1 coding region of isolate 7/b/97
| Region | Nucleotide
|
Amino acid
|
Amino acid location (reference[s]) | ||||
|---|---|---|---|---|---|---|---|
| Position | Sabin 1 | 7/b/97 | Position | Sabin 1 | 7/b/97 | ||
| 5′ UTR | 471 | C | T | ||||
| 583 | C | G | |||||
| VP4 | 790 | A | G | ||||
| 793 | T | C | |||||
| 814 | C | T | |||||
| 856 | T | C | |||||
| VP2 | 1060 | T | C | ||||
| 1174 | G | A | |||||
| 1354 | T | C | |||||
| 1357 | C | T | |||||
| 1477 | G | A | |||||
| 1624 | T | C | |||||
| VP3 | 1864 | T | C | ||||
| 1942 | A | G | |||||
| 1944 | A | C | 60 | Lys | Thr | N-AgIII (4, 43) | |
| 2243 | C | T | |||||
| 2332 | C | T | |||||
| 2438 | A | T | 225 | Met | Leu | Protomer interface (16, 40) | |
| 2476 | A | G | |||||
| VP1 | 2542 | A | G | ||||
| 2617 | A | G | |||||
| 2623 | A | G | |||||
| 2645 | G | A | 56 | Val | Ile | Capsid interior (15) | |
| 2749 | A | G | 90 | Ile | Met | N-AgI (43) | |
| 2767 | T | C | |||||
| 2774 | A | G | 99 | Lys | Glu | N-AgI, VP1 BC loop (20, 26, 27, 44) | |
| 2779 | T | C | |||||
| 2789 | C | T | |||||
| 2795 | A | G | 106 | Thr | Ala | VP1 BC loop (16, 26, 36) | |
| 2905 | C | T | |||||
| 3067 | A | G | |||||
| 3070 | G | A | |||||
| 3133 | G | A | |||||
| 3157 | C | T | |||||
| 3188 | T | A | 237 | Phe | Ile | Canyon floor (4) | |
| 3349 | G | A | |||||
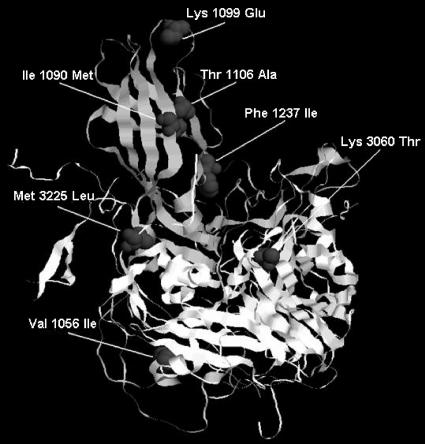
The mutational pattern of the capsid was characterized by a preponderance of synonymous mutations over nonsynonymous mutations. Of the 34 mutations identified in the capsid region, 27 were synonymous (∼80%) and only seven were nonsynonymous (Table 2). Nonsynonymous mutations identified in the capsid are shown in Fig. 1.
FIG. 1.
Amino acid substitutions in the capsid protomer of isolate 7/b/97. Amino acids of the VP1 and VP3 capsid proteins are numbered starting at positions 1001 and 3001, respectively. For this visualization the capsid protomer structure of poliovirus type 1 Mahoney strain was used (Protein Data Bank accession number 1ASJ.pdb) (55).
In VP4 and VP2 10 synonymous mutations were identified. In the VP3 capsid protein, only two of seven mutations that were identified resulted in amino acid substitutions. Specifically, an A→C transition at nucleotide 1944 resulted in a lysine-to-threonine substitution at amino acid 60 of VP3, while an A→T transversion at nucleotide 2438 resulted in a methionine-to-leucine change at amino acid 225 of VP3.
In the VP1 genomic region, 12 of the 17 nucleotide changes identified were substitutions at synonymous positions (∼70%), and only five nucleotide changes resulted in amino acid substitutions (Table 2). A G→A mutation at nucleotide 2645 resulted in a valine-to-isoleucine substitution at residue 56 of the VP1 capsid protein. Substitutions at nucleotides 2749 (A→G), 2774 (A→G), and 2795 (A→ G) resulted in amino acid substitutions at residues 90 (isoleucine to methionine), 99 (lysine to glutamic acid), and 106 (threonine to alanine), respectively. A mutation at nucleotide 3188 (T→A) resulted in a phenylalanine-to-isoleucine amino acid substitution at residue 237.
