Abstract
Compared to freshwater and the open ocean, less is known about bacterioplankton community structure and spatiotemporal dynamics in estuaries, particularly those with long residence times. The Chesapeake Bay is the largest estuary in the United States, but despite its ecological and economic significance, little is known about its microbial community composition. A rapid screening approach, ITS (internal transcribed spacer)-LH (length heterogeneity)-PCR, was used to screen six rRNA operon (16S rRNA-ITS-23S rRNA) clone libraries constructed from bacterioplankton collected in three distinct regions of the Chesapeake Bay over two seasons. The natural length variation of the 16S-23S rRNA gene ITS region, as well as the presence and location of tRNA-alanine coding regions within the ITS, was determined for 576 clones. Clones representing unique ITS-LH-PCR sizes were sequenced and identified. Dramatic shifts in bacterial composition (changes within subgroups or clades) were observed for the Alphaproteobacteria (Roseobacter clade, SAR11), Cyanobacteria (Synechococcus), and Actinobacteria, suggesting strong seasonal variation within these taxonomic groups. Despite large gradients in salinity and phytoplankton parameters, a remarkably homogeneous bacterioplankton community was observed in the bay in each season. Stronger seasonal, rather than spatial, variation of the bacterioplankton population was also supported by denaturing gradient gel electrophoresis and LH-PCR analyses, indicating that environmental parameters with stronger seasonal, rather than regional, dynamics, such as temperature, might determine bacterioplankton community composition in the Chesapeake Bay.
Estuaries rank among the most productive and dynamic aquatic ecosystems on earth. Mixing of fresh and marine waters and significant recycling of nutrients and organic matter production provide strong environmental gradients to the microbes living in these ecosystems. A number of studies have shown that the compositions of freshwater and marine pelagic microbial communities are fundamentally different (20, 28). Analyses of rRNA genes from open-ocean samples show that bacterioplankton generally contains representatives of the SAR11 (Pelagibacter ubique), SAR116, and Roseobacter clades of the Alphaproteobacteria, the SAR86 clade of the Gammaproteobacteria, the Synechococcus group of Cyanobacteria, and members of the phylum Bacteroidetes (12, 20, 29, 58, 75). In contrast, among 34 clades identified as typical freshwater bacteria are species associated with Alphaproteobacteria, Betaproteobacteria, Bacteroidetes, Actinobacteria, and Verrucomicrobia, with relatively little overlap with the marine clades (28, 82). As an ecotone between the freshwater and marine realms, estuaries contain typical taxa from both environments (20) and bacterioplankton appear to undergo strong physiological stresses that can result in variations in biomass, activities, and population composition (42, 69, 73, 78).
The composition of bacterioplankton communities has been studied in a number of estuaries, mostly relatively small systems with short residence times. These include the Columbia River estuary (20), the San Francisco Bay (39), the Weser River estuary in Germany (68), the Parker River estuary (21), the Rhone River estuary in France (78), the Ria de Aveiro estuary in Portugal (35, 36), the Changjiang River estuary in China (67), the Moreton Bay estuary in Australia (37), and the Delaware River estuary (19, 46). We have also recently studied the spatial and temporal variations of Chesapeake Bay bacterioplankton (44, 45). The majority of these studies used fingerprinting approaches (i.e., denaturing gradient gel electrophoresis [DGGE], terminal restriction fragment length polymorphism [RFLP], and automated ribosomal intergenic spacer analysis) or FISH (fluorescence in situ hybridization) to monitor bacterial community changes at a relatively broad resolution along the salinity gradient or over time. However, the results of these studies do not result in a consistent picture of estuarine bacterioplankton spatial and temporal dynamics. In some studies, dominant populations shifted from Betaproteobacteria in freshwater to Alpha- and Gammaproteobacteria in marine sections (10, 36, 67). However, Selje and Simon (68) reported that Alpha-, Beta-, and Gammaproteobacteria each constituted about 10% of the community, with no pronounced changes among the various sections of the Weser River estuary. In terms of temporal variation, the picture is similar. For instance, the middle Ria de Aveiro estuary and the Chesapeake Bay exhibited pronounced temporal variations (36, 44) but, in contrast, no significant temporal variation was observed in the Weser River estuary (68). Combined, these observations suggest that the factors that determine the community composition of estuarine bacterioplankton might have a strong regional or physiographic component.
Although the composition of estuarine bacterial communities at a high taxonomic resolution has not yet been well defined, unique estuarine populations have been reported for the Columbia, Parker, and Weser River estuaries and the Plum Island Sound (20, 21, 68). Most of the bacterioplankton defined as typically estuarine was closely related to typical freshwater or marine bacterial groups and belongs to the phyla Proteobacteria, Bacteroidetes, and Actinobacteria (21), with these estuarine phylotypes occurring within a wider salinity range than what is considered typical for freshwater or marine biota (20, 68). Finally, it has been hypothesized that the development of local estuarine bacterial communities depends on both the residence time and growth rate of the bacterial populations in an estuary (21). For example, Crump and colleagues showed that unique estuarine bacterial communities formed in the Parker River estuary and the Plum Island Sound only in summer and fall, when residence times were longer than bacterial doubling times (21).
The Chesapeake Bay is the largest and one of the most well-studied estuaries in the United States, with a long average residence time (i.e., 7 months) (54). In general, the main freshwater inputs to the bay (ca. 82%) are from the Susquehanna (52%), Potomac (18%), and James (12%) Rivers (14). Extensive surveys in the main stem of the bay have shown that bacterioplankton biomass, production, growth rates, and respiration vary over time and space (42, 69). In general, bacterioplankton biomass and activity peak in the middle bay region (69, 73) and in nonsummer seasons, when the temperature is below 20°C, bacterial activities are highly correlated with temperature (69). In summer, bacterioplankton appears to be resource limited in the northern and southern regions of the bay, as bacterioplankton activity responded to experimental enrichments with organic substrates and inorganic nutrients (70, 73). However, a lack of response to these enrichments in the middle bay region led to the suggestion that bacterivores control the bacterioplankton biomass and activity in this region (23).
Whether these seasonal and spatial variations in bacterioplankton bulk activities are a function of differences in bacterioplankton community composition remains an open question. The community composition of the Chesapeake Bay bacterioplankton has been investigated by using a variety of molecular tools, including analysis of 5S rRNA patterns (9, 55), FISH (34), and 16S rRNA-based DGGE analysis (44, 45), and community composition appeared to have a higher temporal than spatial variation (44, 45). However, in these studies community composition was described at broad phylum and class levels or only single genera or species were considered. Remarkably, to date, cloning and sequencing of 16S rRNA genes have not been applied to bacterioplankton from the Chesapeake Bay or other large estuaries with long (>6-month) average residence times.
Typically, clone libraries are constructed from environmental samples and 16S rRNA genes are sequenced to determine the phylogenetic origins of the clones recovered. Preliminary screening for identical or closely related clones is typically performed by using RFLP analysis to avoid redundant sequencing. This type of analysis is labor intensive and time-consuming, and inferences of clone phylogenetic identity from RFLP patterns are, in general, not possible. Recently, a novel high-throughput analysis, internal transcribed spacer (ITS) length heterogeneity (LH) PCR, was developed for the screening of rRNA-containing clones in large-insert genomic libraries (76). Based on the length of the entire ITS region (53) and the location of the tRNA-alanine in these regions (76), identification of environmental clones to the subclade level was possible. This approach has been so far only applied to multiplex screening of genomic libraries of marine bacterioplankton or freshwater environments (53, 76), and thus, here we evaluated its applicability to the screening of PCR-based ribosomal operon libraries of estuarine bacterioplankton that were likely to contain clades from both freshwater and marine environments. Six rRNA operon clone libraries were constructed from the northern, middle, and southern Chesapeake Bay in the cold and warm seasons, respectively. Temporal and spatial dynamics of bacterioplankton were determined on the basis of the compositions of these clone libraries. DGGE and LH-PCR were also applied to the same water samples, and the results were compared to those of the clone library analyses.
MATERIALS AND METHODS
Sample collection.
Water samples were collected at three stations along the longitudinal axis of the Chesapeake Bay on 26 to 30 September 2002 and 4 to 8 March 2003. Stations 908 (39°08′N, 76°20′W), 818 (38°18′N, 76°17′W), and 707 (37°07′N, 76°07′W) were chosen to represent the northern, middle, and southern areas of the bay, which covered the salinity gradient in the Chesapeake Bay (Fig. 1). The Chesapeake Bay is a partially mixed estuary, and the bottom layers are anoxic in the summer. In order to evaluate primarily the diversity in the estuarine transition, we used near-surface samples (taken at a 2-m depth) in the mid-stem of the bay. At each station, 500-ml subsamples were taken from a 10-liter Niskin bottle aboard the research vessel Cape Henlopen and filtered immediately through 0.2-μm-pore-size polycarbonate filters (47-mm diameter; Millipore, Billerica, MA). The filters were stored at −20°C prior to DNA extraction. Water temperature, salinity, and the dissolved oxygen concentration were recorded with a Sea-Bird 911 CTD.
FIG. 1.
Map of the Chesapeake Bay showing sampling stations (modified from reference 72 with permission of the publisher).
Chl a and nutrients analyses.
Chlorophyll a (Chl a) data were only collected during the March 2003 cruise and were kindly provided by Wayne Coats of the Smithsonian Environmental Research Center. Duplicate samples (100 ml) from each station were vacuum filtered (<150 mm Hg) onto 25-mm Whatman GF/C filters, and Chl a was extracted in 90% acetone for 24 h at 4°C in the dark. Chl a concentration was determined fluorometrically with a Turner Designs 10-AU fluorometer. Concentrations of inorganic nutrients, including ammonia, nitrite and nitrate, and phosphate, were determined with a Technicon AutoAnalyzer II at the Horn Point Analytical Services Laboratory (www.hpl.umces.edu/services/as.html). The analysis conformed to the standard methods for chemical analysis of water and wastes proposed by the U.S. Environmental Protection Agency (79).
Synoptic data.
The Chesapeake Bay is a highly monitored system, and we were able to retrieve synoptic data from a variety of online data repositories. Historical salinity values were retrieved from the Chesapeake Bay Program (CBP) water quality (WQ) database (http://www.chesapeakebay.net/wqual.htm). Monthly to biweekly Chl a concentrations estimated from in vivo fluorescence, as well as cell extracts, were retrieved from the CBP-WQ and fluorescence databases. Although the CBP stations do not have the exact same coordinates as our stations, they are within 2 to 3 km, mostly on the main axis of the bay. Thus, for clarity, we will refer hereafter to CBP southern bay stations CB7.3, CBI-707O, and CBI-707P interchangeably as station 707; to middle bay stations CB5.1 and CBI-818P interchangeably as station 818; and to northern bay stations 3.2 and CBI-908 interchangeably as station 908.
