Abstract
The influx of enterococcal antibiotic resistance (AR) and virulence genes from ready-to-eat food (RTEF) to the human digestive tract was assessed. Three RTEFs (chicken salad, chicken burger, and carrot cake) were sampled from five fast-food restaurants five times in summer (SU) and winter (WI). The prevalence of enterococci was significantly higher in SU (92.0% of salad samples and 64.0% of burger samples) than in WI (64.0% of salad samples and 24.0% of burger samples). The overall concentrations of enterococci during the two seasons were similar (∼103 CFU/g); the most prevalent were Enterococcus casseliflavus (41.5% of isolates) and Enterococcus hirae (41.5%) in WI and Enterococcus faecium (36.8%), E. casseliflavus (27.6%), and Enterococcus faecalis (22.4%) in SU. Resistance in WI was detected primarily to tetracycline (50.8%), ciprofloxacin (13.8%), and erythromycin (4.6%). SU isolates were resistant mainly to tetracycline (22.8%), erythromycin (22.1%), and kanamycin (13.0%). The most common tet gene was tet(M) (35.4% of WI isolates and 11.9% of SU isolates). The prevalence of virulence genes (gelE, asa1, cylA, and esp) and marker genes for clinical isolates (EF_0573, EF_0592, EF_0605, EF_1420, EF_2144, and pathogenicity island EF_0050) was low (≤12.3%). Genotyping of E. faecalis and E. faecium using pulsed-field gel electrophoresis revealed that the food contamination likely originated from various sources and that it was not clonal. Our conservative estimate (single AR gene copy per cell) for the influx of tet genes alone to the human digestive tract is 3.8 × 105 per meal (chicken salad). This AR gene influx is frequent because RTEFs are commonly consumed and that may play a role in the acquisition of AR determinants in the human digestive tract.
The emergence and spread of antibiotic resistance (AR) determinants are a growing problem in clinical intensive care units, due to the declining number of effective antimicrobial agents for treatment of bacterial diseases (28, 51). The relationship between antibiotic use and the development and spread of AR has been studied extensively in clinical as well as environmental isolates (1, 18, 20, 25, 28, 29, 45).
The microbial community of the human digestive tract, especially of the colon, likely represents an important reservoir of AR genes as well as a site for horizontal intra- and interspecies gene transfer (10, 38, 40). It has been shown that horizontal transfer of AR genes from the ingested bacteria to the microbial community of the human digestive tract is possible and likely represents an important aspect of the ecology of AR determinants (27, 38, 39, 40). The ubiquity of enterococci in mammalian digestive tracts, their medical importance, frequent multiple AR, and great capacity for horizontal gene transfer (16) make the enterococci an ideal bacterial group for investigating the ecology of AR genes. The transfer of the transposon Tn1546 conferring vancomycin resistance from Enterococcus faecalis to a clinical isolate of Staphylococcus aureus (49) highlighted the importance of horizontal gene transfer among bacteria from the clinical as well as ecological perspective.
While many studies have assessed the diversity and AR of enterococci in food, the majority have focused on food before preparation and cooking (12, 17, 21, 24, 50), during which many microorganisms and associated genes are likely destroyed. Only a few studies have evaluated enterococcal contamination in ready-to-eat foods (RTEFs), and these included cheese (13, 17), fermented sausages (14), and produce (12, 22, 31). However, RTEFs such as meals from fast-food restaurants that are very commonly consumed in developed countries have not been assessed for the frequency and level of enterococcal contamination nor as a source of a possible influx of AR and virulence genes to the resident microbial community in the human digestive tract.
In this study, the prevalence and diversity of enterococci in RTEFs (chicken salad, chicken burger, and carrot cake) from fast-food restaurants as well as the influx of enterococcal AR and virulence genes to the human digestive tract were evaluated.
MATERIALS AND METHODS
Isolation of enterococci from RTEF.
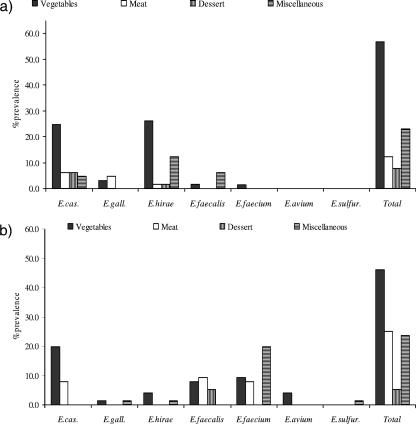
Three types of RTEFs (chicken salad meal, chicken sandwich with French fries, and carrot cake) were purchased five times in both summer (SU; June to August) and winter (WI; November to January) from five fast-food restaurants (R1 to R5) in a town in northeastern Kansas. Each meal was brought to the laboratory, and the total weight of each meal was determined. To screen for enterococcal contamination, each meal was aseptically taken apart to separate the individual ingredients. Overall, meals consisted of (i) chicken salad (chicken meat, lettuce, tomatoes, red cabbage, peppers, onions, dressing, croutons, cheese, and chips), (ii) chicken burger/sandwich (chicken fillet, lettuce, tomatoes, pickles, bread, cheese, French fries, and barbecue sauce), or (iii) carrot cake (cake with frosting). Some minor ingredients such as croutons, chips, dressing, and type of vegetables varied between restaurants depending on the recipe as well as within restaurants based on availability. Five to 10 grams of each ingredient was separately homogenized in 100 ml of phosphate-buffered saline (pH 7.2; ICN Biomedicals, OH) for 5 min using a stomacher (Stomacher 400 Circulator). From this sample, 100 μl was plated in triplicate on mEnterococcus Agar (Becton Dickinson, MA) and incubated at 37°C for 48 h. Red, pink, and burgundy colonies were counted, and the concentration of enterococci was calculated in CFU per gram of RTEF (meal and its main ingredients) (Table 1). Up to four presumptive enterococcal colonies with different colony morphologies per RTEF sample were streaked on Trypticase soy agar (Becton Dickinson, MA), incubated at 37°C for 24 h, and stored at 4°C for further analysis. The genus-level identification was confirmed by the esculin hydrolysis test using Enterococcossel broth (Becton Dickinson, MA) and by growth at 44.5°C in Trypticase soy broth (Becton Dickinson, MA) with 5% sodium chloride. Results for prevalence and concentration of enterococci in RTEF were analyzed and presented as values per each meal (the meal was considered positive if it contained one or more contaminated individual ingredients) as well as per its three main groups of ingredients: vegetables, meat, and miscellaneous ingredients (chips, fries, sauce, bread, croutons, dressing, and cheese) (Table 1). The groups of ingredients were created because of the variation in availability of some ingredients (vegetables and miscellaneous) between and within restaurants over time (resulting in a low number of replicates). The diversity of enterococci is reported per each of the main ingredient groups of three different RTEF meals (chicken salad, chicken burger, and carrot cake) (Fig. 1).