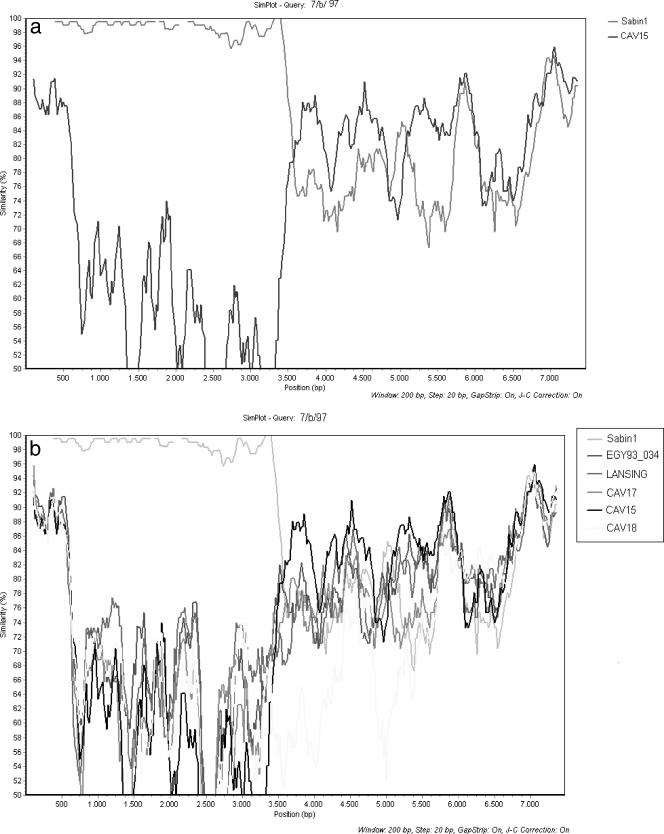
Genomic sequencing revealed a recombination event in the 2A region of isolate 7/b/97. Isolate 7/b/97 was the product of recombination between Sabin 1 and non-Sabin viruses. Simplot analysis revealed that in most genomic regions the strain most closely related to isolate 7/b/97 was CAV15, with levels of homology fluctuating between 72 and 92%. Sabin 1 and CAV18 were the strains with the highest levels of homology (∼85%) in parts of the 2C and 3D genomic regions (Fig. 2a and 2b). After a comprehensive search of the GenBank database, no strain with a level of homology greater than 95% was found. Furthermore, analysis with the MEGA program (version 3.1) revealed that our strain is more closely related to Enterovirus species C than to Enterovirus species A, B, or D (data not shown) (35). These results suggest that the donor of noncapsid sequences of isolate 7/b/97 was an unidentified strain probably belonging to Enterovirus species C.
FIG. 2.
(a) Simplot analysis of the complete genome of isolate 7/b/97 compared with the genome of isolate Sabin 1 (accession number AY184219) and CAV15 strain G-9 (accession number AF465512). (b) Simplot analysis of the complete genome of isolate 7/b/97 compared with the genomes of isolate Sabin 1 (accession number AY184219), CAV15 strain G-9 (accession number AF465512), CAV17 strain G12 (accession number AF499639), CAV18 strain G-13 (accession number AF465513), and strain EGY93_034 (accession number AF448783).
RCT marker test.
The RCT marker assay showed that isolate 7/b/97 had lost the temperature-sensitive phenotype. Specifically, the titer of isolate 7/b/97 was the same at 36°C and at 39.5°C (4.7 log TCID50/0.1 ml), which means that the virus can efficiently replicate at elevated temperatures. For the Sabin 1 reference strain, the difference between the titers at 36°C (4.5 log TCID50/0.1 ml) and at 39.5°C (1.5 log TCID50/0.1 ml) was greater than 2 logarithms, a result consistent with the temperature-sensitive phenotype of Sabin strains.
DISCUSSION
In the current study retrospective isolation and characterization of a type 1 VDPV isolate, 7/b/97, isolated from sewage in Athens, Greece, in 1997 is reported. Genomic sequencing revealed that this isolate originated from recombination between Sabin 1 and an unidentified enterovirus belonging to species C. Moreover, amino acid substitutions frequently found in previous studies were scattered throughout the capsid. The important findings of the present report are that highly divergent vaccine derivatives can be found even in communities with high immunization levels and that environmental surveillance for poliovirus should be permanent even in countries considered polio free.
Two mutations were identified in the 5′ UTR. Nucleotide 471, where a C→T transition was identified, is located in a single-stranded part of domain V, while nucleotide 583, where a C→G transversion was observed, is located in a double-stranded region of domain VI (50). The C→G substitution at nucleotide 583 was not accompanied by a substitution at nucleotide 616 of domain VI in the frame of covariation in the 5′ UTR.
