Abstract
We investigated a destructive pathogenic variant of the plant pathogen Ralstonia solanacearum that was consistently isolated in Martinique (French West Indies). Since the 1960s, bacterial wilt of solanaceous crops in Martinique has been caused primarily by strains of R. solanacearum that belong to either phylotype I or phylotype II. Since 1999, anthurium shade houses have been dramatically affected by uncharacterized phylotype II strains that also affected a wide range of species, such as Heliconia caribea, cucurbitaceous crops, and weeds. From 1989 to 2003, a total of 224 R. solanacearum isolates were collected and compared to 6 strains isolated in Martinique in the 1980s. The genetic diversity and phylogenetic position of selected strains from Martinique were assessed (multiplex PCRs, mutS and egl DNA sequence analysis) and compared to the genetic diversity and phylogenetic position of 32 reference strains covering the known diversity within the R. solanacearum species complex. Twenty-four representative isolates were tested for pathogenicity to Musa species (banana) and tomato, eggplant, and sweet pepper. Based upon both PCR and sequence analysis, 119 Martinique isolates from anthurium, members of the family Cucurbitaceae, Heliconia, and tomato, were determined to belong to a group termed phylotype II/sequevar 4 (II/4). While these strains cluster with the Moko disease-causing strains, they were not pathogenic to banana (NPB). The strains belonging to phylotype II/4NPB were highly pathogenic to tomato, eggplant, and pepper, were able to wilt the resistant tomato variety Hawaii7996, and may latently infect cooking banana. Phylotype II/4NPB constitutes a new pathogenic variant of R. solanacearum that has recently appeared in Martinique and may be latently prevalent throughout Caribbean and Central/South America.
The bacterial wilt pathogen, Ralstonia solanacearum (Smith) Yabuuchi et al., is widespread in tropical, subtropical, and warm temperate regions where it is a major constraint to the production of diverse crop species, including both monocots and dicots (41). R. solanacearum causes bacterial wilt on many host plants, and the disease is called by many different names, including Moko and Bugtok diseases of bananas and brown rot disease of potato. The host range of R. solanacearum is unusually wide for a plant pathogen, including over 450 host species in 54 botanical families (14). R. solanacearum is a highly heterogeneous bacterial species; based on phenotypic properties, the species has been divided into five races (related to the ability to wilt members of the family Solanaceae (race 1), banana (race 2), potato and tomato in temperate conditions (race 3), ginger (race 4), and mulberry (race 5) (2, 17, 30) and six biovars (metabolic profiles, related to the ability to metabolize three sugar alcohols and three disaccharides) (15-17). The assessment of the genetic diversity of R. solanacearum employing restriction fragment length polymorphism analysis resulted in the identification of two clusters of strains denoted divisions 1 (“Asiaticum”) and 2 (“Americanum”) (3, 4). Recently, Fegan and Prior (9) proposed a new hierarchical classification scheme, based on sequence analysis of the internal transcribed spacer region, the endoglucanase (egl) gene, and the hrpB gene, that defines phylotypes as “a monophyletic cluster of strains revealed by phylogenetic analysis of sequence data” (8). Four phylotypes were distinguished. Phylotype I corresponds to the “Asiaticum” division 1 of Cook et al. (3) and contains strains belonging to biovars 3, 4, and 5. Phylotype II corresponds to the “Americanum” division 2 of Cook and Sequeira (4) and contains strains belonging to biovar 1/race 1, biovar 1/race 2 (Moko disease-causing strains), biovar 2/race 3, and biovar 2T strains. Phylotype III contains strains from Africa and the Indian Ocean, which belong to biovars 1 and 2T. Phylotype IV contains strains from Indonesia, some strains from Japan, and a single strain from Australia, belonging to biovars 1, 2, and 2T. Phylotype IV also contains the closely related species Ralstonia syzygii and the blood disease bacterium. Each phylotype can be further subdivided into sequevars based on differences in the sequence of a portion of the endoglucanase (egl) gene. The phylotyping scheme proposed by Fegan and Prior (9) is broadly consistent with the former phenotypic and molecular typing schemes and adds valuable information about the geographical origin and in some cases the pathogenicity of strains. In a recent study (13) assessing hybridization of genomic DNA from 18 strains, representative of the known diversity within the R. solanacearum species complex, to a microarray representative of 99% of the genes from reference strain GMI1000, the hierarchical clustering of strains perfectly matched the previous classification in four phylotypes (13).
From this hierarchical scheme, a phylotype-specific multiplex PCR (Pmx-PCR) was derived (9) and a Musa-specific multiplex PCR (Mmx-PCR) which distinguishes three subgroups within R. solanacearum Moko disease-causing strains, sequevar 3 (MLG 24 sensu (3), sequevar 4 (MLG 25), and sequevar 6 (MLG 28). Within sequevar 4, Moko disease-causing strains could be distinguished from strains not pathogenic on banana (34).
In the French West Indies, bacterial wilt has been the most damaging disease of solanaceous crops for decades (33). R. solanacearum was first found on tomato plants in Martinique in 1965 (6) and was responsible for great yield losses on eggplant crops during the early 1980s (24). However, although bananas are an extensive agroindustry crop, R. solanacearum has never been isolated from banana, and Moko disease has never been reported in Martinique. Prior and Steva reported in 1990 (36) that the R. solanacearum populations consisted of a majority of biovar 3 strains (phylotype I) and a minority of biovar 1 strains (phylotype II), all being previously typed as race 1. In April 1999, unusual strains of R. solanacearum were isolated from diseased anthurium (Anthurium andreanum) plants developing leaf yellowing and necrosis, as well as rotting of the rhizome with abundant bacterial ooze. This bacterial disease has caused severe damage in the main anthurium production areas in the central humid highland regions of the island of Martinique (25). In December 2001, R. solanacearum was identified as the causal agent of the first outbreaks of wilting cantaloupe (Cucumis melo L.) in the southeastern part of the island of Martinique. Other cases of bacterial wilt were then reported in 2002 on cucumber (Cucumis sativus L.), pumpkin (Cucurbita moschata L.), zucchini (Cucurbita pepo L.), and in 2003 on watermelon (Citrullus lunatus) (45). Two R. solanacearum strains (ANT307 and ANT1121) isolated from the first outbreaks on anthurium have been previously found to cluster into phylotype IIB sequevar 4 (IIB/4) (34), which contains Moko disease-causing strains (8). Employing the phylotype classification scheme and DNA-based molecular tools (9, 34), we examined the genetic diversity and phylogeny of these strains. We have also examined the pathogenicity of these strains of R. solanacearum causing bacterial wilt to anthurium and cucurbits in Martinique.
(A portion of these results has previously been presented elsewhere [44].)
MATERIALS AND METHODS
Bacterial strains.
Strains (n = 262) used in this study are listed in Table 1 and in Table S1 in the supplemental material. Strains were provided by authors and obtained from collections (Collection Française de Bactéries Phytopathogènes [CFBP], Angers, France; Coleção de Culturas de Fitobactérias do Instituto Biológico [IBSBF], São Paulo, Brazil). Among the non-Martinique strains (called foreign strains in Table 1), one strain from Trinidad was included, as it was isolated in 2006 from a diseased anthurium in the northern range of the island (source, W. Elibox, University of West Indies, St. Augustine, Trinidad). Strains from Brazil that were also isolated from cucurbits and Musa were included as well. The Martinique strains (n = 232) consisted of six strains described by Prior and Steva (36) in 1990, and 225 strains collected during (i) routine analyses (1989 to 2002) of the SPV (Plant Protection Service), and (ii) surveys carried out from 1999 to 2003 by FREDON (Regional Pest Management Federation), and CIRAD (International Center of Agronomic Research for Development). Strains were collected from the highland central humid areas of the island of Martinique (FREDON-SPV surveys 1999, 2000, 2001, and early 2002), from the main production zone for anthurium, from contaminated anthurium shade houses, from weeds (to search for alternative hosts), and from vegetable crops throughout the island (CIRAD survey 2002 to 2003). Diseased plants (leaf, stem, and rhizome) were sampled and cultured to isolate R. solanacearum. Samples were taken from the oozing stems of rotted rhizomes, surface-disinfected leaves of anthurium, stems of wilted cucurbits and other solanaceous plants, and roots and stems of symptomless weeds. The following culture media were used: K+ medium, modified from Kelman's triphenyltetrazolium chloride (TZC) medium (21) (Bacto Peptone, 10 g·liter−1; glucose, 5 g·liter−1; Casamino Acids, 1 g·liter−1; yeast extract, 1 g·liter−1; agar, 15 g·liter−1; TZC, 50 mg·liter−1; pH 7.4), and modified Granada and Sequeira (MGS) medium (12), as modified by Poussier et al. (32) (agar, 18 g·liter−1; tryptone, 1 g·liter−1; Bacto Peptone, 10 g·liter−1; glycerine, 6.3 ml·liter−1; pH 7.2; crystal violet, 3 mg·liter−1; polymyxin B sulfate, 10 mg·liter−1; tyrothricine, 20 mg·liter−1; chloramphenicol, 5 mg·liter−1; TZC, 25 mg·liter−1; propiconazole, 0.04 ml·liter−1; and penicillin G, 20 U·liter−1). Strains were purified by streaking single colonies on K+ medium which was incubated at 28°C for 48 h. Three loopfuls of bacterial culture were then transferred in 3 ml of double-distilled sterile water, and the cultures were stored at room temperature. Strains from the CIRAD collection (CIR strains) were cryopreserved in glycerol-CPG broth as described by Denny and Hayward (5) and stored in a −70°C freezer. A set of 31 strains were deposited at the CFBP under the accession numbers 6776 to 7017 (Table 1 and Table S1 in the supplemental material). For PCR diagnosis, strains were incubated for 1 or 2 days on CPG medium (Casamino Acids, 1 g·liter−1; Bacto Peptone, 10 g·liter−1; glucose, 5 g·liter−1; agar, 15 g·liter−1; pH 7.2) at 28°C, one loopful of bacterial culture was added per ml of double-distilled sterile water, and the suspension was homogenized by vortexing and used directly as the template for PCR.
TABLE 1.