In order to increase the temporal resolution of Chl a around the sampling times, 8-day, 9-km resolution, level 3, merged aqua-MODIS/SeaWIFS remote-sensed Chl a concentrations were retrieved from the National Aeronautics and Space Administration OceanColor Web (http://oceancolor.gsfc.nasa.gov) (24) and plotted with the SeaDAS package (http://oceancolor.gsfc.nasa.gov/seadas) (7). We are aware of the problems with overestimation of Chl a in estuarine systems by SeaWIFS (32) and likely by aqua-MODIS (L. W. Harding, personal communication), and thus these data were only used in a relative fashion.
Enumeration of bacteria and viral particles.
Subsamples of 50 ml of water were fixed in 1% glutaraldehyde and stored at 4°C. For bacterial cell counts, 1 ml of fixed sample was filtered onto a 0.2-μm-pore-size black polycarbonate membrane filter (Osmonics, Minnetonka, MN). For viral particle counts, 200 μl of fixed sample was mixed with 800 μl of Tris-EDTA-sucrose buffer and filtered onto a 0.02-μm-pore-size 25-mm Anodisc membrane filter (Whatman, Maidstone, United Kingdom). Samples on filters were stained with 2.5× SYBR gold solution for 15 min in the dark as previously described (16). Viral particles were enumerated under blue excitation (485 nm) on a Zeiss Axioplan epifluorescence microscope (Zeiss, Jena, Germany). At least 200 bacterial cells or viral particles were counted per sample.
Extraction of nucleic acids.
Bacterial genomic DNA was extracted by a phenol-chloroform protocol as previously described (45). DNA concentration was measured by determining absorbance at 260 nm with a SmartSpec 3000 spectrophotometer (Bio-Rad, Hercules, CA) and assuming that 50 ng/μl corresponds to an absorbance of 1 optical density unit.
Clone library construction.
Clone libraries containing a large portion of the rRNA operon (16S rRNA gene-ITS-23S rRNA) of bacterioplankton from the six samples described above were constructed by PCR with primers 16S-27F (26) and 23S-1933R (5) (see Table S1 in the supplemental material) as previously described (77), except that (i) Platinum HIFI polymerase mix (Invitrogen, Carlsbad, CA) was used and provided hotstart amplification, (ii) PCR products were A tailed with the QIAGEN A addition kit (QIAGEN, Chatsworth, CA), and (iii) products were cloned with the TOPO TA cloning kit (Invitrogen, Carlsbad, CA) by following the manufacturer's instructions. A total of 576 clones from six libraries were picked for further analysis.
Library screening by ITS-LH-PCR and sequencing.
For two libraries, CB1 and CB2, clones were grown overnight in U-bottom microtiter plates (Corning, Acton, MA) and cells were pelleted at 3,000 rpm for 10 min in a Legend T plate centrifuge (Sorvall, Asheville, NC). Plasmids were isolated by a standard alkaline-lysis protocol (6) with a Hydra 96 microfluidic dispenser (Matrix Co., Hudson, NH). For the remaining libraries, 50 μl of cells grown overnight were pelleted in 96-well PCR plates as described above, the supernatant was withdrawn, and the cells were resuspended in 20 μl of 10× Platinum Taq PCR buffer. The cells were lysed for 5 min at 94°C, cell debris was pelleted as described above, and 1 μl of the supernatant was used for ITS-LH-PCR.
ITS-LH-PCR was performed as previously described (75), with some modifications. Briefly, for 96 clones of each library, two separate reaction mixtures were set up with different primer pairs; 6-carboxyfluorescein-labeled 16S-1406F (47) and unlabeled 23S-66R were used to amplify the ITS region, and 6-carboxy-2′,4,4′,5′,7,7′-hexachlorofluorescein-labeled 16S-1406F and unlabeled tRNAalaR were used to amplify the tRNA fragment (47, 75) (see Table S1 in the supplemental material). After the analysis of CB1 and CB2 revealed that a number of clones did not produce an ITS amplicon because of mismatches to Actinobacteria and Bacteroidetes by the original 23S-66R primer, modified 23S-66R primer versions targeting these groups were designed. Thus, the new 23S-66R primer represents a mixture of these different primer versions (see Table S1 in the supplemental material).
Labeled fragments were discriminated with an Applied Biosystems 3100 Genetic Analyzer (Applied Biosystems, Foster City, CA) in Genescan mode. Sizes of ITS and tRNA fragments were determined with the Genescan software (Applied Biosystems) and the GS2500 size standard (Applied Biosystems). Escherichia coli ITS and tRNA fragments amplified from the residual E. coli genomic DNA during the template preparations were used as positive controls for PCRs. The phylogenetic origin of clones represented by different combinations of fragment pairs was determined in comparison to previously measured fragment sizes (75). Clones representing novel combinations, as well as those without amplified fragments with both primer pairs, were identified by sequencing of 16S rRNA genes by the dideoxy termination reaction with the Big Dye V3.1 kit (Applied Biosystems, Foster City, CA). A thorough phylogenetic analysis of the clones retrieved was performed and will be presented elsewhere (J. Kan et al., submitted for publication).
LIBSHUFF and ∫-LIBSHUFF.
In order to compare the compositions of the six clone libraries, LIBSHUFF (71) and ∫-LIBSHUFF (66) analyses were performed with 592 homologous positions between positions 48 and 720 of the E. coli numbering system and the Jukes and Cantor distance metric (43). After we got the unique pair size by ITS-LH-PCR, sequences of five of six clones from randomly chosen operational taxonomic units (OTUs) were compared and high similarity (equal to or greater than 98.7%) was observed within OTUs. Therefore, 98.7% was our cutoff to define our OTUs. Since only a subset of clones belonging to each of the OTUs defined by ITS-LH-PCR was sequenced, virtual sequences were created for nonsequenced clones by assigning (copying) clone sequences in each OTU to nonsequenced clones in the same OTU, observing the criterion that these virtual sequences represented a copy of a clone sequence from the same library.
DGGE.
Partial 16S rRNA genes from each microbial community were amplified by PCR with primers 16S-1070F and 16S-1392R (25, 52) (see Table S1 in the supplemental material). PCR amplicons were subjected to DGGE analysis by a previously described protocol (44). Briefly, PCR products were loaded onto polyacrylamide gels with a gradient of 40 to 55%. Electrophoresis was run at 60°C in 1× TAE buffer and 70 V for 16 h, and the gel was stained with SYBR gold (Invitrogen, Carlsbad, CA). Representative DNA bands were excised from the gel and reamplified, and a second DGGE was performed. PCR products were excised from reamplified bands, purified with the QIAGEN PCR purification kit (QIAGEN, Chatsworth, CA) according to the manufacturer's protocol, and sequenced as described above with primer 16S-1070F.
LH-PCR.
Two hypervariable regions of the 16S rRNA gene (V1, E. coli 16S rRNA gene positions 72 to 101; V2, E. coli 16S rRNA gene positions 176 to 221) were analyzed by LH-PCR (74) with primers 16S-27F (labeled with 6-carboxyfluorescein) and 16S-355R (see Table S1 in the supplemental material). A modified protocol was used in which 0.25 U of Platinum Taq DNA polymerase (Invitrogen) and 5 pmol of each primer were used in 10-μl (final volume) reaction mixtures run with 20 amplification cycles. One-microliter volumes of products were combined with 9 μl of a GeneScan 2500 ROX size standard (5% vol/vol) in highly deionized formamide and denatured at 94°C for 5 min. Amplicons and standards were discriminated in an AB3100 genetic analyzer (Applied Biosystems). Amplicon sizes and relative fluorescence were determined with the Genescan 3.7 software (Applied Biosystems). Peaks with less than five times the baseline fluorescence intensity were excluded from the analysis. The relative abundance of each peak was estimated by dividing the integrated fluorescence of an individual peak by the total integrated fluorescence of all of the peaks.
Diversity analysis.
Clone library coverage (C, the fraction of the population represented by the phylotypes that have been discovered in each clone library) was calculated by the equation C = 1 − (n/N) × 100, where n is the number of unique clones and N is the total number of clones examined (61). OTUs were defined and confirmed by the ITS-LH-PCR data and sequencing of clones with unique paired sizes (with more than 98.7% similarity). Rarefaction curves were interpolated with the freeware program aRarefactWin (38) and the analytical approximation algorithm (40) and 95% confidence intervals (33). The statistical methods used for the estimation of species richness and diversity indices were based on coverage. Coverage-based estimations of species richness, the Shannon-Wiener index (H) and Simpson's index (D) were calculated by the software SPADE (15). For DGGE and LH-PCR, species richness (presence or absence) was estimated on the basis of the number of DGGE bands or LH-PCR peaks.
RESULTS
Distinct environmental conditions between September 2002 and March 2003.
During the two cruises, samples were collected in warm and cold seasons and the average water temperature was 23.8°C in September 2002 and 2.5°C in March 2003 (Table 1). The salinity in September 2002 (15.5 to 27.0 ppt) was higher than in March 2003 (10.0 to 23.0 ppt). Concentrations of nitrate, nitrate, and dissolved oxygen in March 2003 were higher than in September 2002. No significant differences in ammonia and phosphate concentrations were observed between these two cruises (Table 1, paired t test, P = 0.05). Abundances of bacterioplankton, cyanobacteria, and viruses in September 2002 were higher than those in March 2003 (Table 1).
TABLE 1.