Molecular characterization of strain 7/b/97 in the capsid coding region revealed amino acid substitutions in key spots of the virion. Amino acid substitutions were clustered in spots affecting the virion stability (Val-56-Ile of VP1 and Met-225-Leu of VP3), in virus antigenic sites (Lys-60-Thr of VP3 in N-AgIII and Ile-90-Met and Lys-99-Glu of VP1 in N-AgI), and in residues affecting virus-receptor interactions (Thr-106-Ile in the VP1 BC loop and Phe-237-Ile in the canyon floor).
Amino acid 56 of VP1 is located at the internal side of the capsid and probably participates in hydrophobic interactions between the VP3 and VP1 capsid proteins. These interactions are crucial for the stability of the capsid, as proposed by Cherkasova et al. (15). Amino acid 225 of VP3 is situated in the internal part of the virion at the interface between protomers, a region important for the regulation of virus-receptor interactions and for virion assembly (16, 40).
Amino acid 60 of VP3 is located at the exterior of the virion, at antigenic site III of the virus (44). Furthermore, amino acid 60 is located in an insertion in β-strand B of VP3, a part of the viral surface which is in contact with the D1 domain of the CD155 receptor during virus cell entry (4). Residues 90 and 99 of VP1 are highly exposed in the capsid exterior and are located at antigenic site I (44). Residue 99 of VP1 is localized in the VP1 BC loop, a region which is involved in virus-receptor interactions, as part of the north rim of the canyon (26, 27, 45). Furthermore, the Lys→Glu amino acid substitution at residue 99 eliminates a trypsin cleavage site, which is characteristic of Sabin 1 strains (20).
Residue 106 of VP1 is also located in the VP1 BC loop and participates in virus-receptor interactions. Amino acid 237 leans towards the inner surface of the capsid and is located next to the GH loop of VP1, a region which participates in formation of the canyon floor (4, 16, 26, 37).
It has been proved that certain amino acids located in the antigenic sites of Sabin 1 strains (amino acid 60 of VP3 and amino acids 106, 99, and 90 of VP1) change very rapidly once the viruses replicate in the human intestine into the corresponding wild-type amino acids or into amino acids homotypic to the corresponding residues of the wild-type virus. The very early fixation of these substitutions probably indicates that they are not the result of immune evasion and they might reflect an effort of the virus to adapt in the human intestine (60). Considering the fact that many of these substitutions were identified in the capsid coding region of isolate 7/b/97 and also in many previous studies (3, 15, 17, 23, 34, 43) in distinct evolving lineages of OPV strains, we suggest that they might confer a selective advantage to isolate 7/b/97. It can be hypothesized that these changes, which occur in the virus antigenic sites, may facilitate the virus-receptor interactions. Our hypothesis is reinforced by the fact that a large part of the region in contact with the poliovirus receptor CD155 comprises simultaneously all the virus antigenic sites. This is especially the case for residue 60, which is located in β-strand B of the VP3 part of the viral surface in contact with the D1 domain of receptor CD155, and for residues 99 and 106 of the VP1 BC loop, which are implicated in the formation of the north rim of the canyon (4, 26, 27, 28). Furthermore, the limitation of proteins' conformations in regions crucial for capsid stability is reflected in the conservative nature of amino acid substitutions observed in our isolate, especially the substitutions in residues 56 (Val→Ile) of VP1 and 225 (Met→Leu) of VP3. Both of these residues participate in hydrophobic interactions between the VP3 and VP1 capsid proteins, which contribute to capsid stabilization (15).
Mutations that result in loss of the attenuated and temperature-sensitive phenotype of Sabin 1 strains have been identified in the genome of strain 7/b/97. Mutations at nucleotides 2438 (A→T [Met-225-Leu]) and 2795 (A→G [Thr-106-Ala]) are well-known determinants of attenuation (7), and both were identified in the capsid coding region of isolate 7/b/97. Reversion of one of the major determinants of attenuation, located at nucleotide 480 of the 5′ UTR, was not identified. Mutations at nucleotides 1944 (A→C [Lys-60-Thr]) and 2438 (A→T [Met-225-Leu]) and an amino acid change at residue 73 of 3Dpol from histidine to tyrosine were also present in isolate 7/b/97 and contributed to the loss of the temperature-sensitive phenotype (7), as confirmed by the RCT marker assay.