Ralstonia solanacearum strains used in this study
| Straina | Alternate name | Yr of isolation | Host | Originb | Biovarc | Phylotype/sequevar determined byd:
|
Plant pathogenicity tested one: | Reference(s)f | GenBank accession no. for:
|
||
|---|---|---|---|---|---|---|---|---|---|---|---|
| PCR | Trees | egl | mutS | ||||||||
| Strains from Martinique | |||||||||||
| CFBP2955 | MT3 | 1984 | Tomato | Le Lamentin | 3 | I | 36 | ||||
| CFBP2957 | RUN27 | 1987 | Tomato | Case Pilote | 1 | II | IIA/5 | 34, 36 | AF295265 | EF371845 | |
| CFBP2964 | MA4 | 1983 | Eggplant | Schoelcher | 3 | I | 36 | ||||
| CFBP2970 | MP1 | 1985 | Capsicum sp. | Fort de France | 3 | I | 36 | ||||
| CFBP2972 | RUN30 | 1986 | Potato | Le Lamentin | 1 | II | IIA/5 | 34, 36 | AF295264 | AY756807 | |
| CFBP2976 | Mb1 | 1987 | Ensete ventricosum | Le Lamentin | 3 | I | 36 | ||||
| RS25 | 1997 | Tomato | Le Lamentin | II | Tomato | TS | |||||
| CFBP6786 | SPV98-1537 | 1998 | Tomato | Fort de France | II/4NPB | IIB/4 | TS | EF371823 | EF371858 | ||
| CFBP6784 | ANT307 | 1999 | Anthurium sp. | Saint Joseph | 1 | II/4NPB | IIB/4 | AAA banana | 35; TS | EF371813 | AY756742 |
| SPV99-1119 | RUN283 | 1999 | Anthurium sp. | Morne Rouge | 1 | II/4NPB | IIB/4 | TS | EF371829 | EF371863 | |
| SPV99-1541 | RUN280 | 1999 | Anthurium sp. | Gros Morne | 1 | II/4NPB | IIB/4 | TS | EF371822 | EF371857 | |
| CFBP6801 | RUN279 | 2001 | Heliconia caribea | Gros Morne | II/4NPB | IIB/4 | TS | EF371836 | EF371873 | ||
| CFBP6800 | RUN281 | 2001 | Anthurium sp. | Gros Morne | II/4NPB | IIB/4 | TS | EF371825 | EF371865 | ||
| SPV02-60144 | RUN284 | 2002 | Cucumber | Fond Saint Denis | II/4NPB | IIB/4 | TS | EF371832 | EF371867 | ||
| CFBP6795 | SPV02-60152 | 2002 | Pumpkin | Saint Esprit | 1 | II/4NPB | IIB/4 | TS | EF371837 | EF371874 | |
| SPV02-60037 | RUN285 | 2002 | Cantaloupe | Vauclin | II/4NPB | IIB/4 | TS | EF371815 | EF371850 | ||
| CFBP6797 | SPV02-30308 | 2002 | Solanum americanum | Saint-Joseph | II/4NPB | IIB/4 | TS | EF371820 | EF371855 | ||
| CIR02-005(1) | 2002 | Anthurium ferrierense | Saint-Joseph | II/4NPB | AAB and AAA banana, Solanaceae | TS | |||||
| CIR02-035 | 2002 | Sweet pepper | Marigot | I | Solanaceae | TS | |||||
| CIR02-045 | 2002 | Eggplant | Morne des Esses | I | Solanaceae | TS | |||||
| CFBP6783 | RUN17 | 2002 | Heliconia caribea | Morne Rouge | II/4NPB | IIB/4 | AAB and AAA banana, Solanaceae | TS | EF371817 | EF371852 | |
| CIR02-080 | RUN18 | 2002 | A. andreanum | Morne Rouge | II/4NPB | IIB/4 | AAB and AAA banana, Solanaceae | TS | EF371819 | EF371854 | |
| CFBP6782 | CIR02-083 | 2002 | Pandanus sp. | Morne Rouge | II/4NPB | IIB/4 | TS | EF371827 | EF371861 | ||
| CIR02-136(4) | 2002 | A. ferrierense | Le Vauclin | II | AAA banana | TS | |||||
| CIR02-141 | 2002 | Tomato | Ste Anne | II | AAA banana | TS | |||||
| CIR02-204 | 2002 | Tomato | Le Lamentin | II | AAA banana | ||||||
| CFBP6781 | CIR02-142(1) | 2002 | Cucumber | Ste Anne | II/4NPB | AAB and AAA banana, Solanaceae | TS | ||||
| CFBP6780 | RUN287 | 2002 | Tomato | Ste Anne | II/4NPB | IIB/4 | Solanaceae | TS | EF371830 | EF371864 | |
| CFBP6779 | RUN288 | 2002 | Canna indica | Le Lamentin | II | IIA | AAB and AAA banana | TS | EF371835 | EF371872 | |
| CFBP6778 | RUN289 | 2002 | Tomato | Le Lorrain | II/4NPB | IIB/4 | Solanaceae | TS | EF371816 | EF371851 | |
| CFBP7012 | CIR02-229(1) | 2002 | Pumpkin | Le Marin | II/4NPB | IIB/4 | AAB and AAA banana, Solanaceae | TS | EF371818 | EF371853 | |
| CFBP6777 | RUN291 | 2002 | Watermelon | Basse-Pointe | II/4NPB | IIB/4 | Solanaceae | TS | EF371821 | EF371856 | |
| CFBP7013 | RUN292 | 2003 | Tomato | Saint-Esprit | II/4NPB | IIB/4 | Solanaceae | TS | EF371824 | EF371859 | |
| CIR03-076 | 2003 | Tomato | Macouba | I | Tomato | TS | |||||
| CIR03-077 | 2003 | Tomato | Macouba | II/4NPB | AAB and AAA banana, Solanaceae | TS | |||||
| CFBP7017 | RUN293 | 2003 | Watermelon | Macouba | II/4NPB | IIB/4 | Solanaceae | TS | EF371826 | EF371860 | |
| CFBP7016 | RUN294 | 2003 | A. andreanum | Gros Morne | II | IIA/5 | TS | EF371828 | EF371870 | ||
| CFBP7015 | RUN295 | 2004 | Plantain | Trinité | II/4NPB | IIB/4 | TS | EF371842 | EF371866 | ||
| CIR04-006/16F | RUN296 | 2004 | Plantain | Trinité | II/4NPB | IIB/4 | TS | EF371834 | EF371862 | ||
| Foreign strains | |||||||||||
| CFBP7014 | CIR06-024 | 2006 | A. andreanum | Trinidad | II/4NPB | IIB | TS | AF371831 | EF371875 | ||
| IBSBF1503 | RUN302 | 1999 | Cucumis sativus | Brazil | II/4NPB | IIB/4 | TS | EF371840 | EF371868 | ||
| IBSBF1546 | RUN298 | 2001 | Fuchsia sp. | Brazil | II | IIA | TS | EF371838 | EF371877 | ||
| IBSBF1454 | RUN303 | 1999 | Cucurbita pepo | Brazil | II/4NPB | IIB/4 | TS | EF371844 | EF371876 | ||
| IBSBF1712 | RUN299 | 2002 | Pelargonium × hortorum | Brazil | II | IIB | TS | EF371833 | EF371869 | ||
| IBSBF1900 | RUN301 | 2000 | Musa sp. | Brazil | II/6 | IIA/6 | TS | EF371839 | EF371871 | ||
| Reference strains | |||||||||||
| GMI1000 | RUN54 | Tomato | French Guyana | I | I/13 to 18 | 9 | AF295251 | AY756804 | |||
| CFBP2968 | RUN58 | Eggplant | Guadeloupe | I | I/13 to 18 | 34 | EF371806 | AY756800 | |||
| JT523 | RUN333 | Potato | Reunion Island | I | I/13 to 18 | 34 | AF295252 | AY756803 | |||
| CFBP765 | RUN34 | Tobacco | Japan | I | I/13 to 18 | 34 | EF371810 | AY756740 | |||
| NCPPB3190 | RUN78 | Tomato | Malaysia | I | I/13 to 18 | 34 | AF295253 | AY756738 | |||
| ACH92 | RUN158 | Ginger | Australia | I | I/13 to 18 | 34 | AF295254 | AY756764 | |||
| R292 | RUN91 | Morus alba | China | I | I/12 | 34 | AF295255 | AY756801 | |||
| ICMP7963 | RUN55 | Potato | Kenya | II | IIA/7 | 34 | AF295263 | AY766776 | |||
| K60 | CFBP2047 | Tomato | United States | II | IIA/7 | 34 | AF295262 | AY756799 | |||
| A3909 | RUN9 | Heliconia | Hawaii | II/6 | IIA/6 | 34 | EF371812 | AY756753 | |||
| CFBP2958 | RUN28 | Tomato | Guadeloupe | II | IIA/5 | 34 | AF295266 | AY756806 | |||
| IPO1609 | RUN1 | Potato | The Netherlands | II | IIB/1 and 2 | 34 | EF371814 | EF371849 | |||
| JT516 | RUN160 | Potato | Reunion Island | II | IIB/1 and 2 | 34 | AF295258 | AY756783 | |||
| NCPPB3987 | RUN81 | Potato | Brazil | II | IIB/ND | 34 | AF295261 | AY756785 | |||
| UW477 | RUN110 | Potato | Peru | II | IIB/ND | 34 | AF295260 | AY756821 | |||
| MOLK2 | RUN74 | Musa sp. | Philippines | II/3 | IIA/3 | 34 | EF371841 | EF371848 | |||
| UW9 | JT644 | Heliconia | Costa Rica | II/3 | IIA/3 | 34 | AF295257 | AY756744 | |||
| CFBP1409 | JS775 | Musa sp. | Honduras | II/3 | IIB/3 | 34 | EF371808 | AY756751 | |||
| CFBP1183 | JS793 | Heliconia | Costa Rica | II/3 | IIB/3 | 34 | EF371805 | AY756749 | |||
| UW70 | RUN99 | Musa sp. (plantain) | Colombia | II/4SFR | IIB/4 | 34 | DQ011550 | AY756794 | |||
| UW129 | Musa sp. (plantain) | Peru | II/4A | IIB/4 | 34 | EF371811 | AY756782 | ||||
| UW162 | JT648 | Musa sp. (plantain) | Peru | II/4A | IIB/4 | 35 | AF295256 | AY756795 | |||
| ICMP6782 | Musa sp. | Brazil | II/6 | IIB/6 | 8 | DQ011553 | NAg | ||||
| ICMP9600 | Musa sp. | Brazil | II/6 | IIB/6 | 8 | DQ011554 | NA | ||||
| ANT307 | RUN16 | A. andreanum | Martinique | II/4NPB | IIB/4 | 35 | EF371813 | AY756742 | |||
| UW21 | RUN97 | Musa sp. | Honduras | II/6 | IIB/6 | 35 | DQ011546 | AY756758 | |||
| CFBP3059 | RUN39 | Eggplant | Burkina Faso | III | III/19 to 23 | 9 | AF295270 | AY756766 | |||
| CFBP734 | JS767 | Potato | Madagascar | III | III/19 to 23 | 34 | AF295274 | AY756746 | |||
| NCPPB332 | RUN75 | Potato | Zimbabwe | III | III/19 to 23 | 34 | AF295276 | AY756760 | |||
| JT525 | RUN60 | Pelargonium asperum | Reunion Island | III | III/19 to 23 | 34 | AF295272 | AY756786 | |||
| MAFF301558 | RUN71 | Potato | Japan | IV | IV/10 | 34 | DQ011558 | AY756752 | |||
| PSI7 | RUN83 | Tomato | Indonesia | IV | IV/10 | 34 | EF371804 | AY756732 | |||
CFBP, Collection Françaises de Bactéries Phytopathogènes, Angers, France; IBSBF, Coleção de Culturas de Fitobactérias do Instituto Biológico, São Paulo, Brazil.