TABLE 1.
Prevalence of contamination and concentration (CFU/g) of enterococci in RTEF per meal and per the main group of ingredientsa per meal in five fast-food restaurants in WI and SU
| RTEF meal and ingredient(s) | WI
|
SU
|
||||||
|---|---|---|---|---|---|---|---|---|
| Total no. of samples | % Prevalenceb | No. of positive samples | Mean concn ± SEM (CFU/g) | Total no. of samples | % Prevalenceb | No. of positive samples | Mean concn ± SEM (CFU/g) | |
| Chicken salad | 25 | 64 Ac | 16 | (2.1 ± 1.5) × 103 | 25 | 92 B | 23 | (2.1 ± 0.5) × 103 |
| Vegetables | 60 | 28 (40) | 17 | (1.2 ± 3) × 103 | 60 | 43 (84) | 26 | (1.9 ± 6.2) × 103 |
| Meat | 25 | 20 (20) | 5 | (7.5 ± 4.5) × 102 | 25 | 28 (28) | 7 | (2.8 ± 1.8) × 102 |
| Miscellaneous | 45 | 25 (36) | 11 | (8.0 ± 1.5) × 103 | 45 | 20 (32) | 9 | (4.2 ± 3.9) × 102 |
| Chicken burger | 25 | 24 A | 6 | (1.6 ± 0.9) × 103 | 25 | 64 B | 16 | (5.8 ± 1.8) × 102 |
| Vegetables | 50 | 18 (24) | 9 | (0.4 ± 3.0) × 103 | 60 | 17 (28) | 10 | (0.7 ± 1.2) × 103 |
| Meat | 25 | 12 (12) | 3 | (4.8 ± 4.5) × 102 | 25 | 24 (24) | 6 | (1.5 ± 1.1) × 102 |
| Miscellaneous | 65 | 12 (24) | 8 | (0.6 ± 1.5) × 103 | 65 | 14 (16) | 9 | (1.6 ± 1.8) × 102 |
| Carrot cake | 15 | 20 | 3 | (8.4 ± 4.2) × 103 A | 15 | 7 | 1 | (4.1 ± 1.6) × 101 B |
Vegetables, two to three ingredients per meal; meat, one ingredient per meal; miscellaneous, two to four ingredients per meal (chips, fries, sauce, bread, croutons, dressing, and cheese).
Values in parentheses show percent meals (salads and burgers) contaminated due to contamination of a main ingredient.
Different superscript capital letters within the same row indicate significant differences.
FIG. 1.
Prevalence and diversity of enterococci from RTEF from five fast-food restaurants in WI (a) and SU (b). E. cas., E. casseliflavus; E. gall., E. gallinarum; E. sulfur., E. sulfureus. The “miscellaneous” group includes chips, fries, sauce, bread, croutons, dressing, and cheese.
Isolate identification and screening for AR by phenotype.
Multiplex PCR was used to identify four common species: E. faecalis, Enterococcus faecium, Enterococcus casseliflavus, and Enterococcus gallinarum (23). PCR amplification and sequencing of the manganese-dependent superoxide dismutase gene sodA (36) were used for those isolates which could not be identified by multiplex PCR. The strains used as positive and negative controls and the primer sequences have been reported previously (30).
Identified isolates were screened for AR by the disc diffusion method on Mueller-Hinton agar (Becton Dickinson, MA) using six antibiotics: tetracycline, 30 μg/ml; chloramphenicol, 30 μg/ml; ciprofloxacin, 5 μg/ml; erythromycin, 15 μg/ml; vancomycin, 30 μg/ml; and ampicillin, 10 μg/ml. High-level resistance to aminoglycosides was assessed by the agar dilution technique using 2,000 μg/ml of streptomycin and 2,000 μg/ml kanamycin in brain heart infusion agar (Becton Dickinson, MA). E. faecalis ATCC 19433 was used as a quality control strain. The protocols followed the guidelines of the Clinical and Laboratory Standards Institute (34).
Screening for AR and virulence determinants by PCR.
Multiplex or single PCR was used to screen all identified isolates for tetracycline and erythromycin resistance genes. The group I multiplex reaction included tet(A), tet(C), and tet(Q) genes; group II covered tet(M), tet(S), tet(K), and tet(O) as described previously (30). Each reaction mixture consisted of 25 μl Master Mix (Promega, Madison, WI) with 4 mM MgCl2 (group I), 3 mM MgCl2 (group II), and 3 μl of supernatant from freshly boiled cells. The PCR conditions were described previously (35, 47). Single PCRs were used to screen tet(W) (3) and erm(B) (43). All identified isolates were screened for four putative virulence determinants including gelE (gelatinase), asa1 (aggregation substance), cylA (cytolysin), and esp (enterococcus surface protein) using multiplex PCR as described previously (46). To confirm the identity of the determinants, one randomly selected PCR product for each resistance and virulence determinant was purified, sequenced using either the PCR primers or the T7 primer after cloning of the PCR product using the pGEM-T Easy Vector System (Promega, Madison, WI), and compared to the sequences in GenBank using BLAST (Basic Local Alignment Search Tool) (2). Manual sequence alignment was done with the CodonCode Aligner (version 1.5.2; CodonCode Corporation, Dedham, MA). The same positive-control strains were used as reported previously (30).