Genomic sequencing revealed a recombination event in the 2A region of isolate 7/b/97 between a Sabin type 1 strain and an unidentified strain probably belonging to enterovirus species C. Recombination provides evidence of circulation of isolate 7/b/97 in the community. This notion is in agreement with the fact that all circulating VDPVs isolated so far have been found to be recombinants with recombination in the nonstructural region of their genomes with enteroviruses belonging to species C (32, 36, 51, 52). Circulating wild polioviruses containing a block of Sabin 1-derived sequences have also been isolated (38, 39). Furthermore, we suggest that recombination may increase the fitness of isolate 7/b/97 by substitution in the temperature sensitivity determinants mapped to the 3Dpol and 3′ UTR of the isolate. This is actually true for 3Dpol, since amino acid 73 was changed from histidine in the wild-type virus to tyrosine (unfortunately, we have no data for the 3′ UTR since we obtained the genomic sequence of the isolate from nucleotide 48 to nucleotide 7406). This suggestion is also supported by recent phylogenetic studies which proposed that polioviruses and species C enteroviruses are distinct capsid sequences in quest of noncapsid sequences with the highest fitness (8).
Based on WHO criteria, isolate 7/b/97 is characterized as an aVDPV. Nevertheless, genomic evidence presented here could place isolate 7/b/97 in the cVDPV category. First, the variability among antigenic sites is limited despite the high degree of nucleotide divergence of the isolate, and second and most important, the recombination in the nonstructural part of the genome with an unidentified species C enterovirus strain provides evidence of circulation of the isolate in the population (33, 52, 61). However, there were no polio epidemics or suspected poliomyelitis cases, no gaps in OPV coverage, and no other conditions that may have benefited cVDPV emergence and spread in Greece during the time period that the virus was isolated. We hypothesize, therefore, that strain 7/b/97 is more likely to be an iVDPV strain that was excreted from an unidentified source.
Although AFP surveillance is one of the two major cornerstones in poliovirus eradication, environmental surveillance has been proven to be a highly sensitive tool in some situations. Environmental surveillance has the potential to identify poliovirus in the environment, even in the absence of paralytic poliomyelitis cases. For example, highly divergent VDPVs were isolated in Finland, Israel, and Estonia in the absence of suspected poliomyelitis cases (5, 41, 53). Furthermore, indigenous type 1 wild poliovirus was identified in environmental samples in Egypt even after OPV immunization campaigns (19). High-sensitivity environmental surveillance should be implemented during and after poliovirus eradication to prevent cases or even outbreaks of paralytic poliomyelitis that otherwise would be inevitable (54).
In conclusion, we report genomic and phenotypic characterization of an environmental aVDPV isolate (7/b/97). Extensive genomic sequencing revealed 1.87% VP1 sequence divergence and a vaccine/nonvaccine recombination event in the 2A genomic region of isolate 7/b/97. The genomic features of isolate 7/b/97 may place this strain in the cVDPV category, but as strain 7/b/97 was not connected with any polio case, it is more likely that it belongs in the iVDPV category.
The preponderance of countries certified to be polio free have switched from the OPV vaccine to the IPV vaccine. However, the question now is what would happen after importation of a highly divergent VDPV isolate in a population. Taking into consideration the fact that IPV does not induce the same immunity levels as OPV, importation of a highly virulent strain would have serious implications for public health. For this reason, increased laboratory surveillance is necessary even after eradication of poliomyelitis. Environmental surveillance should also be implemented as a supportive tool in the polio surveillance system in communities that can support this procedure both financially and professionally. Furthermore, extensive genomic characterization of such evolved derivatives of vaccine strains would help to further elucidate the genomic properties of these isolates, since there is a significant risk of prolonged circulation in populations with a low immunity level and subsequent conversion of the isolates into epidemic strains.
Intensive research for optimal vaccination strategies for the final stages of polio eradication and the posteradication era should be urgently promoted.
Acknowledgments
This work was funded by a research grant from the Greek General Secretariat for Research and Technology, project EPAN-M.4.3.6.1.g (code 172-g).
Footnotes
Published ahead of print on 7 September 2007.