Strains were all isolated in Martinique (French West Indies) except for the foreign strains and reference strains.
The biovars were determined in microplates by the method of French et al. (10).
The phylotypes and sequevars determined by PCR were those determined by the Pmx-PCR and Mmx-PCR (9, 34). The strains marked II gave no signal when tested by the Mmx-PCR. The phylotypes and sequevars determined by trees were determined by egl and mutS sequence analyses.
Pathogenicity was tested as described in the text on plantain banana (AAB), Cavendish banana (AAA), members of the family Solanaceae, and/or tomato genotypes only.
Reference numbers are given with one exception. TS, this study.
NA, not available.
DNA typing.
Two mx-PCR procedures were applied to characterize strains based on the hierarchical classification scheme described previously (9, 34). Pmx-PCR (11) was performed to identify which phylotype a strain belonged to. When the strain belonged to phylotype II, Mmx-PCR (35) was performed to identify whether the strain belonged to sequevar 3, 4, or 6 (Moko disease-inducing strains). Briefly, Pmx-PCR includes the primers 759/760 (29) as an internal marker providing the 280-bp reference PCR product which is specific for the R. solanacearum species and a set of four phylotype-specific forward primers (Nmult:21:1F [5′-CGTTGATGAGGCGCGCAATTT-3′], Nmult:21:2F [5′-AAGTTATGGACGGTGGAAGTC-3′], Nmult:22:InF [5′-ATTGCCAAGACGAGAGAAGTA-3′], and Nmult:23:AF [5′-ATTACGAGAGCAATCGAAAGATT-3′]), with a unique and conserved reverse primer (Nmult:22:RR [5′-TCGCTTGACCCTATAACGAGTA-3′]), targeted in the 16S-23S intergenic spacer region (internal transcribed spacer). This Pmx-PCR produces the following phylotype-specific PCR products: a 144-bp amplicon from phylotype I strains, a 372-bp amplicon from phylotype II strains, a 91-bp amplicon from phylotype III strains, and a 213-bp amplicon from phylotype IV strains. Taq polymerase and deoxynucleoside triphosphates (dNTPs) were from Promega France (Charbonnières-les-Bains, France), and primers (11) were obtained from Eurogentec (Seraing, Belgium). PCRs were performed in a PCR Express thermocycler (Hybaid, Teddington, United Kingdom). PCR products were resolved through 2% agarose gels in Tris-acetate-EDTA (TAE) buffer and revealed by staining in 0.5 μg·ml−1 ethidium bromide. Amplicon sizes were estimated by comparison to a 100-bp DNA ladder (Invitrogen, Life Technologies, Cergy-Pontoise, France). Mmx-PCR includes four sets of forward and reverse primers amplifying genomic DNA fragments by a subtractive hybridization approach (35): Mus35-F (5′-GCAGTAAAGAAACCCGGTGTT-3′) and Mus35-R (5′-TCTGGCGAAAGACGGGATGG-3′), Mus20-F (5′-CGGGTGGCTGAGACGAATATC-3′) and Mus20-R (5′-GCCTTGTCCAGAATCCGAATG-3′), Mus06-F (5′-GCTGGCATTGCTCCCGCTCAC-3′) and Mus06-R (5′-TCGCTTCCGCCAAGACGC-3′), and Si28-F (5′-CGTTCTCCTTGTCAGCGATGG-3′) and Si28-R (5′-CCCGTGTCACCCCGATAGC-3′). This Mmx-PCR provides sequevar-specific PCR products: a 400-bp product indicates the strain belongs to sequevar 3 (equivalent to MLG 24 of Cook and Sequeira [3]); a 220-bp product indicates the strain belongs to sequevar 6 (equivalent to MLG 28 of Cook and Sequeira [3]); a 351-bp product indicates the strain belongs to sequevar 4 (equivalent to MLG 25 of Cook and Sequeira [3]). Within sequevar 4, strains of the SFR ecotype (insect-transmitted strains whose colony morphology is small, fluidal, and round sensu French and Sequeira [11]) produce an additional amplicon of 167 bp as do strains of the A ecotype (insect-transmitted strains from the Amazon basin, colonies are round and mucoid with no or very little formazan coloration; sensu French and Sequeira (11). Prior and Fegan (34) found that the sequevar 4 strains ANT307 and ANT1121 from Martinique (CFBP6784 and 6785 in our study), did not produce this additional amplicon. These strains have since been designated phylotype II/sequevar 4 NPB (II/4NPB), as they were also found to be nonpathogenic for banana (P. Prior and D. Dufeal, personal communication in 1999). Strains that do not belong to the Musa group (sequevar 3, 4, or 6) do not produce any PCR product when subjected to Mmx-PCR.
PCR amplification and DNA sequencing of portions of the endoglucanase (egl) and DNA repair (mutS) genes.
PCR amplification of a 750-bp region of the egl gene was performed by using the primer pair Endo-F (5′-ATGCATGCCGCTGGTCGCCGC-3′) and Endo-R (5′-GCGTTGCCCGGCACGAACACC-3′). The reaction mixture (total volume, 100 μl) contained PCR buffer (supplied by the manufacturer), 1.5 mM MgCl2, 200 μM of each dNTP, 25 pmol of each primer, 2 μl of a turbid bacterial suspension as the template, and 1 U of AmpliTaq Gold DNA polymerase (Applied Biosystems, Foster City, CA). PCR was performed using a MJ Research PTC200 thermocycler (Bio-Rad Laboratories, Inc., Hercules, CA) using the following protocol: (i) initial denaturation at 96°C for 9 min; (ii) 30 cycles, with 1 cycle consisting of 95°C for 1 min, 70°C for 1 min, and 72°C for 2 min; and (iii) a final extension step of 72°C for 10 min. PCR amplification of a 758-bp fragment of the terminal portion of the mutS gene was performed using a MJ Research PTC200 thermocycler. Reactions were carried out by the method of Prior and Fegan (34) in a total volume of 25 μl containing 1× PCR buffer (Promega), 1.5 mM MgCl2, 200 μM of each dNTP, 0.8 μM dimethyl sulfoxide, 1.25 U of Taq polymerase (Promega), and 9 pmol each of the primers mutS-RsF.1570 (5′-ACAGCGCCTTGAGCCGGTACA-3′) and mutS-RsR1926 (5′-GCTGATCACCGGCCCGAACAT-3′). Reaction mixtures were heated to 96°C for 5 min and then cycled through 35 cycles of 94°C for 60 s, 66°C for 60 s, and 72°C for 90 s, followed by a final extension period of 5 min at 72°C. Samples (5 μl) of reaction mixtures were examined by electrophoresis through 2% agarose gels in TAE buffer, and bands were revealed by staining in 0.5 μg ml−1 ethidium bromide. PCR products were purified, and sequencing reaction was performed on both strands by Macrogen sequencing services (Kumchun-ku, Seoul, Korea) with the mutS and Endo primers mentioned above.
Sequence analysis.
Portions of the sequences of the housekeeping DNA repair mutS gene located on the chromosome and of the endoglucanase (egl) virulence-related gene located on the megaplasmid were analyzed by using the ARB software environment (23). Sequences were manually aligned using the ARB sequence editor. Sequence data were analyzed using the three major phylogenetic approaches offered by the ARB software. Evolutionary relationships between sequences were computed by the following methods: (i) by using the algorithm of Jukes and Cantor (20), phylogenetic trees were constructed from the genetic distance data by using the neighbor-joining (NJ) method (37) with 5,000 bootstrap resamplings of the data to test the tree topologies; (ii) maximum likelihood methods using the FastDNAML program; and (iii) the maximum parsimony methods using the Phylip DNAPARS package implemented in the ARB software. From the pool of mutS and egl sequences included in this study (61 and 63 sequences, respectively), sequences from reference strains were retrieved, and sequences from newly described strains were deposited into the GenBank database (Table 1).
Pathogenicity tests.
A subset of R. solanacearum strains was assembled to represent all phylotypes, with particular attention to phylotype II/4NPB strains. This subset of strains was tested for pathogenicity for different hosts: banana, tomato, sweet pepper, and eggplant (Table 1). Pathogenicity was assessed on Cavendish (AAA genome, cv. Grande Naine) and plantain (cooking banana) (AAB genome, cv. Dominico-Harton) banana, and different solanaceous hosts. Strains were transferred from glycerol-CPG broth onto K+ medium or Sequiera medium South Africa (SMSA) (5) and grown for 2 or 3 days at 28°C. Single mucoid colonies were then selected, streaked on CPG medium, and grown at 28°C for 48 h. Bacterial growth on a petri dish was harvested into 5 ml Tris buffer (0.01 M Tris, pH 7.2). Cell suspensions were spectrophotometrically adjusted to 108 CFU·ml−1 (optical density at 600 nm of 0.1) prior to use as a plant inoculum.