All E. faecalis isolates (n = 21) were screened for six marker genes (EF_0573, EF_0592, EF_0605, EF_1420, EF_2144, and pathogenicity island EF_0050) for potentially clinically relevant strains. In addition, we analyzed 50 isolates of E. faecalis isolated in the previous study (30) from houseflies collected from the same five fast-food restaurants for these marker genes. PCRs were conducted under conditions described previously (26).
Screening for virulence genes by phenotype.
Trypticase soy agar with 3% skim milk was used for detection of gelatinase activity. All isolates were streaked and after 24 h of incubation at 37°C were examined for a clearance zone surrounding the colonies (15).
For the phenotypic expression of the asa1 gene, E. faecalis JH2-2 was grown for 6 h at 37°C in Todd-Hewitt broth (Becton Dickinson, MA). Broth was then centrifuged at 6,000 rpm for 10 min on a Sorvall RC-5B refrigerated (4°C) Superspeed centrifuge with the SS-34 rotor, and the pheromone-containing supernatant that induces pheromone-responsive plasmids was removed and autoclaved for 15 min. Tested isolates were grown in Todd-Hewitt broth (5 ml) for 6 h at 37°C. After incubation, 1 ml of the supernatant from E. faecalis JH2-2 was added to each tube and incubated at 37°C overnight on a shaker (150 rpm). Isolates that showed clumping (examined by naked eye and under a compound microscope) were considered positive for aggregation substance expression (7). E. faecalis OG1RF:pCF10 was used as a positive control.
Phenotypic assays for cytolysin were conducted using Columbia blood agar base (Becton Dickinson, MA) with 5% cattle blood. Isolates were streaked and incubated at 37°C for 48 to 72 h. Isolates showing a complete clearance zone around the colonies (β-hemolysis) were considered positive for cytolysin expression (15).
Genotyping by PFGE.
Relationships of E. faecalis and E. faecium were analyzed by pulsed-field gel electrophoresis (PFGE) using the CHEF Mapper (Bio-Rad, Hercules, CA) and the restriction enzyme AluI as previously described (33, 44). Cluster analysis was performed with BioNumerics (Applied Maths, Kortrijk, Belgium) by using the Dice correlation coefficient (6) and the unweighted-pair group mathematical average algorithm (41).
Statistical analysis.
Data for the prevalence of enterococci in different RTEFs were analyzed by t test (bacterial concentration) and chi-square test (frequency of contamination) using the PopTools plug-in for Excel (G. Hood, 2000; http://www.cse.csiro.au/poptools/).
RESULTS
Prevalence, concentration, and diversity of enterococci in RTEF.
The prevalence of contaminated RTEF in SU was significantly higher than that in WI, increasing from 64 to 92% in chicken salads and from 24 to 64% in chicken burgers (P = 0.001) (Table 1). More specifically, the increase in frequency of contamination in chicken salads in SU was mainly due to contaminated vegetables (from 28% in WI to 43% in SU per vegetable ingredients), making 84% of chicken salads contaminated in SU due to one or more contaminated vegetable ingredients (Table 1). In contrast, the contamination frequency increase in chicken burgers in SU was due to higher prevalence of contaminated meat (from 12% in WI to 24% in SU per ingredient and meal) and also because of a relatively even distribution of contaminated ingredients (17% vegetables, 24% meat, and 14% miscellaneous) across meals, making 64% of chicken burgers contaminated (Table 1). The frequency of contamination in miscellaneous ingredients was similar between the seasons within different meals (Table 1), although the frequency of contamination in chicken burgers during WI (12% of 65 miscellaneous ingredients) was more evenly spread across the meals (making 24% of chicken burgers contaminated due to one or more miscellaneous ingredients) than that in SU (Table 1). The prevalence of enterococci in carrot cakes decreased from 20% in WI to 7% in SU; however, the number of samples was lower than that of other foods due to low availability of carrot cakes in two of the selected restaurants (Table 1).
The overall concentrations of enterococci in contaminated RTEF (CFU/g) during the two seasons were similar (chicken salad, 2.1 × 103 ± 1.5 × 103 [WI] and 2.1 × 103 ± 0.5 × 103 [SU]; chicken burger, 1.6 × 103 ± 0.9 × 103 [WI] and 5.8 × 102 ± 1.9 × 102 [SU]) while enterococcal concentration in carrot cakes decreased in SU from 8.4 × 103 (WI) to 4.1 × 101 CFU/g (Table 1). In salads and burgers, the concentrations of enterococci in WI and SU were not significantly different (P = 0.56 and P = 0.96, respectively), while the concentration in carrot cakes decreased significantly in SU (P = 0.03) (Table 1). This represents a relatively high concentration of enterococci per each meal from both sampling seasons (total weight of each meal): 7.6 × 105 CFU/chicken salad (362.2 g), 4.8 × 105 CFU/chicken burger (443.1 g), and 6.6 × 105 CFU/carrot cake (156.2 g).
A total number of 145 enterococcal colonies from RTEF were isolated and further characterized. After the identification by sequencing of the sodA gene, four isolates were found not to be enterococci and were not analyzed further. Thus, 65 WI and 76 SU isolates were selected for further analysis (Table 2) (3).
TABLE 2.