REFERENCES
- 1.Abraham, R., P. D. Minor, G. Dunn, J. F. Modlin, and P. L. Ogra. 1993. Shedding of virulent poliovirus revertants during immunization with oral poliovirus vaccine after prior immunization with inactivated poliovirus vaccine. J. Infect. Dis. 168:1105-1109. [DOI] [PubMed] [Google Scholar]
- 2.Balanant, J., S. Guillot, A. Candrea, F. Delpeyroux, and R. Crainic. 1991. The natural genomic variability of poliovirus analyzed by a restriction fragment length polymorphism assay. Virology 184:645-654. [DOI] [PubMed] [Google Scholar]
- 3.Bellmunt, A., G. May, R. Zell, P. Pring-Akerblom, W. Verhagen, and A. Heim. 1999. Evolution of poliovirus type 1 during 5.5 years of prolonged enteral replication in an immunodeficient patient. Virology 265:178-184. [DOI] [PubMed] [Google Scholar]
- 4.Belnap, D. M., B. M. McDermott, D. J. Filman, N. Cheng, B. L. Trus, H. J. Zuccola, V. R. Racaniello, J. M. Hogle, and A. C. Steven. 2000. Three-dimensional structure of poliovirus receptor bound to poliovirus. Proc. Natl. Acad. Sci. USA 97:73-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blomqvist. S., C. Savolainen, P. Laine, P. Hirttiö, E. Lamminsalo, E. Penttilä, S. Jöks, M. Roivainen, and T. Hovi. 2004. Characterization of a highly evolved vaccine-derived poliovirus type 3 isolated from sewage in Estonia. J. Virol. 78:4876-4883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blomqvist, S., A. Skyttä, M. Roivainen, and T. Hovi. 1999. Rapid detection of human rhinoviruses in nasopharyngeal aspirates by a microwell reverse transcription-PCR-hybridization assay. J. Clin. Microbiol. 37:2813-2816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bouchard, M. J., D. H. Lam, and V. R. Racianello. 1995. Determinants of attenuation and temperature sensitivity in the type 1 poliovirus Sabin vaccine. J. Virol. 69:4972-4978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brown, B., M. S. Oberste, K. Maher, and M. A. Pallansch. 2003. Complete genomic sequencing shows that polioviruses and members of human enterovirus species C are closely related in the noncapsid coding region. J. Virol. 77:8973-8984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Buttinelli, G., V. Donati, S. Fiore, J. Marturano, A. Plebani, P. Balestri, A. R. Soresina, R. Vivarelli, F. Delpeyroux, J. Martin, and L. Fiore. 2003. Nucleotide variation in Sabin type 2 poliovirus from an immunodeficient patient with poliomyelitis. J. Gen. Virol. 84:1215-1221. [DOI] [PubMed] [Google Scholar]
- 10.Cammack, N., A. Phillips, G. Dunn, V. Patel, and P. D. Minor. 1988. Intertypic genomic rearrangements of poliovirus strains in vaccinees. Virology 167:507-514. [PubMed] [Google Scholar]
- 11.Caro, V., S. Guillot, A. Candrea, F. Delpeyroux, and R. Crainic. 2001. Molecular strategy of ‘serotyping’ of human enterovirus. J. Gen. Virol. 82:79-91. [DOI] [PubMed] [Google Scholar]
- 12.Casas, I., L. Powell, P. E. Klapper, and G. M. Cleator. 1995. New method for the extraction of viral RNA and DNA from cerebrospinal fluid for use in the polymerase chain reaction assay. J. Virol. Methods 53:25-36. [DOI] [PubMed] [Google Scholar]
- 13.Centers for Disease Control and Prevention. 1997. Poliomyelitis prevention in the United States: introduction of a sequential vaccination schedule of inactivated poliovirus vaccine followed by oral polovirus vaccine. Recommendations of the Advisory Committee on Immunization Practices (ACIP). Morb. Mortal. Wkly. Rep. 46:1-25. [PubMed] [Google Scholar]
- 14.Centers for Disease Control and Prevention. 2006. Update on vaccine-derived polioviruses. Morb. Mortal. Wkly. Rep. 55:1093-1097. [PubMed] [Google Scholar]
- 15.Cherkasova, E. A., E. A. Korotkova, M. L. Yakovenko, O. E. Ivanova, T. P. Eremeeva, K. M. Chumakov, and V. I. Agol. 2002. Long-term circulation of vaccine-derived poliovirus that causes paralytic disease. J. Virol. 76:6791-6799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Colston, E. M., and V. R. Racianello. 1994. Soluble receptor-resistant poliovirus mutants identify surface and internal capsid residues that control interaction with the cell receptor. EMBO J. 13:5855-5862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dedepsidis, E., I. Karakasiliotis, E. Paximadi, Z. Kyriakopoulou, D. Komiotis, and P. Markoulatos. 2006. Detection of unusual mutation within the VP1 region of different re-isolates of poliovirus Sabin vaccine. Virus Genes 33:183-191. [DOI] [PubMed] [Google Scholar]