Environmental parameters at sampling stations in the Chesapeake Bay
| Date and station | Water temp (°C) | Salinity (ppt) | Dissolved oxygen concn (mg/liter) | Chl a concn (μg/liter) | Ammonia concn (μM) | Nitrite and nitrate concn (μM) | Phosphate concn (μM) | Bacterial abundance (106 cells/ml) | Cyanobacteria (104 cells/ml) | Viral abundance (107 particles/ml) |
|---|---|---|---|---|---|---|---|---|---|---|
| Sept. 2002 | ||||||||||
| 908 | 23.3 | 15.5 | 6.85 | 11.1 | 1.23 | 7.96 | 1.36 | 6.42 ± 0.41a | 23.0 ± 6.7 | 3.78 ± 0.73 |
| 818 | 23.9 | 19.4 | 6.71 | 6.9 | 0.79 | 4.27 | 0.46 | 4.96 ± 0.18 | 28.6 ± 6.2 | 5.38 ± 0.95 |
| 707 | 24.2 | 27 | 6.45 | 3.4 | 0.95 | 1.37 | 0.36 | 4.11 ± 0.2 | 36.3 ± 7.8 | 5.21 ± 0.94 |
| Mar. 2003 | ||||||||||
| 908 | 1.2 | 10 | 14.07 | 41.6 | 1.15 | 42 | 0.58 | 1.24 ± 0.51 | 0.09 ± 0.02 | 0.98 ± 0.25 |
| 818 | 1.8 | 15.8 | 12.08 | 22.5 | 0.59 | 17.6 | 0.48 | 0.57 ± 0.2 | 0.088 ± 0.01 | 0.81 ± 0.14 |
| 707 | 4.4 | 23 | 11.93 | 14.9 | 2.83 | 0.33 | 0.45 ± 0.17 | 0.11 ± 0.03 | 0.64 ± 0.1 |
Mean ± standard deviation.
The two sampled seasons represented hydrographically distinct conditions in the Chesapeake Bay. The eastern U.S. region experienced a prolonged drought from 1999 to the end of 2002, which was particularly acute in the summer of 2002 and was followed by the snowiest winter on record in the Baltimore-Washington region (3). The hydrography of the bay in September 2002 reflected these conditions, as salinity values were the highest on record since 1954 in the CBP-WQ data set. In contrast, because of the sustained high flows in late February and early March, the salinity in March 2003 in the CBP-WQ database was 2 to 5 ppt lower than normal.
Phytoplankton also showed distinct distributions between the two seasons and among the three sampling stations. On average, the concentration of Chl a in March 2003 was nearly fourfold higher than that in September 2002 (Table 1). In general, Chl a values decreased from the northern bay to the southern bay, following the same trend as inorganic nutrients (ammonia, nitrate, and phosphate). Satellite imagery and CBP-WQ data showed that phytoplankton at station 908 in March 2003 was in an early bloom stage, while at stations 818 and 707 the main spring bloom occurred later in the season (see Fig. S1 and S2 in the supplemental material). Total microphytoplankton counts reached 2.5 × 104 cells/ml, mostly dominated by diatoms (1.4 ×104 cells/ml) and dinoflagellates (1.0 × 104 cells/ml), in the northern bay (Table 2). Diatoms accounted for approximately 88% of the total microphytoplankton in the middle bay (station 804) and continued to dominate the phytoplankton community (ca. 84%) in the southern bay (Table 2).
TABLE 2.
Microscopic counts of microphytoplankton at the northern, middle, and southern Chesapeake Bay stations during the September 2002 and March 2003 cruises
| Phytoplankton | Microphytoplankton count (cells/ml)
|
|||||
|---|---|---|---|---|---|---|
| September 2002
|
March 2003
|
|||||
| Station 858a | Station 818 | Station 707 | Station 858a | Station 818 | Station 707 | |
| Diatoms | 3,691 | 2,296 | 390 | 14,085 | 21,744 | 7,571 |
| Dinoflagellates | 128 | 94 | 13 | 10,476 | 1,056 | 1,232 |
| Other | 13 | 0 | 1 | 881 | 2,025 | 264 |
| Total | 3,832 | 2,390 | 404 | 25,442 | 24,825 | 9,067 |
Data for station 908 were not available, and station 858 (38°58′N, 76°23′W) here served as a reference to station 908.
ITS-LH-PCR.
High variability in the length of the ITS region and tRNA content between bacterial clades was observed (see Table S2 in the supplemental material). Sizes of ITS fragments varied significantly among the typical marine, freshwater, and estuarine bacterioplankton groups and ranged from 347 bp for planktonic marine Actinobacteria to 1,275 bp for Ahrensia sp. strain DFL-42. Sizes of tRNA fragments ranged from 260 bp for unclassified Bacteroidetes strain CB11B01 to 655 bp for the deltaproteobacterium Desulfotalea arctica (see Table S2 in the supplemental material). Very few overlaps occurred among members of different major groups, and this allowed a putative phylogenetic identification of clones with combined ITS and tRNA lengths. Genes coding tRNA-alanine were absent in several groups, including SAR86, SAR116, and Actinobacteria. In some cases, tRNA-alanine-encoding regions were found after sequencing but no corresponding ITS fragments were detected, likely because of primer mismatch. The modified versions of primer 23S-66R (including 23S-66R-Actino and 23S-66R-Bactero; see Table S1 in the supplemental material) greatly improved the detection of clones associated with Bacteroidetes and Actinobacteria, and thus we used these primers to analyze all of the libraries and we recommend its use for future analyses.
ITS-LH-PCR fragment sizes of bacterioplankton with marine origin agreed well with sizes measured in a previous study (76) and estimated sizes from sequences deposited in GenBank. However, Chesapeake Bay clones contained a much broader spectrum of ITS-LH-PCR fragment sizes, indicating a high diversity of bacterioplankton in the Chesapeake Bay estuary. This finding extended the current ITS-LH-PCR size database to estuarine bacterioplankton, allowing putative identification of a wider diversity of environmental clones from aquatic environments without sequencing and will facilitate the future screening of large-insert genomic clone libraries.
Different bacterial community compositions seen in the warm and cold seasons.
The clonal composition and distribution frequency of major bacterial clades (phyla and classes) varied considerably between the warm and cold seasons (Table 3). Alpha-, Beta-, and Gammaproteobacteria accounted for approximately 20.9, 2.0, and 10.2% of the bacterial libraries, respectively, in September 2002 and 49.2, 16.2, and 2.0% in March 2003. The Bacteroidetes phylum accounted for 11.2% of the total number of bacterial clones in September 2002 and 3.6% in March 2003. Cyanobacteria made up 9.4% of the September 2002 libraries but were not detected in March 2003. Actinobacteria accounted for 40.2 and 26.6% of the bacterial clone libraries in September 2002 and March 2003, respectively.
TABLE 3.
Clone composition and distribution of bacterioplankton ribosomal operon libraries from the Chesapeake Bay
| Bacterial group | No. (%) of clones
|
|||||
|---|---|---|---|---|---|---|
| September 2002
|
March 2003
|
|||||
| Station 908, CB01 | Station 804, CB11 | Station 707, CB22 | Station 908, CB31 | Station 804, CB41 | Station 707, CB51 | |
| Alphaproteobacteria | 13 (14.3) | 18 (21.7) | 23 (26.7) | 51 (60.0) | 33 (38.8) | 40 (48.8) |
| SAR11 | ||||||
| SAR11-I | 8 (8.8) | 7 (8.4) | 9 (10.5) | |||
| SAR11-II | 3 (3.3) | 2 (2.4) | 5 (5.8) | |||
| SAR11-III | 2 (2.4) | 1 (1.2) | ||||
| Rhodospirillalles | ||||||
| SPOTSAUG01_5m94 | 1 (1.2) | 1 (1.2) | ||||
| SAR116-IIIa | 3 (3.5) | |||||
| Roseobacter | ||||||
| Chesapeake Roseobacter I (ChesI)b | 1 (1.1) | 5 (6.0) | 4 (4.7) | |||
| Chesapeake Roseobacter II (ChesII)b | 1 (1.2) | 1 (1.2) | 5 (6.1) | |||
| Chesapeake Roseobacter III (ChesIII)b | 2 (2.4) | 5 (5.9) | 6 (7.3) | |||
| Chesapeake Roseobacter IV (ChesIV)b | 4 (4.7) | 5 (5.9) | 2 (2.4) | |||
| Chesapeake Roseobacter V (ChesV)b | 1 (1.1) | 1 (1.2) | ||||
| DG1128 | 2 (2.4) | 2 (2.4) | 1 (1.2) | |||
| GAI-37 | 1 (1.2) | 1 (1.2) | 3 (3.7) | |||
| AS26 | 1 (1.2) | |||||
| Slope strain DI4b | 5 (5.9) | 14 (16.5) | 13 (15.9) | |||
| Arctic sea ice strain ARK9990b | 15 (17.6) | 2 (2.4) | ||||
| Sulfitobacter mediterraneus | 1 (1.2) | 1 (1.2) | ||||
| Rhodobacter | ||||||
| Pseudorhodobacter ferrugineus | 15 (17.6) | 1 (1.2) | 1 (1.2) | |||
| Other | 5 (5.9) | |||||
| Sphigomonas | 1 (1.2) | |||||
| Other | ||||||
| Ahrensia sp. strain DFL-42 | 1 (1.2) | 6 (7.3) | ||||
| Defluvibacter lusatiae | 1 (1.2) | |||||
| Betaproteobacteria | 0 | 3 (3.6) | 2 (2.3) | 14 (16.5) | 21 (24.7) | 6 (7.3) |
| OM 156c | 1 (1.2) | 1 (1.2) | ||||
| OM 43c | 2 (2.4) | |||||
| SPOTS_APR01_5m110 | 1 (1.2) | |||||
| GKS16d | 1 (1.2) | |||||
| Polynucleobacter necessariusd | 3 (3.5) | |||||
| GKS98d | 10 (11.8) | 21 (24.7) | 6 (7.3) | |||
| Gammaproteobacteria | 4 (4.4) | 12 (14.5) | 10 (11.6) | 2 (2.4) | 0 | 3 (3.7) |
| Gamma strain AGG47a | 1 (1.1) | 1 (1.2) | 1 (1.2) | |||
| Novel clade strain SAR86-IV | 1 (1.1) | 4 (4.8) | 3 (3.5) | |||
| SDF1-40 | 1 (1.1) | |||||
| CHAB-III-7 | 1 (1.2) | |||||
| OM60c | 1 (1.2) | |||||
| SPOTSOCT00_5m87 | 1 (1.2) | |||||
| Marine gamma strain HTCC2188 | 1 (1.2) | 1 (1.2) | ||||
| Acinetobacter junii | 1 (1.2) | |||||
| RedeBAC7D11 | 1 (1.2) | |||||
| KTC1119a | 1 (1.2) | |||||
| SAR86-II | 2 (2.3) | |||||
| Pseudomonas syringae | 1 (1.2) | |||||