Cavendish and plantain bananas were tested in separate experiments. Banana and plantain plants were grown in a quarantine nursery in heat-sterilized potting mix. When they reached the four-leaf stage (approximately 40 cm high), they were infected by employing two techniques: injection of 0.1 ml of inoculum into the base of the pseudostem and root scarification prior to adding 10 ml inoculum to the soil (1 liter of soil per pot) surface (drenching inoculation). Seven strains (six phylotype II/4NPB strains and one phylotype II non-sequevar 3, 4, or 6 strain) were inoculated on Cavendish and plantain bananas, whereas four other strains (one phylotype II/4NPB strain and three phylotype II non-sequevar 3, 4, or 6 strains) were inoculated on Cavendish bananas only. Six plants were inoculated with each strain (three plants by each inoculation technique), and six control plants were inoculated with sterile water. All 48 plantain and 72 Cavendish plants were maintained on plastic trays 1 m above the ground in a randomized experimental design at 29°C ± 6°C in a closed quarantine greenhouse, watered as needed with tap water, and observed every 2 days for 50 to 54 days. Finally, plants were uprooted, and microbiological isolations were done on three sections of each plant: roots, pseudocorm, and the top of the pseudostem (at the insertion of the leaf blades). Pseudostem and pseudocorm pieces were surface sterilized by dipping the pieces in 95% ethanol and flamed; roots were first thoroughly washed under tap water, then surface sterilized by dipping the roots in ethanol, and dried in a laminar hood on sterile filter paper. Plant material was then chopped and crushed with a pestle in 5 ml Tris buffer (pH 7.2) and then macerated for 15 min. Afterwards, macerates were streaked (50 μl per petri dish) on MGS medium and SMSA (5) and incubated 28°C for 2 or 3 days to allow development of characteristic R. solanacearum colonies (mucoid egg-shaped colonies, with pink-reddish center). Typical colonies, chosen at random, were identified as R. solanacearum by Pmx-PCR.
The pathogenicity of 10 phylotype II/4NPB strains chosen at random (Table 1) was tested on solanaceous commercial varieties, namely, tomato cv. Heat Master (Seminis), sweet pepper cv. Narval (Tezier), and eggplant cv. Kalenda (Technisem). Plants were inoculated at the two to three expanded leaf stage in a complete block design with three blocks and 10 plants per block. Inoculation was done by drenching inoculation by adding 10 ml of inoculum to the soil (108 CFU·ml−1) of each plant, after the roots had been scarified with a sterile scalpel. Incubation was carried out in a greenhouse (29°C ± 6°C), and plants were monitored for wilting symptoms every 2 or 3 days for 41 days. Disease incidence was assessed by the final wilting rate. Disease severity was estimated by the area under the disease progression curve (AUDPC) (39), calculated with the formula:
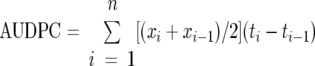 |
where n is the number of times the plants were evaluated, xi is the wilting rate at each evaluation time, and (ti − ti−1) is how long wilting lasted (39).
The virulence/aggressiveness of strains that represent phylotypes I (CIR03-076), IIA (RS25), and IIB/4NPB (CIR03-077, CFBP6783, and CFBP7013) on tomato plants was assessed on a range of cultivars carrying some degree of genetic resistance to bacterial wilt (38, 43): Roma (susceptible control, Technisem), R3034 (University of the Philippines, Los Baños), CRA84 (AVRDC, Taiwan), Hawaii7996 (University of Florida), and BF-Okitsu (NIVOT, Japan). Plants were inoculated as described above at the two or three fully expanded leaf stage in a complete randomized design with two replicates and 10 plants per replicate. Incubation was carried out in a climatic chamber with a 14-h photoperiod (28°C day/24°C night), and wilt symptoms were monitored every 2 or 3 days for 30 days. Disease incidence was assessed by the final wilting rate, and disease severity was estimated by the AUDPC (38).
Data analysis.
Bliss transformation (arcsin √x) was applied to wilt rate data prior to statistical analysis. Analysis of variance (ANOVA) was used to determine the effects of isolate, species, and isolate-species interactions on the final wilting incidence and AUDPC scores. Statistical analysis was performed with the software STATBOX Pro 2.5 (Grimmersoft, Paris, France), and the ANOVA requirements (homogeneity of the intratreatment variances, normality of the residuals, and independence of the residuals) were fulfilled for both arcsin-transformed wilting data and AUDPC. When the ANOVA assumptions were not validated for AUDPC, AUDPC values were transformed by the function √x. The chi-square test (Microsoft Excel 2000) was applied to analyze the distribution of the different phylotypes within the different host groups (members of the family Solanaceae, anthurium, and cucurbits) and the distribution of the different host groups for the different types of strains (phylotypes I, II, and II/4NPB).
Nucleotide sequence accession numbers.
The sequences from newly described strains were deposited into the GenBank database (Table 1).
RESULTS
Strain characterization.
Pmx-PCR showed that all biovar 3 strains belonged to phylotype I and biovar 1 strains belonged to phylotype II (Table 1). Mmx-PCR showed that strains isolated from cucurbit and anthurium hosts predominantly belonged to phylotype II sequevar 4 and displayed the 351-bp amplicon only. Strains with this pattern were designated phylotype II/4NPB (not pathogenic to banana) as explained in the introduction. The phylotyping results for the 232 R. solanacearum strains from Martinique are reported in Table 2. Strains from the collection of Prior and Steva (36) which were collected in the 1980s belonged to phylotype I or phylotype II (Table 1), but the strains belonging to phylotype II did not produce amplicon when used as the template in the Mmx-PCR, indicating that they did not belong to sequevar 3, 4, or 6. Within strains collected in Martinique from 1989 to early 2002 (SPV collection) and late 2002 to 2003 (CIRAD collection), three groups of strains were identified: phylotype I; phylotype II non-sequevar 3, 4, or 6; and phylotype II/4NPB. In the SPV collection, 64.7% strains were phylotype II/4NPB, 21.2% were phylotype I, and 14.1% were phylotype II non-sequevar 3, 4, or 6 strains. In the CIRAD collection, 46% were phylotype II/4NPB strains, 28.8% were phylotype I, and 25.2% were phylotype II non-sequevar 3, 4, or 6 strains.
TABLE 2.
Distributions of the different phylotypes in the collections of Ralstonia solanacearum strains from Martinique based upon DNA typing employing the Pmx- and Mmx-PCR tests
| Collection (yr) (reference) | No. (%) of strains of collection
|
Total no. of strains | ||
|---|---|---|---|---|
| Phylotype I | Phylotype II subcluster/sequevar
|
|||
| A/Non-Musa | B/4NPB | |||
| Prior and Steva (1990) (36) | 4 (66.7) | 2 (33.3) | 0 (0) | 6 |
| SPV (1989-2002) | 18 (21.2) | 12 (14.1) | 55 (64.7) | 85 |
| CIRAD survey (2002-2003) | 40 (28.8) | 35 (25.2) | 64 (46) | 139 |
| Total no. of strains | 62 (26.7) | 51 (22) | 119 (51.3) | 232 |
Relationship of R. solanacearum genotype to host.
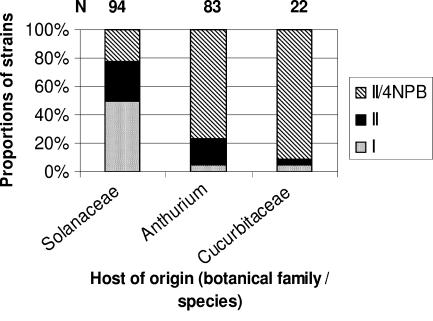
In Martinique, R. solanacearum strains were mostly isolated from the family Solanaceae, Anthurium andreanum, and the family Cucurbitaceae (Fig. 1). Strains isolated from Solanaceae were 50% phylotype I, approximately 28% phylotype II non-sequevar 3, 4, or 6, and 22% phylotype II/4NPB. In contrast, strains isolated from anthurium were 5% phylotype I, 18% phylotype II non-sequevar 3, 4, or 6, and 77% phylotype II/4NPB. Strains from cucurbits were 4.5% phylotype I, 4.5% phylotype II (not sequevar 3, 4, or 6), and 91% phylotype II/4NPB. The frequency distributions of each host group within the different phylotypes were significantly different from their expected values (χ2 test, P = 2.21 × 10−9 for Solanaceae, P = 7.67 × 10−6 for anthurium, and P = 0.0018 for cucurbits). The frequency distributions of phylotype I strains within the different host groups were significantly different from their expected values (χ2 test, P = 1.54 × 10−8), much higher within the strains isolated from the Solanaceae and much lower within the strains isolated from anthurium and cucurbits. The frequency distributions of phylotype II/4NPB strains were also significantly higher than expected (χ2 test, P = 6.03 × 10−6) within the strains derived from anthurium and cucurbits and significantly lower than expected within the Solanaceae strains. Thus, these data suggested that phylotype I strains were predominantly isolated from Solanaceae, whereas phylotype II/4NPB strains were predominantly isolated from anthurium and cucurbits.
FIG. 1.
Distribution of phylotypes I, II, and II/4NPB of Ralstonia solanacearum in Martinique as related to the host of origin of the strains.
Pathogenicity to Musa spp.