- 18.Domingo, E., and J. J. Holland. 1997. RNA virus mutations and fitness for survival. Annu. Rev. Microbiol. 51:151-178. [DOI] [PubMed] [Google Scholar]
- 19.El Bassioni, L., I. Barakat, E. Nasr, E. M. deGourville, T. Hovi, S. Blomqvist, C. Burns, M. Stenvik, H. Gary, O. M. Kew, M. A. Pallnsch, and M. H. Wahdan. 2003. Prolonged detection of indigenous wild polioviruses in sewage from communities in Egypt. Am. J. Epidemiol. 158:807-815. [DOI] [PubMed] [Google Scholar]
- 20.Fricks, C. E., J. P. Icenogle, and J. M. Hogle. 1985. Trypsin sensitivity of the Sabin strain of type 1 poliovirus: cleavage sites in virions and related particles. J. Virol. 54:856-859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Furione, M., S. Guillot, D. Otelea, J. Balanant, A. Candrea, and R. Crainic. 1993. Polioviruses with natural recombinant genomes isolated from vaccine-associated paralytic poliomyelitis. Virology 196:199-208. [DOI] [PubMed] [Google Scholar]
- 22.Gavrilin, G. V., E. A. Cherkasova, G. Y. Lipskaya, O. M. Kew, and V. I. Agol. 2000. Evolution of circulating wild poliovirus and of vaccine-derived poliovirus in an immunodeficient patient: a unifying model. J. Virol. 74:7381-7390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Georgescu, M. M., J. Balanant, A. Macadam, D. Otelea, M. Combiescu, A. A. Combiescu, R. Crainic, and F. Delpeyroux. 1997. Evolution of the Sabin type 1 poliovirus in humans: characterization of strains isolated from patients with vaccine-associated paralytic poliomyelitis. J. Virol. 71:7758-7768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Georgopoulou, A., P. Markoulatos, N. Spyrou, and N. C. Vamvakopoulos. 2000. Improved genotyping vaccine and wild-type poliovirus strains by restriction fragment length polymorphism analysis: clinical diagnostic implications. J. Clin. Microbiol. 38:4337-4342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guillot, S., V. Caro, N. Cuervo, E. Korotkova, M. Combiescu, A. Persu, A. Aubert-Combiescu, F. Delpeyroux, and R. Crainic. 2000. Natural genetic exchanges between vaccine and wild poliovirus strains in humans. J. Virol. 74:8434-8443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Harber, J., G. Bernhardt, H. H. Lu, J. Y. Sgro, and E. Wimmer. 1995. Canyon rim residues, including antigenic determinants, modulate serotype-specific binding of polioviruses to mutants of the poliovirus receptor. Virology 214:559-570. [DOI] [PubMed] [Google Scholar]
- 27.He, Y., V. D. Bowman, S. Mueller, C. M. Bator, J. Bella, X. Peng, T. S. Baker, E. Wimmer, R. J. Kuhn, and M. G. Rossmann. 2000. Interaction of the poliovirus receptor with poliovirus. Proc. Natl. Acad. Sci. USA 97:79-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.He, Y., S. Mueller, P. R. Chipman, C. M. Bator, X. Peng, V. D. Bowman, S. Mukhopadhyay, E. Wimmer, R. J. Kuhn, and M. G. Rossmann. 2003. Complexes of poliovirus serotypes with their common cellular receptor, CD155. J. Virol. 77:4827-4835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hidalgo, S., M. Garcia Erro, D. Cisterna, and M. C. Freire. 2003. Paralytic poliomyelitis caused by vaccine-derived polio virus in an antibody-deficient Argentinean child. Infect. Dis. J. 22:570-572. [PubMed] [Google Scholar]
- 30.Hovi, T., M. Stevnik, H. Partanen, and A. Kangas. 2001. Poliovirus surveillance by examining sewage specimens: quantitative recovery of virus after introduction into sewerage at remote upstream location. Epidemiol. Infect. 127:101-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hull, H. F., N. A. Ward, J. B. Milstien, and C. de Quadros. 1994. Paralytic poliomyelitis: seasoned strategies, disappearing disease. Lancet 343:1331-1337. [DOI] [PubMed] [Google Scholar]