| Psychrobacter submarinus | 1 (1.2) | |||||
| Gamma strain Sva0091 | 1 (1.2) | |||||
| Psychromonas antarctica | 1 (1.2) | |||||
| Unidentified Gammaproteobacteria | 1 (1.1) | 3 (3.6) | 1 (1.2) | |||
| Deltaproteobacteria | 1 (1.1) | 1 (1.2) | 2 (2.3) | 0 | 0 | 1 (1.2) |
| Bacteroidetes | 11 (12.1) | 12 (14.5) | 6 (7.0) | 2 (2.4) | 4 (4.7) | 3 (3.7) |
| Flavobacteria | ||||||
| Flavobacteriaceae UC1 | 4 (4.4) | |||||
| Unidentified strain CB22B12 | 1 (1.2) | |||||
| Antarctic bacterium strain R-9033 | 1 (1.2) | 1 (1.2) | ||||
| ATAM173_A3 | 1 (1.2) | |||||
| Cellulophaga sp. | 3 (3.5) | |||||
| Sphingobacteria | ||||||
| Unclassified strain CB11C03 | 1 (1.1) | 1 (1.2) | ||||
| Unidentified strain CB22A10 | 1 (1.2) | |||||
| Antarctic bacterium strain R-9286 | 1 (1.2) | 1 (1.2) | ||||
| TM18_28 | 1 (1.2) | |||||
| Unclassified Bacteroidetes | ||||||
| Unclassified strain CB11B01 | 1 (1.1) | 6 (7.2) | ||||
| Unidentified strain CB11D01 | 2 (2.2) | 2 (2.4) | 1 (1.2) | |||
| RW262e | 3 (3.3) | 1 (1.2) | ||||
| Bacteroidetes strain OM 273 | 2 (2.4) | 1 (1.2) | ||||
| Bacteroidetes strain AGG58e | 1 (1.2) | |||||
| Unidentified strain CB22H06 | 1 (1.2) | |||||
| Cyanobacteria/Synechococcus | 5 (5.5) | 12 (14.5) | 7 (8.1) | 0 | 0 | 0 |
| Plastids | 0 | 0 | 8 (9.3) | 0 | 1 (1.2) | 0 |
| Actinobacteria | 57 (62.6) | 22 (26.5) | 27 (31.4) | 15 (17.6) | 25 (29.4) | 27 (32.9) |
| Freshwater acIf | 3 (3.3) | 4 (4.8) | 2 (2.4) | 11 (12.9) | 6 (7.3) | |
| Freshwater acIIf | 7 (7.7) | 1 (1.2) | 9 (10.6) | 5 (5.9) | 5 (6.1) | |
| Freshwater acIIIf | 2 (2.2) | 1 (1.2) | 4 (4.7) | 2 (2.4) | 8 (9.4) | 13 (15.9) |
| Freshwater acIVf | 7 (7.7) | 2 (2.4) | 1 (1.2) | 1 (1.2) | 2 (2.4) | |
| Plankton marine Actinobacterium | 37 (40.7) | 12 (14.5) | 23 (26.7) | |||
| Sediment marine Actinobacterium | 2 (2.4) | |||||
| Novel Actinobacterium strain CB31D05 | 1 (1.1) | 1 (1.2) | 1 (1.2) | |||
| Fibrobacters | 0 | 1 (1.2) | 0 | 0 | 0 | 0 |
| Verrucomicrobia | 0 | 2 (2.4) | 1 (1.2) | 1 (1.2) | 1 (1.2) | 2 (2.4) |
| Total valid clones | 91 | 83 | 86 | 85 | 85 | 82 |
| Chimeras or short inserts | 5 | 13 | 10 | 11 | 11 | 14 |
More dramatic shifts in community composition between the cold and warm seasons were seen at the subgroup level (Table 3). Clones associated with the SAR11 clade were only present in libraries constructed from September 2002 samples. Five novel subclusters of roseobacters (ChesI to -V; Kan et al., submitted) were found in the Chesapeake Bay. Subclusters ChesII to -IV were only present in March 2003, while ChesI and V were only found in September 2002 (Table 3). Clones associated with Beta- and Gammaproteobacteria and the Bacteroidetes group also exhibited distinct distribution patterns in the cold and warm seasons. Freshwater-like Actinobacteria made up 97% of the total Actinobacteria group in March 2003. In contrast, planktonic marine Actinobacteria represented up to 68% of the total number of Actinobacteria clones in September 2002 (Table 3). Of the 85 clones recovered at station 908 in the cold season (March 2003), 15 were closely related to strain ARK9990, which was isolated from Arctic Sea ice (11), and 20 were closely related to a psychrophilic species, Pseudorhodobacter ferrugineus (64) (Table 3). These two phylotypes were prevalent in the northern bay in the cold season but were not detected in the warm season.
Low spatial variations in bacterial communities.
Despite the strong environmental gradients, LIBSHUFF and ∫-LIBSHUFF analyses indicated low spatial heterogeneity of the bacterial community composition in the Chesapeake Bay. Overall, the results of LIBSHUFF and ∫-LIBSHUFF were comparable except for one case (March 2003, station 908 versus station 818), where LIBSHUFF analysis indicated that two libraries were different while ∫-LIBSHUFF did not (Table 4). In September 2002, the bacterial composition of the northern bay (station 908) was not significantly different from that of the middle bay (station 818) (both P values, 0.2995 and 0.0723, are greater than the critical P value, 0.0085), while that of the southern bay (station 707) was significantly different from those of the other two stations (both P values, 0.0004 and 0.0073, are less than the critical P value, 0.0085). Likely the presence of some clones associated with Gammaproteobacteria (such as Acinetobacter junii, RedeBAC7D11, KTC1119, and SAR86-II) and Bacteroidetes, as well as the absence of freshwater Actinobacteria I, II (80), and IV (Kan et al., submitted), contributed to this difference (Table 3). In March 2003, the middle bay (station 818) and the southern bay (station 707) were not significantly different (both P values, 0.1585 and 0.7037, are greater than the critical P value), while the northern bay (station 908) was significantly different (P = 0.05, Table 4) from the middle and southern bay stations (both P values, 0.0011 and 0.0000, are less than the critical value). In March 2003, the predominance of members of the Rhodobacter (P. ferrugineus) and Roseobacter (Arctic Sea ice ARK9990) clades in the northern bay (each group made up 18% of the clone libraries, Table 3) contributed to the spatial difference seen in the LIBSHUFF and ∫-LIBSHUFF analyses. Gammaproteobacteria and Bacteroidetes in the northern bay were also different from those in the middle and southern bay (Table 3).
TABLE 4.
P values of pairwise comparisons of Chesapeake Bay clone libraries by LIBSHUFFa
| Analysis method and station no. | September 2002
|
March 2003
|
||||
|---|---|---|---|---|---|---|
| Station 908 | Station 818 | Station 707 | Station 908 | Station 818 | Station 707 | |
| LIBSHUFF | ||||||
| 908 | NAc | 0.1320 | 0.0010b | NA | 0.0010b | 0.0001b |
| 818 | 0.0530 | NA | 0.0050b | 0.0460 | NA | 0.0230 |
| 707 | 0.0490 | 0.3990 | NA | 0.0001b | 0.0220 | NA |
| ∫-LIBSHUFF | ||||||
| 908 | NA | 0.1520 | 0.0005b | NA | 0.0242 | 0.0000b |
| 818 | 0.0500 | NA | 0.0085b | 0.0201 | NA | 0.1566 |
| 707 | 0.0545 | 0.5390 | NA | 0.0008b | 0.7940 | NA |
Clonal compositions from different seasons were distinct, and thus LIBSHUFF and ∫-LIBSHUFF were only performed between libraries from the same season.
Libraries are significantly different if equal or below the critical P value for three paired comparisons (0.0085).
NA, not applicable.
Spatial and temporal patterns revealed by DGGE and LH-PCR.
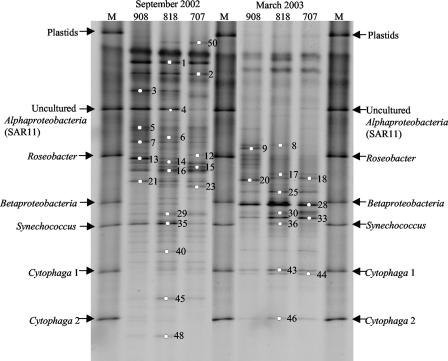
Distinct seasonal patterns of bacterioplankton populations were also seen on the basis of the DGGE analysis (Fig. 2). The majority of the bands present in September 2002 were absent in March 2003, and vice versa. However, within the same cruise, the northern, middle, and southern bay samples had many bands in common. Alphaproteobacteria (bands 1, 2, 3, 13, 29, and 40), Gammaproteobacteria (bands 7, 12, 14, and 21), Cyanobacteria (bands 15 and 35), and Actinobacteria (bands 16 and 48) were present in September 2002, while Alphaproteobacteria (bands 8, 9, 20, 30, and 33), Betaproteobacteria (bands 17 and 18), and Actinobacteria (bands 28, 43, and 44) were commonly seen in March 2003. Although Alphaproteobacteria and Actinobacteria were present in both seasons, the band positions and sequences were different (44).
FIG. 2.
DGGE fingerprints of bacterial communities from the Chesapeake Bay. Representative bands were excised and sequenced (44). A total of 53 bands were excised, and the missing band numbers (4, 10, 11, 19, 22, 24, 26, 27, 31, 32, 34, 37 to 39, 41, 42, 47, 49, and 51 to 53) are from other sampling months (44). The markers (M) covered a broad range of GC contents and represent a mixture of bands excised and reamplified from other environmental samples.
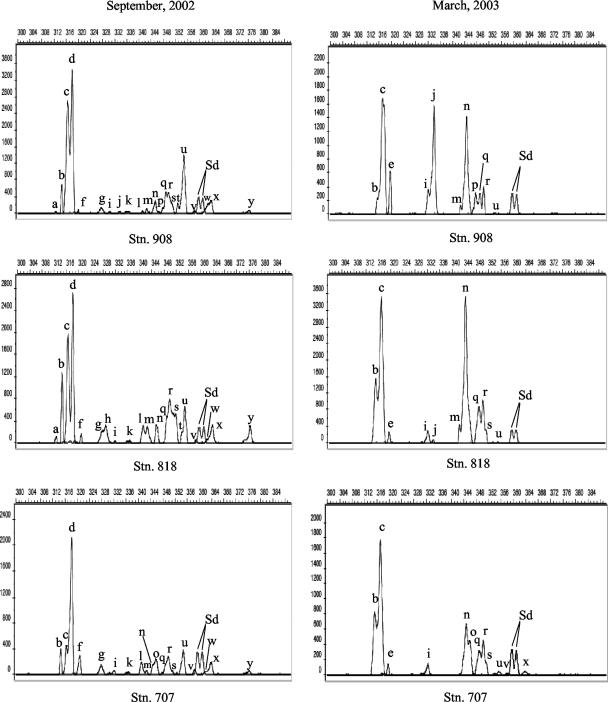
In agreement with clone library and DGGE analyses, samples collected in different regions during the same cruise showed similar LH-PCR electropherogram profiles (Fig. 3). LH-PCR yielded more peaks in September 2002 than in March 2003 (Fig. 3). On the basis of the calculated values of the LH from clones sequenced in this study and those previous published (60, 74), major groups and their relative abundances were putatively assigned to each peak. Overall, the LH of PCR products varied from 313 to 376 bp (Table 5). While it was difficult to assign origins to the peaks because of size overlaps, semiquantitative information could be obtained on the basis of the presence and relative intensities of the peaks identified (Table 5). For instance, peaks assigned to SAR11-IA and planktonic marine Actinobacteria had higher relative fluorescence intensities in September 2002 than in March 2003 while peaks assigned to the Arctic Sea ice ARK9990 and Roseobacter VI groups were exclusively found in March 2003.