No Moko disease or any external symptom of disease was observed on Cavendish and plantain bananas 51 and 54 days after inoculation with any strain. After the plants were cut into pieces, no bacterial oozing was observed, although occasionally purple discoloration of the vascular tissues within the pseudocorm was observed. No bacterium of any genus was recovered from the control plants (inoculated with water). On plantain banana, following injection inoculation, all the phylotype II/4NPB strains were recovered from pseudocorm, pseudostem, and roots, except CIR02-080 which was recovered from pseudostem and pseudocorm only. In contrast, strain CFBP6779 (a phylotype II non-sequevar 3, 4, or 6 strain) was recovered only from pseudocorm, near the inoculation point (Table 3). Following drenching inoculation, recoveries were more variable, even though three out of six strains (CFBP6781, CFBP7012, and CIR03-077) were recovered from all points on the plant, indicating that they invaded the plant from roots to leaf top. On Cavendish banana, R. solanacearum recovery was observed only on plants inoculated by injection with strain CIR03-077 and by drenching inoculation with strain CFBP6783. Both strains were phylotype II/4NPB. No recovery of R. solanacearum was found for reference strain CFBP6784 (ANT307 (34) inoculated on Cavendish banana from any banana plant. The only successful recovery of a phylotype II non-sequevar 3, 4, or 6 strain from an inoculated plant was obtained from the pseudocorm of one plantain plant inoculated with strain CFBP6779 from Canna indica. With the exception of this case, no phylotype II non-sequevar 3, 4, or 6 strain was ever recovered from any infected banana plant.
TABLE 3.
Recovery of Ralstonia solanacearum from plantain (AAB) and Cavendish (AAA) bananas
| Strain, host, and phylotype/sequevar | Tissue or organa | Bacterial recoveryb from banana type by inoculation technique
|
|||
|---|---|---|---|---|---|
| Plantain
|
Cavendish
|
||||
| Injection | Drenching | Injection | Drenching | ||
| CIR02-005(1), Anthurium andreanum, II/4NPB | Roots | + | − | − | − |
| Pseudocorm | + | + | − | − | |
| Pseudostem | + | − | − | − | |
| CFBP6783, Heliconia caribea, II/4NPB | Roots | + | + | − | + |
| Pseudocorm | + | − | − | − | |
| Pseudostem | + | − | − | − | |
| CIR02-080, A. andreanum, II/4NPB | Roots | − | − | − | − |
| Pseudocorm | + | − | − | − | |
| Pseudostem | + | + | − | − | |
| CFBP6781, Cucumis sativus, II/4NPB | Roots | + | + | − | − |
| Pseudocorm | + | + | − | − | |
| Pseudostem | + | + | − | − | |
| CFBP7012, Cucurbita moschata, II/4NPB | Roots | + | + | − | − |
| Pseudocorm | + | + | − | − | |
| Pseudostem | + | + | − | − | |
| CIR03-077, Lycopersicon esculentum, II/4NPB | Roots | + | + | − | − |
| Pseudocorm | + | + | − | − | |
| Pseudostem | + | + | + | − | |
| CFBP6779, Canna indica, II | Roots | − | − | − | − |
| Pseudocorm | + | − | − | − | |
| Pseudostem | − | − | − | − | |
| CIR02-136(4), A. ferrierense, II | Roots | NT | NT | − | − |
| Pseudocorm | NT | NT | − | − | |
| Pseudostem | NT | NT | − | − | |
| CIR02-204, L. esculentum, II | Roots | NT | NT | − | − |
| Pseudocorm | NT | NT | − | − | |
| Pseudostem | NT | NT | − | − | |
| CIR02-141, L. esculentum, II | Roots | NT | NT | − | − |
| Pseudocorm | NT | NT | − | − | |
| Pseudostem | NT | NT | − | − | |
| CFBP6784,cA. andreanum, II/4NPB | Roots | NT | NT | − | − |
| Pseudocorm | NT | NT | − | − | |
| Pseudostem | NT | NT | − | − | |
| None (water control) | Roots | − | − | − | − |
| Pseudocorm | − | − | − | − | |
| Pseudostem | − | − | − | − | |
Tissue or organ from which the bacteria were isolated.
Symbols: +, successful recovery of R. solanacearum on at least one plant; −, no recovery of R. solanacearum. NT, not tested.
Reference strain of reference 34.
Pathogenicity on members of the family Solanaceae.
Ten phylotype II/4NPB strains were tested for pathogenicity on tomato, sweet pepper, and eggplant, and the results were compared to plants inoculated with two phylotype I strains (Table 4). All phylotype II/4NPB strains from Martinique were pathogenic to Solanaceae, irrespective of the host from which they were initially isolated. Based on both AUDPC and the final wilting rate, sweet pepper and tomato were the most susceptible species, eggplant being less affected (Table 4). Virulence within strains belonging to phylotype II/4NPB varied according to strain and host, but these variations were not statistically significant. Surprisingly, the phylotype I strains were not ranked as the most aggressive strains on their respective hosts of origin (sweet pepper for CIR02-035, eggplant for CIR02-045).
TABLE 4.
Test for pathogenicity to members of the family Solanaceae
| Strain (phylotype/sequevar) | Pathogenicity on:
|
|||||
|---|---|---|---|---|---|---|
| Tomato
|
Sweet pepper
|
Eggplant
|
||||
| Mean AUDPCa | % Wilting at day 41b | Mean AUDPC | % Wilting at day 41 | Mean AUDPC | % Wilting at day 41 | |
| CFBP6783 (II/4NPB) | 29.5 A | 90.0 A | 29.8 A | 100.0 A | 8.6 AB | 30.0 A |
| CIR02-080 (II/4NPB) | 27.7 AB | 83.0 A | 16.8 AB | 60.0 B | 9.0 AB | 47.0 A |
| CIR02-005 (II/4NPB) | 27.6 AB | 87.0 A | 30.2 A | 100.0 A | 7.6 ABC | 33.0 A |
| CFBP6781 (II/4NPB) | 26.3 AB | 87.0 A | 16.5 AB | 87.0 AB | 3.8 ABCD | 20.0 A |
| CFBP6780 (II/4NPB) | 26.0 AB | 80.0 A | 13.6 AB | 60.0 B | 6.9 ABC | 33.0 A |
| CFBP7017 (II/4NPB) | 24.7 AB | 73.0 A | 20.1 AB | 80.0 AB | 3.9 ABC | 23.0 A |
| CFBP7012 (II/4NPB) | 18.3 AB | 60.0 A | 20.1 AB | 67.0 AB | 2.3 BCD | 20.0 A |
| CFBP6777 (II/4NPB) | 17.3 AB | 57.0 A | 24.4 A | 80.0 AB | 13.5 A | 53.0 A |
| CIR03-077 (II/4NPB) | 16.6 AB | 57.0 A | 23.6 A | 93.0 AB | 5.4 ABC | 23.0 A |
| CIR02-045 (I) | 14.1 ABC | 47.0 A | 25.5 A | 93.0 AB | 9.7 AB | 47.0 A |
| CFBP6778 (II/4NPB) | 10.7 ABC | 40.0 A | 8.5 B | 53.0 B | 0.4 CD | 20.0 A |
| CIR02-035 (I) | 8.7 BC | 30.0 A | 9.8 B | 67.0 AB | 1.3 BCD | 17.0 A |
| None (control) (water) | 0.0 C | 0.0 B | 0.0 C | 0.0 C | 0.0 D | 0.0 B |
| Mean AUDPCc | 19.0 A | 18.4 A | 5.6 B | |||
| Mean % wiltingc | 61.0 B | 71.0 A | 28.0 C | |||
Mean AUDPC, mean area under the disease progression curve (n = 3). See the text for details on its calculation. Values followed by the same letter in the same column are not significantly different (Newman-Keuls test [P = 0.05] done on square root-transformed values).
Mean percentage of wilted plants (n = 3 replicates, 10 plants per replicate) assessed on the 41st day after inoculation. Values followed by the same letter in the same column are not significantly different (Newman-Keuls test [P = 0.05] done on arcsin [square root]-transformed values).
Values followed by the same letter in the same row are not significantly different (Newman-Keuls test, P = 0.05).
Among tomato cultivars tested with phylotype II/4NPB strains CIR03077, CFBP7013, and CFBP6783, phylotype II non-sequevar 3, 4, or 6 strain RS25, and phylotype I strain CIR03076, Roma was the most affected cultivar, followed by Hawaii7996, R3034, CRA84, and BF-Okitsu. Phylotype II/4NPB strains were able to wilt all five cultivars, whereas phylotype II non-sequevar 3, 4, or 6 strains and phylotype I strains were unable to cause disease on Hawaii7996, CRA84, and BF-OKITSU and did not confer wilting (Table 5) or stem colonization (data not shown). The three phylotype II/4NPB were significantly more aggressive than the phylotype II non-sequevar 3, 4, or 6 strains and phylotype I strains, inducing a final wilting rate of 50 to 60% on the universal resistant reference strain Hawaii7996.
TABLE 5.
Test for virulence to tomato
| Strain | Phylotype | Virulence on tomato line
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| % Wilting at day 30a
|
Mean AUDPCb
|
||||||||||||
| Roma | R3034 | Hawaii7996 | CRA84 | BF-Okitsu | Mean | Roma | R3034 | Hawaii7996 | CRA84 | BF-Okitsu | Mean | ||
| CIR03077 | II/4NPB | 70.0 A | 50.0 A | 60.0 A | 30.0 AB | 55.0 A | 53.0 A | 16.0 A | 10.7 A | 12.2 A | 6.5 AB | 12.1 A | 11.4 A |
| CFBP7013 | II/4NPB | 60.0 A | 25.0 B | 50.0 A | 50.0 A | 20.0 B | 42.0 A | 12.7 A | 6.0 A | 8.6 A | 10.0 A | 4.6 B | 9.0 A |
| CFBP6783 | II/4NPB | 57.0 A | 30.0 B | 50.0 A | 45.0 A | 25.0 B | 42.0 A | 14.1 A | 7.0 A | 8.2 A | 9.8 A | 5.0 B | 9.2 A |
| RS25 | II | 44.0 A | 20.0 B | 0.0 B | 0.0 B | 0.0 B | 13.0 B | 9.6 AB | 3.6 A | 0.0 B | 0.0 B | 0.0 B | 2.6 B |
| CIR03076 | I | 24.0 B | 10.0 B | 0.0 B | 0.0 B | 0.0 B | 7.0 B | 3.7 B | 2.9 A | 0.0 B | 0.0 B | 0.0 B | 1.2 B |
| Meanc | 53.0 A | 27.0 B | 32.0 B | 25.0 B | 20.0 B | 11.2 A | 6.0 B | 6.6 B | 5.2 B | 4.3 B | |||
Mean percentage of wilted plants (n = 2 replicates, 10 plants per replicate) Assessed on the 30th day After inoculation. Values followed by the same letter in the same column Are not significantly different (Newman-Keuls test [P = 0.05], done on arcsin [square root]-transformed wilt values and on AUDPC values).