- 32.Kew, O. M., V. M. Glasgow, M. Landaverde, C. Burns, J. Shaw, Z. Garib, J. Andre, E. Blackman, C. J. Freeman, J. Jorba, R. Sutter, G. Tambini, L. Venczel, C. Pedreira, F. Laender, H. Shimizu, T. Yoneyama, T. Mijamura, H. van der Avoort, M. S. Oberste, D. Kilpatrick, S. Cochi, M. Pallansch, and C. de Quadros. 2002. Outbreak of poliomyelitis in Hispaniola associated with circulating type 1 vaccine-derived poliovirus. Science 296:356-359. [DOI] [PubMed] [Google Scholar]
- 33.Kew, O. M., R. W. Sutter, E. M. de Gourville, W. R. Dowdle, and M. A. Pallansch. 2005. Vaccine-derived poliovirus and the endgame strategy for global polio eradication. Annu. Rev. Microbiol. 59:587-635. [DOI] [PubMed] [Google Scholar]
- 34.Kew, O. M., R. W. Sutter, B. K. Nottay, M. J. McDonough, D. R. Prevots, L. Quick, and M. A. Pallansch. 1998. Prolonged replication of a type 1 vaccine-derived poliovirus in an immunodeficient patient. J. Clin. Microbiol. 36:2893-2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kumar, S., K. Tamura, and M. Nei. 2004. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief. Bioinform. 5:150-163. [DOI] [PubMed] [Google Scholar]
- 36.Liang, X., Y. Zhang, W. Xu, N. Wen, S. Zuo, L. A. Lee, and J. Yu. 2006. An outbreak of poliomyelitis caused by type-1 vaccine-derived poliovirus in China. J. Infect. Dis. 194:545-551. [DOI] [PubMed] [Google Scholar]
- 37.Liao, S., and V. Racianello. 1997. Allele-specific adaptation of poliovirus VP1 B-C loop variants target mutant cell receptors. J. Virol. 71:9770-9777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu, H. M., D. P. Zheng, L. B. Zhang, M. S. Oberste, O. M. Kew, and M. A. Palansch. 2003. Serial recombination during circulation of type 1 wild-vaccine recombinant poliovirus in China. J. Virol. 77:10994-11005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu, H. M., D. P. Zheng, L. B. Zhang, M. S. Oberste, M. A. Palansch, and O. M. Kew. 2000. Molecular evolution of a type 1 wild-vaccine poliovirus recombinant during widespread circulation in China. J. Virol. 74:11153-11161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Macadam, A. J., G. Ferguson, C. Arnold, and P. D. Minor. 1991. An assembly defect as a result of an attenuating mutation in the capsid proteins of poliovirus type 3 vaccine strain. J. Virol. 65:5225-5231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Manor, Y., R. Handsher, T. Halmut, M. Neuman, A. Bobrov, H. Rudich, A. Vonsoner, L. Shulman, O. Kew, and E. Mendelson. 1999. Detection of poliovirus circulation by environmental surveillance in the absence of clinical cases in Israel and the Palestinian authority. J. Clin. Microbiol. 37:1670-1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Martin, J., G. Dunn, R. Hull, V. Patel, and P. D. Minor. 2000. Evolution of the Sabin strain of type 3 poliovirus in an immunodeficient patient during the entire 637-day period of virus excretion. J. Virol. 74:3001-3010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Martin, J., K. Odoom, G. Tuite, G. Dunn, N. Hopewell, G. Cooper, C. Fitzharris, K. Butler, W. W. Hall, and P. D. Minor. 2004. Long-term excretion of vaccine-derived poliovirus by a healthy child. J. Virol. 78:13839-13847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Minor, P. D. 1990. Antigenic structure of picornavirus. Curr. Top. Microbiol. Immunol. 161:124-154. [DOI] [PubMed] [Google Scholar]
- 45.Minor, P. D., M. Ferguson, D. M. Evans, J. W. Almond, and J. P. Icenogle. 1986. Antigenic structures of polioviruses of serotypes 1, 2 and 3. J. Gen. Virol. 67:1283-1291. [DOI] [PubMed] [Google Scholar]
- 46.Mulders, M. N., G. Y. Lipskaya, H. G. van der Avoort, M. P. Koopmans, O. M. Kew, and A. M. van Loom. 1995. Molecular epidemiology of wild poliovirus 1 in Europe, the Middle East, and the Indian subcontinent. J. Infect. Dis. 171:1399-1405. [DOI] [PubMed] [Google Scholar]