FIG. 3.
LH-PCR analysis of bacterioplankton communities in the Chesapeake Bay. The x axis shows peak size in base pairs, and the y axis shows peak intensity in relative fluorescence units. Peaks a to y represent different bacterial groups and subgroups. Sd, peaks of size standards.
TABLE 5.
Analysis of LH-PCR amplicon sizes, identities, and percent peak areas
| Amplicon | Size (bp) | Phylogenetic group(s) | Taxonomic affiliationa | % Peak area
|
|||||
|---|---|---|---|---|---|---|---|---|---|
| September 2002
|
March 2003
|
||||||||
| Station 908 | Station 818 | Station 707 | Station 908 | Station 818 | Station 707 | ||||
| a | 313 | Synechococcus | C | 0.8 | 1.3 | ||||
| b | 314 | Roseobacter Ches I, II, and IV and fibrobacters | α, F | 5.9 | 10.8 | 6.5 | 3.2 | 12.7 | 15.2 |
| c | 316 | Ches I and II, DG1128, AS26 Rhodobacter, slope strain D14, Sulfitobacter, SAR11-II and -III, and SAR116 | α | 23.2 | 16.8 | 7.5 | 23.7 | 29.2 | 32.5 |
| d | 317 | SAR11-IA | α | 29.9 | 23.4 | 35.3 | |||
| e | 318 | Arctic sea ice ARK 9990 and GAI-37 | α | 8.8 | 2.1 | 2.6 | |||
| f | 319 | Plastids | P | 0.9 | 1.4 | 4.9 | |||
| g | 328 | Rhodospirillalles | α | 1.3 | 3.6 | 4.5 | |||
| h | 329 | Rhodospirillalles | α | 2.7 | |||||
| i | 330 | Ahrensia sp. strain DFL-42 | α | 0.2 | 0.4 | 2.2 | 5.1 | 2.5 | 2.7 |
| j | 331 | Pseudorhodobacter | α | 0.6 | 22.3 | 1.2 | |||
| k | 334 | Unknown | ND | 1.1 | 1.0 | 1.0 | |||
| l | 339 | Gamma | γ | 0.6 | 2.7 | 3.3 | |||
| m | 340 | Delta | δ | 1.9 | 3.7 | 2.1 | 1.9 | 3.7 | |
| n | 342 | GKS98, SAR86-II, and acI-B and -C | β, γ, A | 2.4 | 2.7 | 2.6 | 19.9 | 29.4 | 12.3 |
| o | 345 | acI-D, acII-B, and Verrucomicrobiales | A, V | 3.9 | 8.2 | ||||
| p | 346 | acII-A and acIV-A | A | 1.3 | 4.4 | ||||
| q | 347 | Bacteroidetes and Verrucomicrobiales | B, V | 4.5 | 4.1 | 2.9 | 4.3 | 7.3 | 5.9 |
| r | 348 | Bacteroidetes and acIII, IV-C | B, A | 4.3 | 6.7 | 4.5 | 5.5 | 8.7 | 13.0 |
| s | 349 | Bacteroidetes | B | 1.9 | 4.5 | 1.8 | 2.7 | 3.1 | |
| t | 350 | Bacteroidetes and unidentified Gamma | B, γ | 1.7 | 1.6 | ||||
| u | 353 | Plankton marine actinobacteria and acIV-B and -D | A | 12.0 | 5.6 | 6.3 | 0.7 | 0.5 | 0.9 |
| v | 358 | Unknown | ND | 0.4 | 0.4 | 0.8 | 1.1 | ||
| w | 360 | SAR86-IV | γ | 2.0 | 1.2 | 1.7 | |||
| x | 361 | Gamma AGG47, HTCC2188, and Sva0091 | γ | 2.7 | 2.8 | 6.2 | 2.4 | ||
| y | 376 | Unknown | ND | 0.4 | 2.6 | 2.0 | |||
α, β, γ, and δ refer to the subdivisions of the Proteobacteria. C, Cyanobacteria; F, fibrobacters; A, Actinobacteria; B, Bacteroidetes group; P, plastids; ND, not determined.
Diversity estimates.
Coverage analysis of the clone libraries indicated that 60.2 to 75.3% of the actual diversity of Chesapeake Bay bacterioplankton was detected (Table 6). When calculating species richness, the estimated coefficients of variation for the six clone libraries were higher than 0.8 and therefore the nonparametric estimator ACE_1 (modified abundance-based coverage estimator) was applied to estimate species richness (15). The estimated species richness of the six clone libraries ranged from 60 to 96, respectively (Table 6). However, no significant difference was observed between the cold and warm seasons (paired t test, P > 0.05) in terms of overall bacterioplankton complexity, as shown by the Shannon-Wiener index and the Simpson index (Table 6).
TABLE 6.
Diversity analyses of Chesapeake Bay bacterioplankton by clone library, DGGE, and LH-PCR
| Station | Date | Clone library | Phylotype richness | % Coverage | Estimated richness (ACE_1) | Shannon-Wiener index | Simpson's index | DGGE band richness | LH-PCR peak richness |
|---|---|---|---|---|---|---|---|---|---|
| 908 | Sept. 2002 | CB01 | 26 | 71.4 | 59.5 (23.9)a | 2.74 (0.36) | 0.19 (0.04) | 37 | 22 |
| 804 | Sept. 2002 | CB11 | 33 | 60.2 | 94.2 (38.6) | 3.42 (0.19) | 0.07 (0.01) | 45 | 21 |
| 707 | Sept. 2002 | CB22 | 31 | 64 | 95.5 (42.8) | 3.22 (0.26) | 0.10 (0.03) | 37 | 19 |
| 908 | Mar. 2003 | CB31 | 26 | 69.4 | 82.6 (40.3) | 3.02 (0.17) | 0.09 (0.02) | 28 | 11 |
| 804 | Mar. 2003 | CB41 | 21 | 75.3 | 56.7 (29.1) | 2.73 (0.16) | 0.11 (0.02) | 32 | 11 |
| 707 | Mar. 2003 | CB51 | 26 | 68.3 | 87.9 (45.7) | 3.05 (0.16) | 0.08 (0.02) | 29 | 12 |
Standard errors for the estimates are in parentheses.
The total number of DGGE bands ranged from 28 to 45, while LH-PCR peak richness ranged from 11 to 22 (Table 6). In contrast to the clone library analysis, both DGGE and LH-PCR analyses showed significantly higher richness in September 2002 than in March 2003 (paired t test, P = 0.027 for DGGE and P = 0.001 for LH-PCR).
DISCUSSION
Cold versus warm bacterial populations in the bay.
The clone library analysis of Chesapeake Bay bacterioplankton added significant new information to the current knowledge of the population structure and seasonal variations of estuarine planktonic bacteria. Alphaproteobacteria was one of the predominant bacterial groups in the Chesapeake Bay and was composed of three major subgroups, SAR11, Roseobacter, and Rhodobacter. SAR11-related bacteria are known to be abundant and ubiquitous in various marine environments (27, 50). SAR11-related clones appeared in the bay only in the warm season, and this was in line with our previous observations based on the DGGE patterns in the Baltimore Inner Harbor and the Chesapeake Bay (44, 45). This observation is also consistent with previous studies of other estuarine ecosystems, with the SAR11 group in very low abundance or absent in winter and early spring (20, 21, 35, 51). Furthermore, populations of SAR11 at the Bermuda Atlantic Time-Series study site (50, 51) increased during the summer and decreased during the winter. Finally, a previous phylogenetic analysis of ITS regions separated the SAR11 clade into distinct clusters that were associated with temporal (e.g., temperature) but also geographic variations in environmental parameters (13). Overall, these results suggest that the distribution of the SAR11 group in coastal and estuarine regions correlates with but is not necessarily a function of water temperature.
The prevalence of roseobacters in the March 2003 clone libraries suggests that they are able to thrive in cold water (average, 2.5°C throughout the bay). Roseobacters (except for Chesapeake Roseobacter I and V; Kan et al., submitted) made up more than one-third of the winter bacterial community in the bay (Table 3). The occurrence of marine roseobacters in winter is also consistent with our multiyear investigation in the bay based on the DGGE analysis (44). A hallmark of cold adaptation of microorganisms is the presence of proteins containing the cold shock domain (31, 48). Cold shock gene homologues have been found in several Roseobacter genomes, including Silicibacter pomeroy (49), Silicibacter strain TM1040, and Jannaschia sp. strain CCS1 (www.jgi.doe.gov). For instance, Silicibacter strain TM1040 contains two cold shock gene homologues, CSP-A1 and CSP-E (R. Belas, personal communication). In contrast, no cold shock gene homologues are found in some “warm-water species” such as Synechococcus spp. (www.ncbi.nlm.nih.gov/genomes/lproks.cgi). Having these cold shock genes may provide a competitive advantage to marine roseobacters in cold seasons. Moreover, a close relationship between roseobacters and phytoplankton has been reported (4, 30, 62, 81). Many roseobacters are able to turn over dimethylsulfoniopropionate released from microalgae (30, 81) and are commonly found associated with phytoplankton blooms (4, 62). 2003 was a high-flow year in the Chesapeake Bay, and the highest river discharge occurred in March 2003 (2). Large amounts of nutrients carried by river runoffs resulted in a major phytoplankton bloom in the upper bay that was dominated by diatoms and dinoflagellates (Table 2). We propose that the cold adaptation and symbiotic relationship with phytoplankton could partially explain the prevalence of marine roseobacters in the cold season.