Mean AUDPC, mean area under the disease progression curve (n = 2). See the text for details on its calculation. Values followed by the same letter in the same column are not significantly different (Newman-Keuls test [P = 0.05], done on arcsin [square root]-transformed wilt values and on AUDPC values).
Values followed by the same letter in the same row are not significantly different (Newman-Keuls test [P = 0.05]).
Phylogenetic analysis.
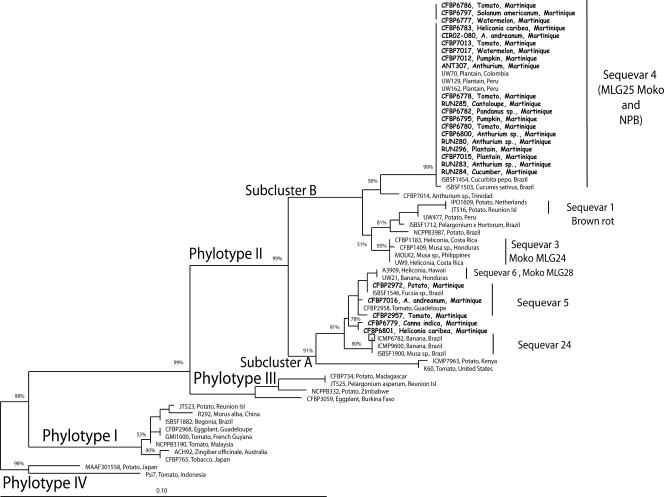
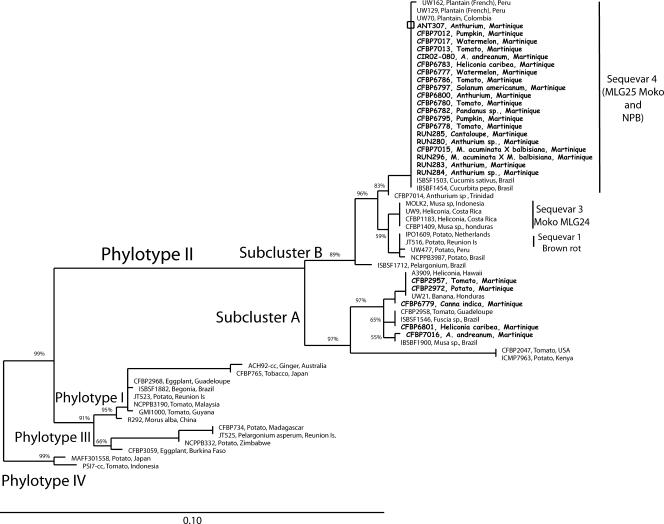
Partial egl and mutS gene sequences were generated from a set of representative strains from Martinique, including phylotype II non-sequevar 3, 4, or 6 strains and phylotype II/4NPB strains, and compared to different reference strains covering the known genetic diversity within the four phylotypes shown in both the endoglucanase (egl) (9) and the mutS (35) phylogenetic trees. Trees constructed with maximum likelihood, maximum parsimony, and distance NJ phylogenetic approaches were totally congruent; therefore, only NJ phylogenetic trees, carrying bootstrap values, are presented (Fig. 2 and 3). NJ trees from egl and mutS gene sequences were in general agreement, showing a similar branching pattern clearly partitioning the four phylotypes as previously described in the literature (Fig. 2 and 3): reference strains from phylotypes I, III, and IV clustered separately with high bootstrap values. Sequences of strains that were Pmx-PCR typed phylotype II resolved in two major and robust branches previously described by Fegan and Prior (8): (i) phylotype IIA, containing strains that were isolated from diverse hosts, and (ii) phylotype IIB, the Moko disease-causing strains from sequevar 3 (MLG24) and 4 (MLG25) and the highland temperate potato brown rot strains (biovar 2/race 3) from sequevars 1 and 2. Phylotype II strains from Martinique clustered in both phylotypes IIA and IIB. The strains that clustered into phylotype IIA were isolated from tomato, potato, heliconia, and anthurium (including CFBP7016), and none of these strains produced amplicons with the Mmx-PCR and were therefore phylotype II non-sequevar 3, 4, or 6 strains. The 21 strains isolated from various hosts in Martinique, which were typed as phylotype II/4NPB based on Mmx-PCR, clustered into phylotype IIB regardless of the egl or mutS sequences analyzed. Phylogenetically, the 20 phylotype II/4NPB strains were undistinguishable from the Moko disease-causing strains UW70, UW129 and UW162 that were included as reference for sequevar 4 (MLG25). Interestingly, Brazilian strains ISBSF1503 from Cucumis sativus and ISBSF1454 from Cucurbita pepo also clustered in phylotype IIB sequevar 4. From both the egl and mutS phylogenetic trees, phylotype IIB sequevar 4 strains had identical sequences, with the exception of strain CFBP7014 which varied by 10 nucleotide positions, while the closely related strain CFBP7014, isolated from anthurium in Trinidad, was distinguishable from sequevar 4 strains, forming a new branch with high bootstrap support (83% and 56% in mutS and egl trees, respectively). The phylogenetic position of strain ISBSF1712 isolated from Pelargonium in Brazil was not congruent between the egl and mutS phylogenetic trees. Based on the egl gene sequence, ISBSF1712 was closely related to phylotype IIB sequevars 1 and 2, in a group that contains biovar 2/race 3 strains that cause potato brown rot and is listed in the United States as a bioterrorism select agent (22). Based on the mutS gene sequence, ISBSF1712 resolved into a distinct and robust cluster (89% bootstrap value). Based on the egl gene sequence analysis of Fegan and Prior (8), the sequences of Brazilian strains ICMP6782 and ICMP 9600 were included as reference sequences for phylotype II subcluster A sequevar 24, a previously unrecognized group of R. solanacearum Moko disease-causing strains. Musa strain IBSBF1900 was the only other strain found to belong to this group (90% bootstrap value), and no Martinique strain was assigned to it.
FIG. 2.
Phylogenetic NJ tree based on the partial endoglucanase (egl) gene sequences of strains from Martinique and reference strains for the R. solanacearum species complex. Strains originating from Martinique are written in bold type. The number at each node is the bootstrap value (5,000 resamplings), and significant bootstrap values less than 100% are indicated at each node. The scale bar represents 1 nucleotide substitution per 10 nucleotides. Reunion Isl, Reunion Island.
FIG. 3.
Phylogenetic NJ tree based on partial sequences of the DNA repair mutS gene from strains from Martinique and reference strains for the R. solanacearum species complex. Strains originating from Martinique are indicated in bold type. The number at each node is the bootstrap value (5,000 resamplings), and significant bootstrap values less than 100% are indicated at each node. The scale bar represents 1 nucleotide substitution per 10 nucleotides. Reunion Is, Reunion Island.
DISCUSSION
Ralstonia solanacearum strains causing bacterial wilt on solanaceous crops and ornamental plants are found in all four phylotypes of the R. solanacearum species complex defined by Fegan and Prior (9). Conversely, strains that have ecologically adapted to a particular host, for example, Moko disease-causing strains pathogenic to banana, have much narrower genetic diversity. R. solanacearum strains causing Moko disease have been resolved into distinct subclusters within phylotype II: sequevar 3 (MLG24), sequevar 4 (MLG25), sequevar 6 (MLG28), and sequevar 24 (8). We report here a new group of strains which were predominantly isolated from diseased anthurium and cucurbit species (Fig. 1 and reference 45) and pathogenic on solanaceous crops as well. Bacterial wilt of anthurium caused by R. solanacearum has been reported in Singapore (46), Reunion Island (31), and Mauritius Island (7) and was caused by biovar 3 strains; in Taiwan (42), it was caused by biovar 4 strains. The positions of these strains in the phylotyping scheme (11) have not been determined, except for the anthurium strains from Reunion Island that have been found to belong to phylotype I (31); however, these strains most likely belong to phylotype I. In Martinique, the R. solanacearum strains from anthurium were shown to belong to phylotype II sequevar 4 according to the DNA molecular tools developed by Prior and Fegan (34). As phylotype II sequevar 4 contains strains that cause Moko disease (10, 35), this finding potentially constituted a great threat to the banana industry in the French West Indies. This paper describes the genetic diversity and pathogenicity of these strains, as well as the phylogenetic position of this new group of R. solanacearum strains in Martinique, which have been named phylotype II/4NPB.
R. solanacearum strains in the French West Indies have previously been isolated from solanaceous plants and ornamental banana (Musa ensete) and classified as biovar 3 and biovar 1, with a dominance of biovar 3 strains (36), whereas Moko disease had never been observed in Martinique. By application of the Pmx- and Mmx-PCR tests in this study on the Prior and Steva strains (36), we confirmed that biovar 3 strains belong to phylotype I and that biovar 1 strains are phylotype II non-sequevar 3, 4, and 6 strains, as would have been expected. The recent (1999 to 2003) extensive survey of isolates of R. solanacearum from Martinique has revealed that the strains belonged to phylotype I, phylotype II non-sequevar 3, 4, and 6, and phylotype II sequevar 4 based on Pmx- and Mmx-PCR-based identification tools. Since the identification of the first phylotype II sequevar 4 strain in Martinique in 1998 from tomato, this group of R. solanacearum strains has been regarded as an emerging group in Martinique. The well-established phylotype I and phylotype II non-sequevar 3, 4, and 6 strains that have been present in Martinique for decades are regarded as the historical population.