- 47.Mulders, M. N., J. H. J. Reimerink, M. Stevnik, I. Alaeddinoglou, H. G. A. M. van der Avoort, T. Hovi, and M. P. G. Koopmans. 1999. A Sabin vaccine-derived field isolate of poliovirus type 1 displaying aberrant phenotypic and genetic features, including a deletion in antigenic site 1. J. Gen. Virol. 80:907-916. [DOI] [PubMed] [Google Scholar]
- 48.Oberste, M. S., K. Maher, M. R. Flemister, G. Marchetti, D. R. Kilpatrick, and M. A. Pallansch. 2000. Comparison of classic and molecular approaches for the identification of untypeable enteroviruses. J. Clin. Microbiol. 38:1170-1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Paximadi, E., I. Karakasiliotis, Z. Mamuris, C. Stathopoulos, V. Krikelis, and P. Markoulatos. 2006. Genomic analysis of recombinant Sabin clinical isolates. Virus Genes 32:203-210. [DOI] [PubMed] [Google Scholar]
- 50.Pilipenko, E. V., V. M. Blinov, L. I. Romanova, A. N. Sinyakov, S. V. Maslova, and V. I. Agol. 1989. Conserved structural domains in the 5′-untranslated region of picornaviral genomes: an analysis of the segment controlling translation and neurovirulence. Virology 168:201-209. [DOI] [PubMed] [Google Scholar]
- 51.Rousset, D., M. R. Andrianarivelo, R. Razafindratsimandresy, B. Randriamanalina, S. Guillot, J. Balanant, P. Mauclère, and F. Delpeyroux. 2003. Recombinant vaccine-derived poliovirus in Madagascar. Emerg. Infect. Dis. 9:885-887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shimizu, H., B. Thorley, F. J. Paladin, K. A. Brussen, V. Stambos, L. Yuen, A. Utama, Y. Tano, M. Arita, H. Yoshida, T. Yoneyama, A. Benegas, S. Roesel, M. Pallansch, O. Kew, and T. Miyamura. 2004. Circulation of type 1 vaccine-derived poliovirus in the Philippines in 2001. J. Virol. 78:13512-13521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shulman, L. M., Y. Manor, R. Handsher, F. Delpeyroux, M. J. McDonough, T. Halmut, I. Silberstein, J. Alfandari, J. Quay, T. Fisher, J. Robinov, O. M. Kew, R. Crainic, and E. Meddelson. 2000. Molecular and antigenic characterization of a highly evolved derivative of the type 2 oral poliovaccine strain isolated from sewage in Israel. J. Clin. Microbiol. 38:3729-3734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vinje, J., N. Gregoricus, J. Martin, H. E. Gary, V. M. Cáceres, L. Venczel, A. Macadam, J. G. Dobbins, C. Burns, D. Wait, G. Ko, M. Landaverde, O. M. Kew, and M. D. Sobsey. 2004. Isolation and characterization of circulating type 1 vaccine-derived poliovirus from sewage and stream waters in Hispaniola. J. Infect. Dis. 189:1168-1175. [DOI] [PubMed] [Google Scholar]
- 55.Wien, M. W., S. Curry, D. J. Filman, and J. M. Hogle. 1997. Structural studies of poliovirus mutants that overcome receptor defects. Nat. Struct. Biol. 4:666-674. [DOI] [PubMed] [Google Scholar]
- 56.Wimmer, E., C. U. T. Hellen, and X. Cao. 1993. Genetics of poliovirus. Annu. Rev. Genet. 27:353-436. [DOI] [PubMed] [Google Scholar]
- 57.World Health Organization. 2002. Expanding contributions of the global laboratory network for poliomyelitis eradication. Wkly. Epidemiol. Rec. 77:133-137. [PubMed] [Google Scholar]
- 58.World Health Organization. 3 March 2003, posting date. Guidelines for environmental surveillance of poliovirus circulation. World Health Organisation, Geneva, Switzerland. http://www.who.int/vaccines-documents/DocsPDF03/www737pdf.
- 59.World Health Organization. 2003. Laboratory surveillance for wild and vaccine-derived polioviruses, January 2002-June 2003. Wkly. Epidemiol. Rec. 78:341-348. [PubMed] [Google Scholar]
- 60.Yakovenko, M. L., E. A. Cherkasova, G. V. Rezapkin, O. E. Ivanova, A. P. Ivanov, T. P. Eremeeva, O. Y. Baykova, K. M. Chumakov, and V. I. Agol. 2006. Antigenic evolution of vaccine-derived polioviruses: changes in individual epitopes and relative stability of the overall immunological properties. J. Virol. 80:2641-2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yang, C. F., T. Naguib, S. J. Yang, E. Nasr, J. Jorba, N. Ahmed, R. Campagoli, H. van der Avoort, H. Shimizu, T. Yoneyama, T. Mijamura, M. Pallansch, and O. M. Kew. 2003. Circulation of endemic type 2 vaccine-derived poliovirus in Egypt from 1983 to 1993. J. Virol. 77:8366-8377. [DOI] [PMC free article] [PubMed] [Google Scholar]