AR profiles of identified enterococcal isolates from WI and SU sampling of RTEFs
| Season and identification | No. (%) of isolates
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Total | Resistant to druga:
|
||||||||
| CIP | CHL | VAN | ERY | AMP | TET | KAN | STR | ||
| WI | |||||||||
| E. casseliflavus | 27 (41.5) | 9 (33.4) | 2 (7.4) | 0 | 1 (3.7) | 0 | 2 (7.4) | 1 (3.7) | 1 (3.7) |
| E. gallinarum | 5 (7.7) | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| E. hirae | 27 (41.5) | 0 | 2 (7.4) | 0 | 1 (3.7) | 0 | 27 (100) | 1 (3.7) | 0 |
| E. faecalis | 5 (7.7) | 0 | 1 (20) | 0 | 1 (20) | 0 | 3 (60) | 0 | 0 |
| E. faecium | 1 (1.5) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Total | 65 | 9 (13.8) | 4 (6.1) | 0 | 3 (4.6) | 0 | 33 (50.8) | 2 (3.1) | 1 (1.5) |
| SU | |||||||||
| E. casseliflavus | 21 (27.7) | 0 | 1 (4.8) | 0 | 3 (14.2) | 0 | 2 (10) | 0 | 1 (4.8) |
| E. gallinarum | 2 (2.6) | 0 | 0 | 0 | 0 | 0 | 2 (100) | 1 (50) | 0 |
| E. hirae | 4 (5.3) | 0 | 0 | 0 | 1 (25) | 0 | 4 (100) | 0 | 0 |
| E. faecalis | 17 (22.4) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| E. faecium | 28 (36.8) | 0 | 0 | 0 | 11 (39.2) | 0 | 4 (14.3) | 8 (28.5) | 0 |
| E. sulfureus | 1 (1.3) | 0 | 0 | 0 | 0 | 0 | 0 | 1 (100) | 0 |
| E. avium | 3 (3.9) | 0 | 0 | 0 | 2 (66.7) | 0 | 1 (33.3) | 0 | 0 |
| Total | 76 | 0 | 1 (1.3) | 0 | 17 (22.3) | 0 | 13 (17.1) | 10 (13.2) | 1 (1.3) |
CIP, ciprofloxacin; CHL, chloramphenicol; VAN, vancomycin; ERY, erythromycin; AMP, ampicillin; TET, tetracycline; KAN, kanamycin; STR, streptomycin.
The diversity of isolates from RTEF is shown in Fig. 1a and b. Overall, five Enterococcus species were found in WI and seven species in SU. The most prevalent species among WI isolates were E. casseliflavus and Enterococcus hirae (41.5% each), followed by E. gallinarum (7.7%), E. faecalis (7.7%), and E. faecium (1.5%) (Fig. 1a). E. faecium (36.8%), E. casseliflavus (27.6%), and E. faecalis (22.4%) were the most common species among isolates from RTEF in SU, followed by E. hirae (5.3%), E. avium (3.9%), E. gallinarum (2.6%), and Enterococcus sulfureus (1.3%) (Fig. 1b).
E. casseliflavus and E. hirae were detected primarily from vegetables (24.6% WI and 19.7% SU and 26.2% WI and 3.9% SU, respectively) (Fig. 1a and b). In WI, E. faecalis was isolated mainly from miscellaneous ingredients while the contamination with E. faecalis increased in SU in meat and vegetables to 9.2% and 7.9%, respectively (Fig. 1a and b). Similarly, E. faecium was more common in SU months, mainly in the miscellaneous group (19.7%), vegetables (9.2%), and meat (7.9%). The prevalence of E. gallinarum was low during both sampling seasons, and Enterococcus avium and Enterococcus sulfuricans were detected at a low frequency in SU only (Fig. 1a and b).
Prevalence and diversity of AR and virulence factors by phenotype and genotype.
Phenotypic analysis of WI isolates showed resistance to several antibiotics including tetracycline (50.8%), ciprofloxacin (13.8%), erythromycin (4.6%), chloramphenicol (6.1%), kanamycin (3.1%), and streptomycin (1.5%) (Table 2). Isolates from SU RTEF were resistant to erythromycin (22.3%), tetracycline (17.1%), and kanamycin (13.2%), followed by chloramphenicol and streptomycin (both 1.3%). Several isolates (7.7%) in WI were resistant to two or more antibiotics (one E. casseliflavus isolate was resistant to six antibiotics), while isolates from RTEF in SU were resistant to a maximum of two antibiotics.
The most common determinant coding for tetracycline resistance was tet(M) (35.4% prevalence in WI and 4.0% in SU). tet(M) was frequent in E. faecalis (80%), followed by E. hirae (66.7%) and E. casseliflavus (3.7%) in WI and E. faecium (3.6%) and E. hirae (50%) in SU (Table 3). The diversity of the tet genes was greater in SU isolates, where tet(S) (3.6%) and tet(O) (3.6%) were detected. The erm(B) gene was detected at low frequency (9.2% in WI and 4.0% in SU).
TABLE 3.
Prevalence of tetracycline and erythromycin resistance genes and virulence determinants (by genotype and phenotype) in identified enterococcal isolates from RTEF
| Season and identification | No. (%) of isolates
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Total | With genotype:
|
With phenotype determined by assay:
|
||||||||
| tet gene(s) | erm(B) | gelE | asa1 | esp | cylA | Gelatinase | Clumping assay | Hemolysisa | ||
| WI | ||||||||||
| E. casseliflavus | 27 | 1 (3.7) [tet(M)] | 2 (7.4) | 2 (7.4) | NAb | 0 | 0 | 1 (3.7) | NA | 11 (40.7) |
| E. gallinarum | 5 | 0 | 2 (40) | 2 (40) | NA | 0 | 0 | 1 | NA | 4 (80) |
| E. hirae | 27 | 18 (66.7) [tet(M)] | 0 | 0 | NA | 0 | 0 | 1 (3.7) | NA | 0 |
| E. faecalis | 5 | 4 (80) [tet(M)] | 2 (40) | 4 (80) | 2 (40) | 0 | 0 | 3 (60) | 0 | 0 |
| E. faecium | 1 | 0 | 0 | 0 | NA | 0 | 0 | 0 | NA | 0 |
| Total | 65 | 23 (35.4) | 6 (9.2) | 8 (12.3) | 2 (3.1) | 0 | 0 | 6 (9.2) | 0 | 15 (23.1) |
| SU | ||||||||||
| E. casseliflavus | 21 | 2 (9.5) [tet(S)] | 0 | 0 | NA | 0 | 0 | 0 | NA | 0 |
| E. gallinarum | 2 | 1 (50) [tet(S)] | 0 | 0 | NA | 0 | 0 | 0 | NA | 0 |
| E. hirae | 4 | 2 (50) [tet(M)]; 1 (25) [tet(O)] | 0 | 0 | NA | 0 | 0 | 0 | NA | 0 |
| E. faecalis | 17 | 0 | 1 (5.9) | 0 | 8 (47.1) | 0 | 0 | 2 (11.8) | 0 | 0 |
| E. faecium | 28 | 1 (3.6) [tet(M)]; 1 (3.6) [tet(S)]; 1 (3.6) [tet(O)] | 0 | 0 | NA | 0 | 0 | 0 | NA | 0 |
| E. sulfureus | 1 | 0 | 0 | 0 | NA | 0 | 0 | 0 | NA | 0 |
| E. avium | 3 | 0 | 2 (66.7) | 0 | NA | 0 | 0 | 0 | NA | 0 |
| Total | 76 | 9 (11.9) | 3 (4.0) | 0 | 8 (10.5) | 0 | 0 | 2 (2.6) | 0 | 0 |
Cattle blood.