Bacterial wilt caused by R. solanacearum biovar 1 strains has been previously reported for summer squash, zucchini summer squash, and cucumber in Brazil, but the phylotype affiliation of the strains involved was not determined (28, 40). This study has shown that some Brazilian strains isolated in 1999 from Cucurbita pepo (ISBSF1454) and Cucumis sativus (ISBSF1503) also clustered into phylotype II/4. As these Brazilian strains were genotyped as phylotype II/4NPB by the Mmx-PCR, we may consider that they would be more closely related to the strains from Martinique than to Moko disease-causing strains. Furthermore, Moko disease-causing strains have never been reported to be pathogenic to cucurbits. However, this hypothesis remains to be validated by pathogenicity tests on banana. While being phylogenetically undistinguishable from other members of phylotype II/4, the pathogenicities of the strains from Martinique within this cluster are clearly different from the Moko disease-causing strains of sequevar 4 (MLG25). Moko disease-causing strains of phylotype II/4 cause wilting of plantain and banana (3, 4, 11, 19). However, the phylotype II/4 strains from Martinique are unable to wilt Cavendish banana (AAA genotype Musa sp.) and plantain (AAB genotype Musa sp.) and were thus designated not pathogenic to banana (NPB) and designated phylotype II/4NPB. These phylotype II/4NPB strains were unable to spread from the inoculation point within the plant tissues of the Cavendish cultivar used, regardless of the inoculation method (pseudostem or roots). For example, reference strain CFBP6784 (34) was not pathogenic to and did not establish any latent infection in Cavendish banana (AAA genotype) tissues. Nevertheless, the phylotype II/4NPB strains, although not pathogenic to banana, had the capacity to establish and move within the vascular tissues of plantain (AAB genotype), whatever the point of entry into the plant. After being injected directly in the pseudostem, strains of R. solanacearum phylotype II/4NPB were shown to invade the tissues both upwards and downwards from the point of inoculation. Following soilborne contamination, these strains were able to enter the root system and spread throughout the plant to establish latent infections in asymptomatic plantain stalks. This feature needs to be confirmed on other plantain cultivars, but if confirmed, this may be a key epidemiological issue, as native plantains are distributed throughout the island of Martinique in Creole gardens, although plantain has never been cultivated as a cash crop in the French West Indies as it has in Central America and especially in Colombia. The ability of phylotype II/4NPB strains to infect plantains was confirmed by the identification of naturally infected plants in 2004 in one of the few industrial plantain plantations (1). Strains CFBP7015 and CIR04-006/16F were isolated from the flower peduncle of a diseased plantain of cv. French in Martinique, whose terminal leaf growth was blocked and bunch was distorted, but showing no typical wilt symptoms. This may indicate the potential of phylotype II/4NPB strains to cause Moko disease. Also, as these strains were isolated from the flower peduncle, it may indicate that these strains were transmitted by insects, as are the Moko disease-causing strains of phylotype II/4 (MLG25). Fortunately, growers of Cavendish bananas on large banana plantations in the French West Indies remove the male bud on each stalk, thus preventing the aerial mode of entry of pathogen into the plant. The phenotype of this last case may lead us to consider these phylotype II/4NPB strains as similar to ecotype “D,” “isolated from Heliconia and causing leaf distortion and slow wilting of bananas” (11), or ecotype “H,” “affecting plantains but not bananas.” However, recent phylogenetic analysis has assigned these two ecotypes to sequevar 3 (34) and French and Sequeira (11) reported these ecotypes to be weakly pathogenic to eggplant and pepper, unlike the strains from Martinique.
Other strains with a genotype similar to those of the phylotype II/4NPB strains have been reported outside of Martinique. In 1998, two strains (P515 and P548) that were isolated in Florida from pothos (Epipremnum aureum) cuttings imported from Costa Rica (26) were reported to cluster with anthurium strains from Martinique in phylotype IIB/4 (35). However, these strains from pothos were described as weakly pathogenic to tomato (unlike the strains from Martinique) and mildly pathogenic to Cavendish banana, inducing “dark veins, leaf necrosis and chlorosis,” but not wilting or death (26). More recently, R. solanacearum strains isolated from anthurium in Florida (27) were reported to induce chlorosis, necrosis, and wilting within 2 weeks on anthurium, tomato, and triploid banana, but their phylogenetic position in the phylotyping scheme has not yet been established. These reports and our study show that extensive comparative studies will be required to assess pathogenicity of strains belonging to phylotype II/4 on the available genetic diversity in triploid banana genomes (AAA, AAB, and ABB) and diploid banana.
Phylotype II/4NPB strains appear to represent a relatively new emerging pathogenic variant within phylotype II sequevar 4. Indeed, this genetic group was absent in R. solanacearum collections from the French West Indies until the first strain was isolated from a wilted tomato during a survey in Martinique in 1998. Furthermore, this group of strains carries a previously unknown host range, including solanaceous, cucurbit, and ornamental plants, while being not pathogenic to Cavendish banana, even in artificial infections. Cantaloupe, watermelon, and pumpkin were not considered hosts of R. solanacearum until the first report by Wicker et al. (45). None of the Moko disease-causing strains belonging to phylotype II/4 (MLG25) have been described as being naturally pathogenic for vegetable crops (11). However, strains can wilt and kill tomato when artificially inoculated into the stem (13). The only Moko disease-causing strains that have been isolated from naturally infected tomato and reported to be both pathogenic to solanaceous and banana plants belong to phylotype II sequevar 6 (MLG28) (34). Although extensively surveyed only in Martinique, it is possible that phylotype II/4NPB strains may be present throughout the entire Caribbean region and even South America. Two phylotype II/4NPB strains were also identified in 1998 from pothos imported from Costa Rica (26), and this study has shown that similar strains are present in Brazil and Trinidad. The factors that may explain the establishment of this new pathogenic variant in Martinique will be discussed in another paper focusing on the epidemiology of this group of strains.
The phylotype II/4NPB strains were also highly aggressive and virulent on tomato, significantly overcoming the resistance carried in tomato strain Hawaii7996 (50 to 60% wilt), whereas the phylotype I and phylotype II non-sequevar 3, 4, and 6 strains tested were unable to wilt this variety. It is therefore possible that phylotype II/4NPB strains thus constitute a new challenge for tomato breeders to develop cultivars in this region; screening tests done recently tend to support this hypothesis (E. Wicker, unpublished 2005 and 2006 data).
The egl and mutS phylogenetic trees presented in this study were in agreement with previous work (8, 9, 13, 34, 35), and all sequences analyzed in this work clustered in previously described phylotypes and sequevars. However, strain CFBP 7014 from Trinidad and strain ISBSF1712 from Brazil did not cluster with other phylotype II/4NPB strains. According to the phylogenetic analysis, strain CFBP7014, isolated from anthurium in Trinidad, resolved in a cluster distinct from the sequevar 4 cluster within subgroup IIB (Fig. 2 and 3), whereas this strain was genotyped as phylotype II/4NPB by Pmx- and Mmx-PCRs. Both egl and mutS sequences from this particular strain placed it in an unique single-isolate cluster, expanding the known genetic diversity within strains of R. solanacearum. Further investigations are needed, particularly for extended knowledge of the R. solanacearum populations and of the epidemiological context in Trinidad. The Brazilian strain ISBSF1712 that was isolated in 2002 from pelargonium also gave inconsistent clusters in egl and mutS phylogenetic trees. Further investigations will be necessary to infer its degree of genetic relatedness with the pelargonium strains reported in the United States (18).
The use of the phylotyping scheme proposed by Fegan and Prior (9) and the PCR-based molecular tools described by Fegan and Prior (9) and Prior and Fegan (34) has allowed the identification and characterization of group of R. solanacearum strains with a novel pathogenicity. We propose that strains described in this paper, belonging to phylotype II/4NPB, should be considered a new pathogenic variant within the phylotype II/4 phylogenetic group. This variant has been recognized in Martinique since 1998, and similar strains have also been isolated in Brazil, Costa Rica, and Trinidad. Many questions remain to be addressed on the epidemiology of this pathogenic variant. It remains unclear whether this variant arose in Martinique or elsewhere and how widespread it may be on the island of Martinique. Further work is required to determine how widespread this pathogenic variant of R. solanacearum has become.
Supplementary Material
Acknowledgments
We thank W. Elibox and A. Ramsubhag (UWI, Trinidad) for kindly providing strains. We thank Gary Burkhardt for editing an early draft of this paper, as well as Olivier Pruvost and Caitylin Allen for very valuable comments.
This work was financially supported by DOCUP-FEOGA (2000 to 2006) and by the Regional Council of Martinique (Conseil Régional de Martinique).
Footnotes
Published ahead of print on 24 August 2007.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Anonymous. 2004. Détection de Ralstonia solanacearum dans un plant de bananier plantain en Martinique. Direction de l'Agriculture et de la Forêt, Service de la Protection des Végétaux, avec le concours du CIRAD-FLHOR.
- 2.Buddenhagen, I. W., L. Sequeira, and A. Kelman. 1962. Designation of races in Pseudomonas solanacearum. Phytopathology 52:726. [Google Scholar]
- 3.Cook, D., E. Barlow, and L. Sequeira. 1989. Genetic diversity of Pseudomonas solanacearum: detection of restriction fragment length polymorphisms with DNA probes that specify virulence and the hypersensitive response. Mol. Plant-Microbe Interact. 2:113-121. [Google Scholar]
- 4.Cook, D., and L. Sequeira. 1994. Strain differentiation of Pseudomonas solanacearum by molecular genetic methods, p. 77-94. In A. C. Hayward and G. L. Hartman (ed.), Bacterial wilt: the disease and its causative agent, Pseudomonas solanacearum. CAB International, Wallingford, United Kingdom.
- 5.Denny, T. P., and A. C. Hayward. 2001. Ralstonia solanacearum, p. 151-173. In N. W. Schaad, J. B. Jones, and W. Chun (ed.), Laboratory guide for identification of plant pathogenic bacteria, 3rd ed. APS Press, St. Paul, MN.
- 6.Digat, B., and A. Escudié. 1967. Reconnaissance du flétrissement bactérien des Solanées aux Antilles Françaises. Phytiatr. Phytopharm. 16:187-197. [Google Scholar]
- 7.Dookun, A., S. Saumtally, and S. Seal. 2001. Genetic diversity in Ralstonia solanacearum strains from Mauritius using restriction fragment length polymorphisms. Phytopatholog. Z. 149:51-55. [Google Scholar]
- 8.Fegan, M., and P. Prior. 2006. Diverse members of the Ralstonia solanacearum species complex cause bacterial wilts of banana. Australas. Plant Pathol. 35:93-101. [Google Scholar]
- 9.Fegan, M., and P. Prior. 2005. How complex is the “Ralstonia solanacearum species complex,” p. 449-462. In C. Allen, P. Prior, and A. C. Hayward (ed.), Bacterial wilt disease and the Ralstonia solanacearum species complex. APS Press, Madison, WI.