It was noteworthy that a high percentage of clones very closely related to Arctic Sea ice strain ARK9990 was seen in the northern bay but not in other stations in March 2003. The northern bay also contained abundant Rhodobacter-related clones in the cold season, and most of these clones were closely affiliated with P. (Agrobacterium) ferrugineus, a psychrophilic bacterium isolated from deep sediments in the North Atlantic Ocean (64). Our previous studies in the Baltimore Inner Harbor, located in the northern Chesapeake Bay area, also identified many winter bacterial isolates that were closely related to other psychrophilic bacteria (45). The retrieval of these putative psychrophilic bacteria in the upper Chesapeake Bay during the winter was intriguing, as is the fate of these putative psychrophiles in summer. Whether these organisms are present in very low abundances that typically escape detection by PCR or are preserved in sediments is a subject for future studies.
Cyanobacterial clones (mostly marine Synechococcus) were detected in September 2002 but not in March 2003, suggesting that these unicellular cyanobacteria are adapted to warm seasons. Concentrations of unicellular cyanobacteria in the Chesapeake Bay are typically low (<103 cells/ml) in winter and high (ca. 105 cells/ml) in summer, and the absence of cyanobacterial clones in March 2003 could be due to the low cyanobacterial abundance (K. Wang and F. Chen, unpublished data). On the other hand, unicellular cyanobacteria were important components of the Chesapeake Bay bacterial communities during the warm season. Cyanobacterial clones accounted for 6 to 15% of the September clone libraries, which is consistent with the fact that picocyanobacteria represented 3.6 to 14.1% of the total bacterial counts in September 2002 samples (Table 2). Chesapeake Bay picocyanobacteria are known to be dominated by a unique group of estuary-adapted Synechococcus bacteria, which are different from oceanic Synechococcus bacteria (17, 18).
Lower spatial than temporal heterogeneity in the Chesapeake Bay.
The LIBSHUFF and ∫-LIBSHUFF analyses showed that the northern bay in March 2003 and the southern bay in September 2002 contained bacterial communities that were different from the other two samples collected during the same cruise (Table 4), even though these differences were not pronounced on the basis of DGGE and LH-PCR analyses. The strong river runoff and consequent phytoplankton bloom in the upper bay in March 2003 and ocean water influx in September 2002 might be the main causes of these spatial differences in bacterial communities. The high frequency of Betaproteobacteria and freshwater Actinobacteria in March 2003 samples also supports the influence of freshwater runoff, while the predominance of Gammaproteobacteria and planktonic marine Actinobacteria indicates the effect of ocean water in the southern bay. Spatial variations in bacterial community structure along the salinity gradients of estuaries have been reported (10, 20, 21, 35, 37, 67). The effect of salinity on microbial diversity appears to be more severe in the low-salinity (<5 ppt) or high-salinity (>30 ppt) end, and 5 ppt has been used in the past as a salinity cutoff for separating freshwater and estuarine bacterial communities (36). The salinities of the six samples analyzed in this study ranged from 10 to 27 ppt (Table 1). Although osmotic stress is known to have negative effects on cell survival (8), the salinity range in this study appeared to have a limited impact on estuarine bacterial diversity patterns.
In general, spatial variations in bacterioplankton communities in the Chesapeake Bay were much less pronounced compared to the temporal variations. LIBSHUFF and ∫-LIBSHUFF analyses (Table 4) indicated that clone libraries represented the same bacterial communities in the middle and southern parts of the bay in March 2003, as well as in the northern and middle parts of the bay in September 2002. This low spatial heterogeneity in bacterial community composition, in stations about 100 km apart, in a physically heterogeneous environment such as the Chesapeake Bay was quite remarkable. This could result from long average residence times, from relatively fast water exchange between these stations through estuarine circulation, or more likely from similar selective pressures exerted by similar environmental parameters among these stations. Temperature and daylight duration are obvious examples of parameters similar at these geographic scales, but many others might exist. Long average residence times are again critical to allow sufficient time for these selective forces to homogenize populations in different regions of the bay. The average residence time of the Chesapeake Bay is on the order of several months and is several orders of magnitude longer than the doubling time of bacteria in the bay (54), thus allowing the development of stable distinct bacterial populations in this estuarine system (21).
Methodological considerations.
Clone library analyses of bacterial spatial and temporal variations in the Chesapeake Bay provide useful information on the interactions between the population structure and environmental parameters. However, with the large number of samples collected in a multiple-year investigation, it is difficult to apply clone library analysis to all of the samples. Therefore, linking the limited data on six clone libraries to environmental parameters might bias the views on the environmental effects on bacterial community changes. In order to investigate the interactions between bacterial community variation and key environmental parameters, we previously applied DGGE to monitor the multiple-year bacterial community fingerprints. Statistical analyses indicated that the community structure variations significantly correlated with water temperature and Chl a, while inorganic nutrients, dissolved oxygen, and viral abundance also contributed (44).
16S-23S rRNA ITS regions display significant heterogeneity in both length and nucleotide sequence, providing higher taxonomic resolution than 16S rRNA genes, and thus have been extensively used to distinguish closely related species or even strains of single species (18, 41, 63). Chesapeake Bay Synechococcus clones, members of different subclades of freshwater Actinobacteria, the Roseobacter clade, and different phylotypes of Gammaproteobacteria and Bacteroidetes could be easily distinguished by ITS-LH-PCR fragments sizes (see Table S2 in the supplemental material), and the phylogeny of ITS sequences of the Synechococcus clones has previously confirmed the divergence between the marine cluster A and B Synechococcus clades (18). However, variable paired sizes of ITS-LH-PCR were commonly observed in very closely related (>98% similarity) phylotypes on the basis of 16S rRNA gene sequences (i.e., Roseobacter DI4 clade, Betaproteobacteria GKS98 clade). The existence of multiple copies of rRNA operons with variable spacer regions in the genomes of these organisms or relatively higher rates of evolution of spacer regions in these groups could explain this observation. Furthermore, not all of the known bacteria contain typically organized rRNA operons. Thus, the operon cloning and ITS-LH-PCR prescreening missed those bacteria lacking the typical operon structures, which might have underestimated the bacterial diversity in the Chesapeake Bay, although comparison to a previous study that used DGGE (44) indicates that this was not likely.
In order to evaluate the effect of virtual sequences in LIBSHUFF and ∫-LIBSHUFF, we estimated the minimal similarity between different sequences in each of the OTUs. The minimal similarity measured (98.7%) indicates that the virtual sequences should only affect the calculation of homologous coverage by LIBSHUFF and ∫-LIBSHUFF above 98.7% sequence similarity. Heterologous coverage, on the other hand, would be impacted less by virtual sequences, since they represent a copy of a clone sequenced from each individual library and thus are less likely to be identical to virtual sequences from other libraries. Delta-C values would, in turn, be overestimated by LISBSUFF and ∫-LIBSHUFF above a 98.7% distance, and the P value for our analyses would be lower (W. B. Whitman, personal communication). For libraries to be statistically significantly different, the lower of the two P values from each pairwise comparison has to be equal to or lower than the critical P value. Most of the P values for the spatial pairwise comparisons in our analyses were higher than the critical P value, even with the “underestimation” of the P value caused by our virtual sequences. Thus, the virtual sequences should make the libraries appear more different than they really are and the observation of low spatial heterogeneity of Chesapeake Bay bacterioplankton should hold.
The three independent methods (clone library analysis, DGGE, and LH-PCR) were consistent for the major bacterial groups despite the fact that different primers were used for each method. Approximately 70% of the DGGE band sequences were similar to clone sequences in the libraries (data not shown). A major discrepancy between DGGE and clone library analyses was the presence of Planctomycetes in DGGE analysis and not in the clone libraries. The absence of Planctomycetes in clone libraries was likely caused by known mismatches between 16S rRNA genes from this clade and the 27F primer used in PCRs for clone library construction. Nevertheless, it was clear that clone library analysis provided the highest “species” richness among the three methods (Table 6). In order to further quantify the dominance of different phylogenetic groups, quantitative PCR or specifically designed FISH must be applied to the Chesapeake Bay.
Conclusion.
This study represents the first clone library analysis performed for bacterioplankton in the Chesapeake Bay or any large estuary with a long average residence time. Seasonal and spatial distributions of Chesapeake Bay bacterial populations in many cases reflected environmental fluctuations. Genetic diversity of the microbial community has been explored with a variety of molecular tools, and recently a new view of bacterial diversity has been uncovered by massive pyrosequencing analyses, which allows the exploration of the “rare biosphere” (70). It has been predicted that the number of different microbial taxa in water samples ranges from a few hundred to one million (1, 22, 65). We believe that estuarine bacterial communities are far more complex than what we learned from the clone library analysis and that, in fact, understanding the diversity and ecology of these systems is in its infancy.
Supplementary Material
Acknowledgments
We thank K. E. Wommack for providing the CTD data, D. W. Coats and his colleagues for Chl a and phytoplankton counting data, and the crew of the research vessel Cape Henlopen for sample collection.
We also acknowledge funding support from the National Science Foundation's Microbial Observatories Program (MCB-0132070, MCB-0238515, and MCB-0537041 to F.C.) and Biological Oceanography Program (OCE-0550547 to M.T.S.).
Footnotes
Published ahead of print on 7 September 2007.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Acinas, S. G., V. Klepac-Ceraj, D. E. Hunt, C. Pharino, I. Ceraj, D. L. Distel, and M. F. Polz. 2004. Fine-scale phylogenetic architecture of a complex bacterial community. Nature 430:551-554. [DOI] [PubMed] [Google Scholar]
- 2.Acker, J. 2 November 2005. Drought & deluge change Chesapeake Bay biology. Earth Observatory features. National Air and Space Administration, Washington, D.C. http://earthobservatory.nasa.gov/Study/ChesapeakeBay/.
- 3.Acker, J. G., L. W. Harding, G. Leptoukh, T. Zhu, and S. Shen. 2005. Remotely-sensed chl a at the Chesapeake Bay mouth is correlated with annual freshwater flow to Chesapeake Bay. Geophys. Res. Lett. 32:L05601. doi: 10.1029/2004GL021852. [DOI] [Google Scholar]
- 4.Alavi, M., T. Miller, K. Erlandson, R. Shneider, and R. Belas. 2001. Bacterial community associated with Pfiesteria-like dinoflagellate cultures. Environ. Microbiol. 3:380-396. [DOI] [PubMed] [Google Scholar]
- 5.Amann, R. I., W. Ludwig, and K. H. Schleifer. 1995. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 59:143-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ausubel, F., R. Brent, R. Kingston, D. Moore, J. Seidman, and K. Struhl. 2003. Current protocols in molecular biology online. John Wiley & Sons, Inc., New York, NY.