NA, not applicable.
The overall prevalence of the putative virulence genes was low: 12.3% for gelE, 3.1% for asa1, and 0% for cylA (Table 3). The gelE gene was found in WI, primarily in E. faecalis (80% of isolates), E. gallinarum (40%), and E. casseliflavus (7.4%). Many of these isolates expressed this gene in the phenotypic assay for gelatinase (Table 3). No isolates from the SU were positive for gelE. Interestingly, one isolate of E. hirae and two of E. faecalis were positive for gelatinase by phenotype but negative for gelE by PCR (Table 3). The gene for the aggregation substance of E. faecalis was detected in 40.0% of WI isolates and 47.1% of SU isolates, but it was not expressed phenotypically (Table 3). The cylA gene was not detected among any of the isolates; however, several isolates of E. casseliflavus and E. gallinarum from WI tested positive for β-hemolysis on cattle blood (Table 3). The identity of selected AR and virulence genes from multiplex PCRs was confirmed by sequencing (data not shown).
Six marker genes for potentially virulent E. faecalis were screened in 21 isolates of E. faecalis from RTEF and an additional 50 isolates of E. faecalis from the houseflies collected from the same five fast-food restaurants in our previous study (30). A single isolate, RTEF59, from the chicken salad (lettuce) in SU was positive for the four of the six marker genes (EF_0573, EF_0592, EF_0605, and EF_0050) (overall prevalence, 4.7%). One E. faecalis isolate, RTEF15, was positive for the EF_1420 gene. From the housefly gut isolates from restaurants R1 and R5, two E. faecalis isolates were positive for EF_0592, another two isolates from R1 were positive for EF_0050, and one isolate from R1 was positive for EF_1420.
Genotypic diversity of E. faecalis and E. faecium assessed by PFGE.
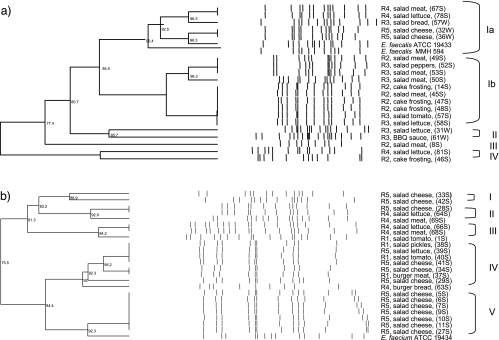
E. faecalis (n = 20) and E. faecium (n = 23) isolates from RTEF were genotyped by PFGE. E. faecalis represented four different genotypes (≤85% similarity) with type I containing the majority of the isolates divided into two subtypes (>85% similarity) (Fig. 2a). Subtype Ia contained isolates from the chicken salad from three different restaurants (R4, meat and lettuce; R3, croutons; and R5, cheese). This subtype clustered together with the positive-control strains E. faecalis ATCC 19433 and E. faecalis MMH583. Subtype Ib contained isolates from restaurants R2 and R3. R2 isolates originated from the chicken salad (meat) and cake frosting, and R3 isolates came from the chicken salad (meat, peppers, tomatoes, and lettuce). The type II isolates were from lettuce and barbecue sauce from restaurant R3. The type III cluster represented one isolate from the chicken salad (meat) from R2. The type IV isolates were from different restaurants: the chicken salad (lettuce) isolate was from R4 and the cake frosting isolate was from R2. Identical or very closely related strains (≥95% similarity) were detected from two restaurants: R2 (chicken salad and carrot cake) and R3 (chicken salad) (Fig. 2a).
FIG. 2.
Dendrogram illustrating relatedness of E. faecalis (a) and E. faecium (b) strains isolated from RTEF based on PFGE patterns of AluI-digested DNA. R, restaurant; S, SU isolate; W, WI isolate.
E. faecium isolates were more divergent and grouped in five genotypes with the closest similarity index being 75.5 (Fig. 2b). Type I, II, and III isolates clustered (81.3% similarity) and originated from R4 and R5 (chicken salad; cheese, lettuce, and meat), and one isolate came from R1 chicken salad (tomato). Types IV and V clustered together (84.4% similarity) and represented chicken salad (vegetables and cheese); chicken burger (meat and bread) from restaurants R5, R1, and R4 (type IV); and the chicken salad cheese from R5 (type V). Identical or very closely related strains (≥95% similarity) were detected from R1 (chicken salad and chicken burger) and R5 (chicken salad) from the SU sampling (Fig. 2b).
DISCUSSION
The recent metagenomic analysis of microbes from the human intestine has shown that the microbial community in this organ, primarily in the colon, is dominated by two bacterial divisions, the Bacteroidetes and the Firmicutes (8, 9, 52), and outnumbers human cells 10-fold and human genes 100-fold (4, 10). This enormous bacterial community represents an important site for gene exchange (intra- and interspecies) (38, 39, 40) among permanent and transient colonizers as well as bacteria ingested with food and drinks and passing through the digestive tract (10, 27, 42). While several studies have assessed the potential for ingested bacteria to colonize the gut of adults and/or horizontally transfer selected genes to the resident microbial community (27, 32, 42), the extent of the actual influx of bacteria and associated resistance and virulence genes from RTEF to the digestive tract has not been extensively studied.
Investigations of the level of contamination of food with antibiotic-resistant strains have been focused on raw food (pork, beef, and poultry) before preparation and cooking (17, 19, 20, 31, 48, 50), during which most of the bacteria and their genes are likely destroyed; milk and milk products (cheese and other fermented products) (12, 13, 17, 48); fresh produce (12, 22, 31, 48); and probiotic strains (11, 12).