- 10.French, E. B., L. Gutarra, P. Aley, and J. Elphinstone. 1995. Culture media for Ralstonia solanacearum isolation, identification and maintenance. Fitopatologia 30:126-130. [Google Scholar]
- 11.French, E. R., and L. Sequeira. 1970. Strains of Pseudomonas solanacearum from Central and South America: a comparative study. Phytopathology 60:506-512. [Google Scholar]
- 12.Granada, G. A., and L. Sequeira. 1983. A new selective medium for Pseudomonas solanacearum. Plant Dis. 67:1084-1088. [Google Scholar]
- 13.Guidot, A., P. Prior, J. Schoenfeld, S. Carrere, S. Genin, and C. Boucher. 2007. Genomic structure and phylogeny of the plant pathogen Ralstonia solancearum inferred from gene distribution analysis. J. Bacteriol. 189:377-387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hayward, A. C. 1991. Biology and epidemiology of bacterial wilt caused by Pseudomonas solanacearum. Annu. Rev. Phytopathol. 29:65-87. [DOI] [PubMed] [Google Scholar]
- 15.Hayward, A. C. 1964. Characteristics of Pseudomonas solanacearum. J. Appl. Bacteriol. 27:265-277. [Google Scholar]
- 16.Hayward, A. C., H. M. El-Nashaar, U. Nydegger, and L. De Lindo. 1990. Variation in nitrate metabolism in biovars of Pseudomonas solanacearum. J. Appl. Bacteriol. 69:269-280. [Google Scholar]
- 17.He, L. Y., L. Sequiera, and A. Kelman. 1983. Characteristics of strains of Pseudomonas solanacearum from China. Plant Dis. 67:1357-1361. [Google Scholar]
- 18.Ji, P., C. Allen, A. Sanchez-Perez, J. Yao, J. Elphinstone, J. B. Jones, and M. T. Momol. 2007. New diversity of Ralstonia solanacearum strains associated with vegetable and ornamental crops in Florida. Plant Dis. 91:195-203. [DOI] [PubMed] [Google Scholar]
- 19.Jones, D. R. (ed.). 2000. Diseases of banana, abaca and enset. CABI Publishing, New York, NY.
- 20.Jukes, T. H., and C. R. Cantor. 1969. Evolution of protein molecules, p. 21-132. In H. N. Munro (ed.), Mammalian protein metabolism. Academic Press, New York, NY.
- 21.Kelman, A. 1954. The relationship of pathogenicity in Pseudomonas solanacearum to colony appearance on tetrazolium medium. Phytopathology 44:693-695. [Google Scholar]
- 22.Lambert, C. D. 2002. Agricultural Bioterrorism Protection Act of 2002: Possession, use and transfer of biological agents and toxins; interim final rule. (7CFR part 331) Fed. Reg. 67:76908-76938. [Google Scholar]
- 23.Ludwig, W., O. Strunk, R. Westram, L. Richetr, H. Meier, Yadhukumar, A. Buchner, T. Lai, S. Steppi, G. Jobb, W. Förster, I. Brettske, S. Gerber, A. W. Ginhart, O. Gross, S. Grumann, S. Hermann, R. Jost, A. König, T. Liss, R. Lüssman, M. May, B. Nonhoff, B. Reichel, R. Strehlow, A. Stamatakis, N. Stuckmann, A. Vilbig, M. Lenke, T. Ludwig, A. Bode, and K. H. Schleifer. 2004. ARB: a software environment for sequence data. Nucleic Acids Res. 32:1363-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Messiaen, C.-M. 1983. Impressions de voyage en Martinique: la culture de l'aubergine va-t-elle disparaître définitivement de ce pays? Bull. Agron. Antilles-Guyane 1:17-22. [Google Scholar]
- 25.Mian, D., R. Coranson Beaudu, D. Dufeal, L. Grassart, and P. Mention. 2002. Ralstonia solanacearum sur anthurium a la Martinique [France]. Une bacteriose inquietante. Phytoma 551:43-45. [Google Scholar]
- 26.Norman, D. J., and J. M. F. Yuen. 1998. A distinct pathotype of Ralstonia (Pseudomonas) solanacearum race 1, biovar 1 entering Florida in pothos (Epipremnum aureum) cuttings. Can. J. Plant Pathol. 20:171-175. [Google Scholar]
- 27.Norman, D. J., and J. M. F. Yuen. 1999. First report of Ralstonia (Pseudomonas) solanacearum infecting pot anthurium production in Florida. Plant Dis. 83:300. [DOI] [PubMed] [Google Scholar]
- 28.Oliveira, J. R. D., and A. B. Moura. 1994. Doenças causadas por bactérias em cucurbitaceas. Inf. Agropecuario (Belo Horizonte) 17:54-57. [Google Scholar]
- 29.Opina, N., F. Tavner, G. Hollway, J.-F. Wang, T.-H. Li, R. Maghirang, M. Fegan, A. C. Hayward, V. Krishnapillai, W. F. Hong, B. W. Holloway, and J. Timmis. 1997. A novel method for development of species and strain-specific DNA probes and PCR primers for identifying Burkholderia solanacearum (formerly Pseudomonas solanacearum). Asia Pac. J. Mol. Biol. Biotechnol. 5:19-30. [Google Scholar]
- 30.Pegg, K., and M. Moffett. 1971. Host range of the ginger strain of Pseudomonas solanacearum in Queensland. Aust. J. Exp. Agric. Anim. Husb. 11:696-698. [Google Scholar]
- 31.Poussier, S., D. Trigalet-Demery, P. Vandewalle, B. Goffiner, J. Luisetti, and A. Trigalet. 2000. Genetic diversity of Ralstonia solanacearum as assessed by PCR-RFLP of the hrp region, AFLP and 16S rRNA sequence analysis, and identification of an African subdivision. Microbiology 146:1679-1692. [DOI] [PubMed] [Google Scholar]
- 32.Poussier, S., P. Vandewalle, and J. Luisetti. 1999. Genetic diversity of African and worldwide strains of Ralstonia solanacearum as determined by PCR-restriction fragment length polymorphism analysis of the hrp gene region. Appl. Environ. Microbiol. 65:2184-2194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Prior, P., P. Cadet, and H. Steva. 1989. Variabilité du pouvoir pathogène de Pseudomonas solanacearum aux Antilles françaises (Martinique et Guadeloupe). Acta Oecol. 10:135-142. [Google Scholar]
- 34.Prior, P., and M. Fegan. 2005. Diversity and molecular detection of Ralstonia solanacearum race 2 strains by multiplex PCR, p. 405-414. In C. Allen, P. Prior, and A. C. Hayward (ed.), Bacterial wilt disease and the Ralstonia solanacearum species complex. APS Press, Madison, WI.
- 35.Prior, P., and M. Fegan. 2005. Recent development in the phylogeny and classification of Ralstonia solanacearum, p. 127-136. In T. Momol, P. Ji, and J. B. Jones (ed.), Proceedings of the First International Symposium on tomato diseases, vol. 695. International Society for Horticultural Science, Bruggen, Belgium. [Google Scholar]
- 36.Prior, P., and H. Steva. 1990. Characteristics of strains of Pseudomonas solanacearum from the French West Indies. Plant Dis. 74:13-17. [Google Scholar]
- 37.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 38.Scott, J. W., J. F. Wang, and P. Hanson. 2005. Breeding tomatoes for resistance to bacterial wilt, a global view, p. 161-172. In M. T. Momol, P. Ji, and J. B. Jones (ed.), Proceedings of the First International Symposium on Tomato Diseases, vol. 695. International Society for Horticultural Science, Bruggen, Belgium. [Google Scholar]
- 39.Silveira, E. B., A. M. A. Gomes, S. J. Michereff, and R. L. R. Mariano. 1998. Variability of Ralstonia solanacearum populations causing wilt on tomato in Agreste of Pernambuco, Brazil. Bacterial Wilt Newsl. 15:8-10. [Google Scholar]
- 40.Sinigaglia, C., M. E. B. M. Lopes, I. M. G. Almeida, and J. R. Neto. 2001. Bacterial wilt of summer squash (Cucurbita pepo) caused by Ralstonia solanacearum in the State of Sao Paulo, Brazil. Summa Phytopathol. 27:251-253. [Google Scholar]
- 41.Smith, J. J., L. C. Offord, M. Holderness, and G. S. Saddler. 1995. Genetic diversity of Burkholderia solanacearum (synonym Pseudomonas solanacearum) race 3 in Kenya. Appl. Environ. Microbiol. 61:4263-4268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Su, C. C., and L. S. Leu. 1995. Bacterial wilt of anthurium caused by Pseudomonas solanacearum. Plant Pathol. Bull. 4:34-38. [Google Scholar]
- 43.Wang, J.-F., P. Hanson, and J. A. Barnes. 1998. Worldwide evaluation of an international set of resistant sources to bacterial wilt in tomato, p. 269-275. In P. Prior, C. Allen, and J. G. Elphinstone (ed.), Bacterial wilt disease—molecular and ecological aspects. Springer-Verlag, Berlin, Germany.
- 44.Wicker, E., L. Grassart, R. Coranson-Beaudu, D. Mian, C. Guilbaud, and P. Prior. 2005. Emerging strains of Ralstonia solanacearum in Martinique (French West Indies): a case study for epidemiology of bacterial wilt, p. 145-152. In M. T. Momol, P. Ji, and J. B. Jones (ed.), Proceedings of the First International Symposium on Tomato Diseases, vol. 695. International Society for Horticultural Science, Bruggen, Belgium. [Google Scholar]
- 45.Wicker, E., L. Grassart, D. Mian, R. Coranson Beaudu, D. Dufeal, C. Guilbaud, and P. Prior. 2002. Cucumis melo, Cucumis sativus, Cucurbita moschata, and Anthurium spp., new hosts of Ralstonia solanacearum in Martinique (French West Indies). Bacterial Wilt Newsl. 17:20-21. [Google Scholar]
- 46.Yik, C. P., A. K. Ong, and R. Ho. 1994. Characterization of Pseudomonas solanacearum strains from Singapore. Singapore J. Prim. Ind. 22:57-62. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.