- 7.Baith, K., R. Lindsay, G. Fu, and C. R. McClain. 2001. Data analysis system for ocean color satellite sensors. EOS 82:202. http://www.agu.org/eos_elec/00289e.html. [Google Scholar]
- 8.Barcina, I., P. Lebaron, and J. VivesRego. 1997. Survival of allochthonous bacteria in aquatic systems: a biological approach. FEMS Microbiol. Ecol. 23:1-9. [Google Scholar]
- 9.Bidle, K. D., and M. Fletcher. 1995. Comparison of free-living and particle-associated bacterial communities in the Chesapeake Bay by stable low-molecular-weight RNA analysis. Appl. Environ. Microbiol. 61:944-952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bouvier, T. C., and P. A. del Giorgio. 2002. Compositional changes in free-living bacterial communities along a salinity gradient in two temperate estuaries. Limnol. Oceanogr. 47:453-470. [Google Scholar]
- 11.Brinkmeyer, R., K. Knittel, J. Jurgens, H. Weyland, R. Amann, and E. Helmke. 2003. Diversity and structure of bacterial communities in arctic versus Antarctic pack ice. Appl. Environ. Microbiol. 69:6610-6619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Britschgi, T. B., and S. J. Giovannoni. 1991. Phylogenetic analysis of a natural marine bacterioplankton population by rRNA gene cloning and sequencing. Appl. Environ. Microbiol. 57:1707-1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brown, M. V., and J. A. Fuhrman. 2005. Marine bacterial microdiversity as revealed by internal transcribed spacer analysis. Aquat. Microb. Ecol. 41:15-23. [Google Scholar]
- 14.Bue, C. D. 1968. Monthly surface-water inflow to Chesapeake Bay. Open-file report. U.S. Geological Survey, Arlington, VA.
- 15.Chao, A., and T.-J. Shen. Last updated November 2006. Program SPADE (Species Prediction And Diversity Estimation). Program and user's guide. http://chao.stat.nthu.edu.tw.
- 16.Chen, F., J. R. Lu, B. Binder, and R. E. Hodson. 2001. Enumeration of viruses in aquatic environments using SYBR gold stain: application of digital image analysis and flow cytometer. Appl. Environ. Microbiol. 67:539-545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen, F., K. Wang, J. Kan, D. Bachoon, J. Lu, S. Lau, and L. Campbell. 2004. Phylogenetic diversity of Synechococcus in the Chesapeake Bay revealed by ribulose-1,5-bisphosphate carboxylase-oxygenase (RuBisCO) large subunit gene (rbcL) sequences. Aquat. Microb. Ecol. 36:153-164. [Google Scholar]
- 18.Chen, F., K. Wang, J. Kan, M. T. Suzuki, and K. E. Wommack. 2006. Diverse and unique picocyanobacteria in Chesapeake Bay, revealed by 16S-23S rRNA internal transcribed spacer sequences. Appl. Environ. Microbiol. 72:2239-2243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cottrell, M. T., L. A. Waidner, L. Yu, and D. L. Kirchman. 2005. Bacterial diversity of metagenomic and PCR libraries from the Delaware River. Environ. Microbiol. 7:1883-1895. [DOI] [PubMed] [Google Scholar]
- 20.Crump, B. C., E. V. Armbrust, and J. A. Baross. 1999. Phylogenetic analysis of particle-attached and free-living bacterial communities in the Columbia river, its estuary, and the adjacent coastal ocean. Appl. Environ. Microbiol. 65:3192-3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Crump, B. C., C. S. Hopkinson, M. L. Sogin, and J. E. Hobbie. 2004. Microbial biogeography along an estuarine salinity gradient: combined influences of bacterial growth and residence time. Appl. Environ. Microbiol. 70:1494-1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Curtis, T. P., W. T. Sloan, and J. W. Scannell. 2002. Estimating prokaryotic diversity and its limits. Proc. Natl. Acad. Sci. USA 99:10494-10499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ducklow, H. W., and C. A. Carlson. 1992. Oceanic bacterial production. Adv. Microb. Ecol. 12:113-181. [Google Scholar]
- 24.Feldman, G. C., and C. R. McClain. 2 November 2006. OceanColor Web. NASA Goddard Space Flight Center, Greenbelt, MD. http://oceancolor.gsfc.nasa.gov.
- 25.Ferris, M. J., G. Muyzer, and D. M. Ward. 1996. Denaturing gradient gel electrophoresis profiles of 16S rRNA-defined populations inhabiting a hot spring microbial mat community. Appl. Environ. Microbiol. 62:340-346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Giovannoni, S. J. 1991. The polymerase chain reaction, p. 177-203. In E. Stackebrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. John Wiley & Sons, Inc., New York, NY.
- 27.Giovannoni, S. J., and M. Rappe. 2000. Evolution, diversity and molecular ecology of marine prokaryotes, p. 47-84. In D. Kirchman (ed.) Microbial ecology of the oceans. John Wiley & Sons, Inc., New York, NY.
- 28.Glöckner, F. O., B. M. Fuchs, and R. Amann. 1999. Bacterioplankton compositions of lakes and oceans: a first comparison based on fluorescence in situ hybridization. Appl. Environ. Microbiol. 65:3721-3726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.González, J. M., and M. A. Moran. 1997. Numerical dominance of a group of marine bacteria in the α-subclass of the class Proteobacteria in coastal seawater. Appl. Environ. Microbiol. 63:4237-4242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.González, J. M., R. Simó, R. Massana, J. S. Covert, E. O. Casamayor, C. Pedrós-Alió, and M. A. Moran. 2000. Bacterial community structure associated with a dimethylsulfoniopropionate-producing North Atlantic algal bloom. Appl. Environ. Microbiol. 66:4237-4246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Goodchild, A., N. F. W. Saunders, H. Ertan, M. Raftery, M. Guilhaus, P. M. G. Curmi, and R. Cavicchioli. 2004. A proteomic determination of cold adaptation in the Antarctic archaeon, Methanococcoides burtonii. Mol. Microbiol. 53:309-321. [DOI] [PubMed] [Google Scholar]
- 32.Harding, L. W., A. Magnuson, and M. E. Mallonee. 2005. SeaWiFS retrievals of chlorophyll in Chesapeake Bay and the mid-Atlantic bight. Estuar. Coast. Shelf Sci. 62:75-94. [Google Scholar]
- 33.Heck, K. L., Jr., G. V. Belle, and D. Simberloff. 1975. Explicit calculation of the rarefaction diversity measurement and the determination of sufficient sample size. Ecology 56:1459-1461. [Google Scholar]
- 34.Heidelberg, J. F., K. B. Heidelberg, and R. R. Colwell. 2002. Seasonality of Chesapeake Bay bacterioplankton species. Appl. Environ. Microbiol. 68:5488-5497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Henriques, I., M. A. Almeida, M. A. Cunha, and A. Correia. 2004. Molecular sequence analysis of prokaryotic diversity in the middle and outer sections of the Portuguese estuary Ria de Aveiro. FEMS Microbiol. Ecol. 49:269-279. [DOI] [PubMed] [Google Scholar]
- 36.Henriques, I. S., A. Alves, M. Tacao, A. Almeida, A. Cunha, and A. Correia. 2006. Seasonal and spatial variability of free-living bacterial community composition along an estuarine gradient (Ria de Aveiro, Portugal). Estuar. Coast. Shelf Sci. 68:139-148. [Google Scholar]
- 37.Hewson, I., and J. A. Fuhrman. 2004. Richness and diversity of bacterioplankton species along an estuarine gradient in Moreton Bay, Australia. Appl. Environ. Microbiol. 70:3425-3433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Holland, S. 1998. aRarefactWin. University of Georgia, Athens.
- 39.Hollibaugh, J. T., P. S. Wong, and M. C. Murrell. 2000. Similarity of particle-associated and free-living bacterial communities in northern San Francisco Bay, California. Aquat. Microb. Ecol. 21:103-109. [Google Scholar]
- 40.Hurlbert, S. H. 1971. The nonconcept of species diversity: a critique and alternative parameters. Ecology 52:577-586. [DOI] [PubMed] [Google Scholar]
- 41.Jensen, M. A., J. A. Webster, and N. Straus. 1993. Rapid identification of bacteria on the basis of polymerase chain reaction-amplified ribosomal DNA spacer polymorphisms. Appl. Environ. Microbiol. 59:945-952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jonas, R. B., and J. H. Tuttle. 1990. Bacterioplankton and organic carbon dynamics in the lower mesohaline Chesapeake Bay. Appl. Environ. Microbiol. 56:747-757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jukes, T. H., and C. R. Cantor. 1969. Evolution of protein, p. 21-132. In H. N. Munro (ed.), Mammalian protein metabolism. Academic Press, Inc., New York, NY.
- 44.Kan, J., B. C. Crump, K. Wang, and F. Chen. 2006. Bacterioplankton community in Chesapeake Bay: predictable or random assemblages. Limnol. Oceanogr. 51:2157-2169. [Google Scholar]
- 45.Kan, J., K. Wang, and F. Chen. 2006. Temporal variation and detection limit of an estuarine bacterioplankton community analyzed by denaturing gradient gel electrophoresis (DGGE). Aquat. Microb. Ecol. 42:7-18. [Google Scholar]
- 46.Kirchman, D. L., L. Y. Yu, and M. T. Cottrell. 2003. Diversity and abundance of uncultured Cytophaga-like bacteria in the Delaware Estuary. Appl. Environ. Microbiol. 69:6587-6596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lane, D. J. 1991. 16S/23S rRNA sequencing, p. 115-175. In E. Stackebrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. John Wiley & Sons, Inc., New York, NY.