Our data showed that RTEFs, mainly chicken salads and chicken burgers (Table 1) from fast-food restaurants, were frequently contaminated with enterococci at relatively high concentrations (∼103 CFU per gram of food) and the contamination frequency increased in SU months. While E. casseliflavus and E. hirae were the dominant species in RTEFs (mainly vegetables) in WI, the enterococcal community was different in RTEF in SU months with an increased diversity and elevated population of E. faecalis (in vegetables and meat) and E. faecium (in vegetables, meat, and miscellaneous ingredients such as cheese, fries, and croutons). The presence of E. casseliflavus and E. hirae on vegetables is not surprising, as they are commonly found associated with plants (12). The source(s) of contamination of RTEF in SU with E. faecalis and E. faecium is unknown; however, the low prevalence of putative virulence genes and virulence markers suggests that the enterococcal contamination in RTEF is environmental and not of clinical origin. It is possible that enterococci originated from people handling the food in restaurants or during harvesting and processing. One of the rationales for screening RTEF in WI and SU in this study was to assess indirectly the potential role of insects, primarily houseflies, in contamination of food with enterococci in SU months. In a previous study (30), we characterized enterococci from houseflies collected in the same five fast-food restaurants and showed that these insects commonly carried a high population of antibiotic-resistant and potentially virulent enterococci. Results from the current study do not directly implicate flies as a source of RTEF contamination, although the frequency of enterococcal contamination as well as the prevalence of E. faecalis (the dominant species detected in houseflies) (30) increased in RTEF in SU months when flies in fast-food restaurants are common. Other factors likely playing a role in the seasonal differences include, for example, possible different geographical origins of RTEF ingredients, various growing practices (e.g., using animal manure as fertilizer), and longer refrigeration and storage times in WI.
Taking into account the total weight of each meal, the concentration of enterococci that would be ingested in each contaminated meal was relatively high (7.6 × 105 CFU/chicken salad, 4.8 × 105 CFU/chicken burger, and 6.6 × 105 CFU/carrot cake), and considering, for example, that 51% of WI isolates were phenotypically resistant to tetracycline, this represents (using a conservative estimate of only a single copy of the tet resistance gene per cell) the influx of 3.8 × 105, 2.5 × 105, and 3.4 × 105 tet genes from the chicken salad, chicken burger, and carrot cake, respectively. Since RTEFs are very popular and commonly consumed in the United States and other parts of the world, the influx of resistance genes is very frequent and may play a role in the acquisition and spread of AR determinants in the human digestive tract.
The most prevalent tetracycline resistance determinants in our food isolates were tet(M), tet(O), and tet(S) genes coding for ribosomal protection proteins (5) which are common in tetracycline-resistant strains reported from various environments including food products (21, 48). No efflux pump genes were identified in our isolates. A seasonal shift in the AR profile was observed: tet(M) dominated in the WI isolates, while SU isolates carried tet(S), tet(M), and tet(O). These genes are commonly carried on mobile genetic elements such as plasmids and conjugative transposons (37); therefore, it is possible that they can be transferred to the resident bacterial microbiota of the human digestive tract by horizontal gene transfer.
The prevalence of putative virulence genes including gelE, asa1, cylA, and esp in our isolates was low, indicating that these isolates were not likely virulent and the influx of virulence genes was not high. Interestingly, a large percentage of E. casseliflavus isolates (40.7%) lysed red blood cells on cattle blood agar but the cylA gene was not detected in any of those isolates. In contrast, several E. faecalis isolates were positive for asa1 but this was not expressed phenotypically. In addition, RTEFs as well as houseflies from restaurants carried enterococcal isolates with a very low prevalence of genes (EF_0573, EF_0592, EF_0605, EF_0050, EF_1420, and EF_2144) predicted to be associated with clinically important isolates (26). This supports the suggestion that these genes are potentially good markers for virulent enterococci and also that our isolates can be considered environmental.
Genotyping with PFGE showed that both E. faecalis and E. faecium overall clustered based on the source (restaurant) and season. E. faecalis represented several types and subtypes from RTEFs across different restaurants, but with the exception of restaurants R2 and R3, no common strains were detected, indicating that the contamination originated from various sources and did not represent a clonal spread. Genotypes of E. faecium were more diverse and similarly grouped by the restaurant, food ingredient, and season. A few common strains were detected from restaurants R4 and R5 (from chicken salad lettuce) as well as R1 and R5 (chicken salad pickles, lettuce, tomatoes, and cheese), indicating a common source of one or more of these ingredients. Clearly, the likelihood of cross-contamination of ingredients within the same meal due to their close physical contact was high.
In conclusion, the results of this study showed that RTEFs such as chicken salads and chicken burgers in fast-food restaurants are frequently contaminated with a relatively high concentration of enterococci. This represents a frequent influx of AR genes (primarily tetracycline and erythromycin resistance genes) to the human digestive tract that can potentially spread by horizontal transfer to the resident microbial community and result in formation of the AR gene reservoir without any selective pressure from antibiotic use. Additional studies are needed to quantify the influx of resistance genes from enterococci and other common food bacterial contaminants and their potential to be transferred to the resident microbial community of the human digestive tract.
Acknowledgments
We thank B. Miles, N. Alderman, and J. Metlevski for technical help and A. Broce and D. Margolies for reviewing the manuscript.
This study was supported by the KS640 grant.
This is contribution no. 07-235-J of the Kansas Agricultural Experiment Station.
Footnotes
Published ahead of print on 31 August 2007.
REFERENCES
- 1.Aarestrup, F. M., A. M. Seyfarth, H.-D. Emborg, K. Pedersen, R. S. Hendriksen, and F. Bager. 2001. Effect of abolishment of the use of antimicrobial agents for growth promotion on occurrence of antimicrobial resistance in fecal enterococci from food animals in Denmark. Antimicrob. Agents Chemother. 45:2054-2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 3.Aminov, R. I., N. Garrigues-Jeanjean, and R. I. Mackie. 2001. Molecular ecology of tetracycline resistance: development and validation of primers for detection of tetracycline resistance genes encoding ribosomal protection proteins. Appl. Environ. Microbiol. 67:22-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Backhed, F., R. E. Ley, J. L. Sonnenburg, D. A. Peterson, and J. I. Gordon. 2005. Host-bacterial mutualism in the human intestine. Science 307:1915-1920. [DOI] [PubMed] [Google Scholar]
- 5.Chopra, I., and M. Roberts. 2001. Tetracycline antibiotics: mode of action, applications, molecular biology, and epidemiology of bacterial resistance. Microbiol. Mol. Biol. Rev. 65:232-260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dice, L. R. 1945. Measures of the amount of ecological association between species. Ecology 26:297-302. [Google Scholar]
- 7.Dunny, G. M., R. Craig, R. Carron, and D. B. Clewell. 1979. Plasmid transfer in Streptococcus faecalis: production of multiple sex pheromones by recipients. Plasmid 2:454-465. [DOI] [PubMed] [Google Scholar]
- 8.Eckburg, P. B., E. M. Bik, C. N. Bernstein, E. Purdom, L. Dethlefsen, M. Sargent, S. R. Gill, K. E. Nelson, and D. A. Relman. 2005. Diversity of the human intestinal microbial flora. Science 308:1635-1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eckburg, P. B., E. M. Bik, C. N. Bernstein, M. Sargent, S. R. Gill, K. E. Nelson, and D. A. Relman. 2005. Multi-site culture-independent analysis of the normal human intestinal microflora using large-scale 16S rDNA sequence analysis. Gastroenterology 128:A194. [Google Scholar]
- 10.Falk, P., L. Hooper, T. Midtvedt, and J. Gordon. 1998. Creating and maintaining the gastrointestinal ecosystem: what we know and need to know from gnotobiology. Microbiol. Mol. Biol. Rev. 62:1157-1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Franz, C. M. A. P., A. B. Muscholl-Silberhorn, N. M. K. Yousif, M. Vancanneyt, J. Swings, and W. H. Holzapfel. 2001. Incidence of virulence factors and antibiotic resistance among enterococci isolated from food. Appl. Environ. Microbiol. 67:4385-4389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Franz, C. M. A. P., M. E. Stiles, K. H. Schleifer, and W. H. Holzapfel. 2003. Enterococci in foods—a conundrum for food safety. Int. J. Food Microbiol. 88:105-122. [DOI] [PubMed] [Google Scholar]
- 13.Gelsomino, R., M. Vancanneyt, T. M. Cogan, and J. Swings. 2003. Effect of raw-milk cheese consumption on the enterococcal flora of human feces. Appl. Environ. Microbiol. 69:312-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gevers, D., M. Danielsen, G. Huys, and J. Swings. 2003. Molecular characterization of tet(M) genes in Lactobacillus isolates from different types of fermented dry sausage. Appl. Environ. Microbiol. 69:1270-1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gilmore, M. S., P. C. Coburn, S. R. Nallapareddy, and B. E. Murray. 2002. Enterococcal virulence, p. 301-354. In M. S. Gilmore (ed.), The enterococci: pathogenesis, molecular biology, and antibiotic resistance. ASM Press, Washington, DC.
- 16.Gilmore, M. S. (ed.). 2002. The enterococci: pathogenesis, molecular biology, and antibiotic resistance. ASM Press, Washington, DC.
- 17.Giraffa, G. 2002. Enterococci from foods. FEMS Microbiol. Rev. 26:163-171. [DOI] [PubMed] [Google Scholar]
- 18.Hayes, J. R., L. L. English, L. E. Carr, D. D. Wagner, and S. W. Joseph. 2004. Multiple-antibiotic resistance of Enterococcus spp. isolated from commercial poultry production environments. Appl. Environ. Microbiol. 70:6005-6011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hayes, J. R., L. L. English, P. J. Carter, T. Proescholdt, K. Y. Lee, D. D. Wagner, and D. G. White. 2003. Prevalence and antimicrobial resistance of Enterococcus species isolated from retail meats. Appl. Environ. Microbiol. 69:7153-7160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heuer, H., A. M. Hammerum, P. Collignon, and H. Wegener. 2006. Human health hazard from animal-resistant enterococci in animals and food. Clin. Infect. Dis. 43:911-916. [DOI] [PubMed] [Google Scholar]
- 21.Huys, G., K. D'Haene, J.-M. Collard, and J. Swings. 2004. Prevalence and molecular characterization of tetracycline resistance in Enterococcus isolates from food. Appl. Environ. Microbiol. 70:1555-1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Johnston, L. M., and L.-A. Jaykus. 2004. Antimicrobial resistance of Enterococcus species isolated from produce. Appl. Environ. Microbiol. 70:3133-3137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kariyama, R., R. Mitsuhata, J. W. Chow, D. B. Clewell, and H. Kumon. 2000. Simple and reliable multiplex PCR assay for surveillance isolates of vancomycin-resistant enterococci. J. Clin. Microbiol. 38:3092-3095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Klein, G. 2003. Taxonomy, ecology and antibiotic resistance of enterococci from food and the gastro-intestinal tract. Int. J. Food Microbiol. 88:123-131. [DOI] [PubMed] [Google Scholar]
- 25.Kummerer, K. 2004. Resistance in the environment. J. Antimicrob. Chemother. 54:311-320. [DOI] [PubMed] [Google Scholar]
- 26.Lepage, E., S. Brinster, C. Caron, C. Ducroix-Crepy, L. Rigottier-Gois, G. Dunny, C. Hennequet-Antier, and P. Serror. 2006. Comparative genomic hybridization analysis of Enterococcus faecalis: identification of genes absent from food strains. J. Bacteriol. 188:6858-6868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lester, C. H., N. Frimodt-Moller, T. L. Sorensen, D. L. Monnet, and A. M. Hammerum. 2006. In vivo transfer of the vanA resistance gene from an Enterococcus faecium isolate of animal origin to an E. faecium isolate of human origin in the intestines of human volunteers. Antimicrob. Agents Chemother. 50:596-599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Levy, S. B., and B. Marshall. 2004. Antibacterial resistance worldwide: causes, challenges and responses. Nat. Med. 10:122-129. [DOI] [PubMed] [Google Scholar]
- 29.Lipsitch, M., R. S. Singer, and B. R. Levin. 2002. Antibiotics in agriculture: when is it time to close the barn door? Proc. Natl. Acad. Sci. USA 99:5752-5754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Macovei, L., and L. Zurek. 2006. Ecology of antibiotic resistance genes: characterization of enterococci from houseflies collected in food settings. Appl. Environ. Microbiol. 72:4028-4035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McGowan, L., C. R. Jackson, J. B. Barrett, L. Hiott, and P. J. Fedorka-Cray. 2006. Prevalence and antimicrobial resistance of enterococci isolated from retail fruits, vegetables, and meats. J. Food Prot. 69:2976-2982. [DOI] [PubMed] [Google Scholar]
- 32.Moubareck, C., N. Bourgeois, P. Courvalin, and F. Doucet-Populaire. 2003. Multiple antibiotic resistance gene transfer from animal to human enterococci in the digestive tract of gnotobiotic mice. Antimicrob. Agents Chemother. 47:2993-2996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Murray, B. E., K. V. Singh, J. D. Heath, B. R. Sharma, and G. M. Weinstock. 1990. Comparison of genomic DNAs of different enterococcal isolates using restriction endonucleases with infrequent recognition sites. J. Clin. Microbiol. 28:2059-2063 (Erratum, 29:418, 1991.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.NCCLS. 2000. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. Approved standard M7-A5. NCCLS, Wayne, PA.