- 48.Methé, B. A., K. E. Nelson, J. W. Deming, B. Momen, E. Melamud, X. Zhang, J. Mouldt, R. Madupu, W. C. Nelson, R. J. Dodson, L. M. Brinkac, S. C. Daugherty, A. S. Durkin, R. T. Deboy, J. F. Kolonay, S. A. Sullivan, L. Zhou, T. M. Davidsen, M. Wu, A. L. Huston, M. Lewis, B. Weaver, J. F. Weidman, H. Khouri, T. R. Utterback, T. V. Feldblyum, and C. M. Fraser. 2005. The psychrophilic lifestyle as revealed by the genome sequence of Colwellia psychrerythraea 34H through genomic and proteomic analysis. Proc. Natl. Acad. Sci. USA 102:10913-10918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Moran, M. A., A. Buchan, J. M. González, J. F. Heidelberg, W. B. Whitman, R. P. Kiene, J. R. Henriksen, G. M. King, R. Belas, C. Fuqua, L. Brinkac, M. Lewis, S. Johri, B. Weaver, G. Pai, J. A. Eisen, E. Rahe, W. M. Sheldon, W. Y. Ye, T. R. Miller, J. Carlton, D. A. Rasko, I. T. Paulsen, Q. H. Ren, S. C. Daugherty, R. T. Deboy, R. J. Dodson, A. S. Durkin, R. Madupu, W. C. Nelson, S. A. Sullivan, M. J. Rosovitz, D. H. Haft, J. Selengut, and N. Ward. 2004. Genome sequence of Silicibacter pomeroyi reveals adaptations to the marine environment. Nature 432:910-913. [DOI] [PubMed] [Google Scholar]
- 50.Morris, R. M., M. S. Rappe, S. A. Connon, K. L. Vergin, W. A. Siebold, C. A. Carlson, and S. J. Giovannoni. 2002. SAR11 clade dominates ocean surface bacterioplankton communities. Nature 420:806-810. [DOI] [PubMed] [Google Scholar]
- 51.Morris, R. M., K. L. Vergin, J. C. Cho, M. S. Rappe, C. A. Carlson, and S. J. Giovannoni. 2005. Temporal and spatial response of bacterioplankton lineages to annual convective overturn at the Bermuda Atlantic Time-Series study site. Limnol. Oceanogr. 50:1687-1696. [Google Scholar]
- 52.Müller, D. G., M. Sengco, S. Wolf, M. Brautigam, C. E. Schmid, M. Kapp, and R. Knippers. 1996. Comparison of two DNA viruses infecting the marine brown algae Ectocarpus siliculosus and E. fasciculatus. J. Gen. Virol. 77:2329-2333. [DOI] [PubMed] [Google Scholar]
- 53.Newton, R. J., A. D. Kent, E. W. Triplett, and K. D. McMahon. 2006. Microbial community dynamics in a humic lake: differential persistence of common freshwater phylotypes. Environ. Microbiol. 8:956-970. [DOI] [PubMed] [Google Scholar]
- 54.Nixon, S. W., J. W. Ammerman, L. P. Atkinson, V. M. Berounsky, G. Billen, W. C. Boicourt, W. R. Boynton, T. M. Church, D. M. Ditoro, R. Elmgren, J. H. Garber, A. E. Giblin, R. A. Jahnke, N. J. P. Owens, M. E. Q. Pilson, and S. P. Seitzinger. 1996. The fate of nitrogen and phosphorus at the land sea margin of the North Atlantic Ocean. Biogeochemistry 35:141-180. [Google Scholar]
- 55.Noble, P. A., K. D. Bidle, and M. Fletcher. 1997. Natural microbial community compositions compared by a back-propagating neural network and cluster analysis of 5S rRNA. Appl. Environ. Microbiol. 63:1762-1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.O'Sullivan, L. A., K. E. Fuller, E. M. Thomas, C. M. Turley, J. C. Fry, and A. J. Weightman. 2004. Distribution and culturability of the uncultivated ‘AGG58 cluster’ of the Bacteroidetes phylum in aquatic environments. FEMS Microbiol. Ecol. 47:359-370. [DOI] [PubMed] [Google Scholar]
- 57.O'Sullivan, L. A., J. Rinna, G. Humphreys, A. J. Weightman, and J. C. Fry. 2005. Fluviicola taffensis gen. nov., sp. nov., a novel freshwater bacterium of the family Cryomorphaceae in the phylum ‘Bacteroidetes’. Int. J. Syst. Evol. Microbiol. 55:2189-2194. [DOI] [PubMed] [Google Scholar]
- 58.Rappé, M. S., and S. J. Giovannoni. 2003. The uncultured microbial majority. Annu. Rev. Microbiol. 57:369-394. [DOI] [PubMed] [Google Scholar]
- 59.Rappé, M. S., P. F. Kemp, and S. J. Giovannoni. 1997. Phylogenetic diversity of marine coastal picoplankton 16S rRNA genes cloned from the continental shelf off Cape Hatteras, North Carolina. Limnol. Oceanogr. 42:811-826. [Google Scholar]
- 60.Rappé, M. S., M. T. Suzuki, K. L. Vergin, and S. J. Giovannoni. 1998. Phylogenetic diversity of ultraplankton plastid small-subunit rRNA genes recovered in environmental nucleic acid samples from the Pacific and Atlantic coasts of the United States. Appl. Environ. Microbiol. 64:294-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ravenschlag, K., K. Sahm, J. Pernthaler, and R. Amann. 1999. High bacterial diversity in permanently cold marine sediments. Appl. Environ. Microbiol. 65:3982-3989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Riemann, L., G. F. Steward, and F. Azam. 2000. Dynamics of bacterial community composition and activity during a mesocosm diatom bloom. Appl. Environ. Microbiol. 66:578-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rocap, G., D. L. Distel, J. B. Waterbury, and S. W. Chisholm. 2002. Resolution of Prochlorococcus and Synechococcus ecotypes by using 16S-23S ribosomal DNA internal transcribed spacer sequences. Appl. Environ. Microbiol. 68:1180-1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ruger, H. J., and M. G. Hofle. 1992. Marine star-shaped-aggregate-forming bacteria: Agrobacterium atlanticum sp. nov.; Agrobacterium meteori sp. nov.; Agrobacterium ferrugineum sp. nov., nom. rev.; Agrobacterium gelatinovorum sp. nov., nom. rev.; and Agrobacterium stellulatum sp. nov., nom. rev. Int. J. Syst. Bacteriol. 42:133-143. [DOI] [PubMed] [Google Scholar]
- 65.Schloss, P. D., and J. Handelsman. 2005. Introducing DOTUR, a computer program for defining operational taxonomic units and estimating species richness. Appl. Environ. Microbiol. 71:1501-1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Schloss, P. D., B. R. Larget, and J. Handelsman. 2004. Integration of microbial ecology and statistics: a test to compare gene libraries. Appl. Environ. Microbiol. 70:5485-5492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sekiguchi, H., M. Watanabe, T. Nakahara, B. Xu, and H. Uchiyama. 2002. Succession of bacterial community structure along the Changjiang River determined by denaturing gradient gel electrophoresis and clone library analysis. Appl. Environ. Microbiol. 68:5142-5150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Selje, N., and M. Simon. 2003. Composition and dynamics of particle-associated and free-living bacterial communities in the Weser estuary, Germany. Aquat. Microb. Ecol. 30:221-237. [Google Scholar]
- 69.Shiah, F.-K., and H. W. Ducklow. 1994. Temperature regulation of heterotrophic bacterioplankton abundance, production, and specific growth rate. Limnol. Oceanogr. 39:1243-1258. [Google Scholar]
- 70.Shiah, F.-K., and H. W. Ducklow. 1994. Temperature and substrate regulation of bacterial abundance, production and specific growth rate in Chesapeake Bay, USA. Mar. Ecol. Prog. Ser. 103:297-308. [Google Scholar]
- 71.Singleton, D. R., M. A. Furlong, S. L. Rathbun, and W. B. Whitman. 2001. Quantitative comparisons of 16S rDNA sequence libraries from environmental samples. Appl. Environ. Microbiol. 67:4373-4376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Smith, D. E., M. Leffler, and G. Mackiernan. 1992. Oxygen dynamics in the Chesapeake Bay—a synthesis of recent research. Maryland and Virginia Sea Grant College Programs, College Park, MD.
- 73.Smith, E. M., and W. M. Kemp. 2003. Planktonic and bacterial respiration along an estuarine gradient: responses to carbon and nutrient enrichment. Aquat. Microb. Ecol. 30:251-261. [Google Scholar]
- 74.Suzuki, M., M. S. Rappe, and S. J. Giovannoni. 1998. Kinetic bias in estimates of coastal picoplankton community structure obtained by measurements of small-subunit rRNA gene PCR amplicon length heterogeneity. Appl. Environ. Microbiol. 64:4522-4529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Suzuki, M. T., O. Beja, L. T. Taylor, and E. F. Delong. 2001. Phylogenetic analysis of ribosomal RNA operons from uncultivated coastal marine bacterioplankton. Environ. Microbiol. 3:323-331. [DOI] [PubMed] [Google Scholar]
- 76.Suzuki, M. T., C. M. Preston, O. Beja, J. R. de la Torre, G. F. Steward, and E. F. DeLong. 2004. Phylogenetic screening of ribosomal RNA gene-containing clones in bacterial artificial chromosome (BAC) libraries from different depths in Monterey Bay. Microb. Ecol. 48:473-488. [DOI] [PubMed] [Google Scholar]
- 77.Suzuki, M. T., L. T. Taylor, and E. F. DeLong. 2000. Quantitative analysis of small-subunit rRNA genes in mixed microbial populations via 5′-nuclease assays. Appl. Environ. Microbiol. 66:4605-4614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Troussellier, M., H. Schafer, N. Batailler, L. Bernard, C. Courties, P. Lebaron, G. Muyzer, P. Servais, and J. Vives-Rego. 2002. Bacterial activity and genetic richness along an estuarine gradient (Rhone River plume, France). Aquat. Microb. Ecol. 28:13-24. [Google Scholar]
- 79.U.S. Environmental Protection Agency. 1983. Method no. 350.1, p. 350-1.1-350-1.4. In Methods for chemical analysis of water and wastes. Report no. EPA-600/4-79-020. Environmental Monitoring and Support Laboratory, U.S. Environmental Protection Agency, Cincinnati, OH.
- 80.Warnecke, F., R. Amann, and J. Pernthaler. 2004. Actinobacterial 16S rRNA genes from freshwater habitats cluster in four distinct lineages. Environ. Microbiol. 6:242-253. [DOI] [PubMed] [Google Scholar]
- 81.Zubkov, M. V., B. M. Fuchs, S. D. Archer, R. P. Kiene, R. Amann, and P. H. Burkill. 2002. Rapid turnover of dissolved DMS and DMSP by defined bacterioplankton communities in the stratified euphotic zone of the North Sea. Deep-Sea Res. Part II Top. Stud. Oceanogr. 49:3017-3038. [Google Scholar]
- 82.Zwart, G., B. C. Crump, M. P. Kamst-van Agterveld, F. Hagen, and S. K. Han. 2002. Typical freshwater bacteria: an analysis of available 16S rRNA gene sequences from plankton of lakes and rivers. Aquat. Microb. Ecol. 28:141-155. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.