- 35.Ng, L. K., I. Martin, M. Alfa, and M. Mulvey. 2001. Multiplex PCR for the detection of tetracycline resistant genes. Mol. Cell. Probes 15:209-215. [DOI] [PubMed] [Google Scholar]
- 36.Poyart, C., G. Quesnes, and P. Trieu-Cuot. 2000. Sequencing the gene encoding manganese-dependent superoxide dismutase for rapid species identification of enterococci. J. Clin. Microbiol. 38:415-418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rice, L. B. 1998. Tn916 family conjugative transposons and dissemination of antimicrobial resistance determinants. Antimicrob. Agents Chemother. 42:1871-1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Salyers, A. 1993. Gene transfer in the mammalian intestinal tract. Curr. Opin. Biotechnol. 4:294-298. [DOI] [PubMed] [Google Scholar]
- 39.Shoemaker, N. B., G. R. Wang, and A. A. Salyers. 1996. NBU1, a mobilizable site-specific integrated element from Bacteroides spp., can integrate nonspecifically in Escherichia coli. J. Bacteriol. 178:3601-3607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shoemaker, N. B., H. Vlamakis, K. Hayes, and A. A. Salyers. 2001. Evidence for extensive resistance gene transfer among Bacteroides spp. and among Bacteroides and other genera in the human colon. Appl. Environ. Microbiol. 67:561-568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sneath, P. H. A., and R. R. Sokal. 1973. Numerical taxonomy: the principles and practice of numerical classification. W. H. Freeman & Co., San Francisco, CA.
- 42.Sorensen, T. L., M. Blom, D. Monnet, N. Frimodt-Moller, R. L. Poulsen, and F. Espersen. 2001. Transient intestinal carriage after ingestion of antibiotic-resistant Enterococcus faecium from chicken and pork. N. Engl. J. Med. 345:1161-1166. [DOI] [PubMed] [Google Scholar]
- 43.Sutcliffe, J., T. Grebe, A. Tait-Kamradt, and L. Wondrack. 1996. Detection of erythromycin-resistant determinants by PCR. Antimicrob. Agents Chemother. 40:2562-2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Turabelidze, D., M. Kotetishvili, A. Kreger, J. G. Morris, Jr., and A. Sulakvelidze. 2000. Improved pulsed-field gel electrophoresis for typing vancomycin-resistant enterococci. J. Clin. Microbiol. 38:4242-4245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.van den Bogaard, A. E., and E. E. Stobberingh. 2000. Epidemiology of resistance to antibiotics links between animals and humans. J. Antimicrob. Agents 14:327-335. [DOI] [PubMed] [Google Scholar]
- 46.Vankerckhoven, V., T. Van Autgaerden, C. Vael, C. Lammens, S. Chapelle, R. Rossi, D. Jabes, and H. Goossens. 2004. Development of a multiplex PCR for the detection of asa1, gelE, cylA, esp, and hyl genes in enterococci and survey for virulence determinants among European hospital isolates of Enterococcus faecium. J. Clin. Microbiol. 42:4473-4479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Villedieu, A., M. L. Diaz-Torres, N. Hunt, R. McNab, D. A. Spratt, M. Wilson, and P. Mullany. 2003. Prevalence of tetracycline resistance genes in oral bacteria. Antimicrob. Agents Chemother. 47:878-882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang, H. H., M. Manuzon, M. Lehman, K. Wan, H. Luo, T. E. Wittum, A. Yousef, and L. O. Bakaletz. 2006. Food commensal microbes as a potentially important avenue in transmitting antibiotic resistance genes. FEMS Microbiol. Lett. 254:226-231. [DOI] [PubMed] [Google Scholar]
- 49.Weigel, L. M., D. B. Clewell, S. R. Gill, N. C. Clark, L. K. McDougal, S. E. Flannagan, J. F. Kolonay, J. Shetty, G. E. Killgore, and F. C. Tenover. 2003. Genetic analysis of a high-level vancomycin-resistant isolate of Staphylococcus aureus. Science 302:1569-1571. [DOI] [PubMed] [Google Scholar]
- 50.Wilcks, A., S. R. Andersen, and T. R. Licht. 2005. Characterization of transferable tetracycline resistance genes in Enterococcus faecalis isolated from raw food. FEMS Microbiol. Lett. 243:15-19. [DOI] [PubMed] [Google Scholar]
- 51.Witte, W. 1999. Antibiotic resistance in gram-positive bacteria: epidemiological aspects. J. Antimicrob. Chemother. 44(Suppl. A):1-9. [DOI] [PubMed] [Google Scholar]
- 52.Zoetendal, E. G., E. E. Vaughan, and W. M. de Vos. 2006. A microbial world within us. Mol. Microbiol. 59:1639-1650. [DOI] [PubMed] [Google Scholar]