Abstract
The growing importance of mass spectrometry for the identification and characterization of bacterial protein toxins is a consequence of the improved sensitivity and specificity of mass spectrometry-based techniques, especially when these techniques are combined with affinity methods. Here we describe a novel method based on the use of immunoaffinity capture and matrix-assisted laser desorption ionization-time of flight mass spectrometry for selective purification and detection of staphylococcal enterotoxin B (SEB). SEB is a potent bacterial protein toxin responsible for food poisoning, as well as a potential biological warfare agent. Unambiguous detection of SEB at low-nanogram levels in complex matrices is thus an important objective. In this work, an affinity molecular probe was prepared by immobilizing anti-SEB antibody on the surface of para-toluene-sulfonyl-functionalized monodisperse magnetic particles and used to selectively isolate SEB. Immobilization and affinity capture procedures were optimized to maximize the density of anti-SEB immunoglobulin G and the amount of captured SEB, respectively, on the surface of magnetic beads. SEB could be detected directly “on beads” by placing the molecular probe on the matrix-assisted laser desorption ionization target plate or, alternatively, “off beads” after its acidic elution. Application of this method to complex biological matrices was demonstrated by selective detection of SEB present in different matrices, such as cultivation media of Staphylococcus aureus strains and raw milk samples.
Protein toxins secreted by bacteria are the most powerful human poisons known (15, 31). Over the last 10 years, both matrix-assisted laser desorption ionization (MALDI) mass spectrometry (MS) and electrospray ionization MS have played more important roles in both the detection and structural characterization of bacterial protein toxins (36), such as botulinum (3, 7, 17), diphtheria (33), tetanus (40), cholera (9, 36), and Shiga-like (42) toxins, as well as staphylococcal enterotoxins (9, 20, 26). MS-based identification of bacterial protein toxins relies on (i) proteomic approaches (5, 38-40), (ii) indirect measurement of target peptides produced as a result of highly specific enzymatic (i.e., endoprotease) activity of the toxin itself (3, 7), and (iii) direct determination of the molecular mass of the toxin after chromatographic or affinity separation (8, 20, 26, 42). MS-based proteomic approaches are used mainly in comparative analyses to obtain an overall picture of the secreted virulence factors, including bacterial protein toxins, in biological samples, such as the exoprotein fraction of culture supernatants (5, 25, 43). One drawback of MS-based proteomic methods for the detection and identification of bacterial protein toxins, however, is the need to perform separation steps before analysis, which limits application of these methods for rapid screening. Also, the dynamic ranges and sensitivities of proteomic techniques are not able to reach the toxin detection limit at the intoxication level. Recently, a rapid method using protein chip technology based on surface-enhanced laser desorption ionization-time of flight (TOF) MS was introduced to study production of the secreted Streptococcus pyogenes cysteine protease (SpeB) in culture supernatant by determination of the molecular mass of the intact protein (8). SpeB was isolated using a hydrophobic surface that binds proteins by hydrophobic interaction.
Staphylococcus aureus is an important human pathogen because of its resistance to environmental stress (10), its increasing antibiotic resistance (34), and its ability to survive extremely well outside the host (41). S. aureus produces a number of virulence factors to counter the host's defense systems (2, 14). Among these are several families of exotoxins, including hemolysins, leukocytolytic toxins, exofoliative toxins, etc.; pyrogenic superantigens, such as toxic shock syndrome toxin 1; and enterotoxins (12). Staphylococcal enterotoxins, a family of at least 16 antigenically distinct medium-molecular-mass (23- to 29-kDa) heat-stable proteins, are known to be potent gastrointestinal toxins. Staphylococcal enterotoxins are superantigens and potent activators of CD4+ T cells, which cause rapid and massive proliferation of cells and cytokine production (22). Staphylococcal enterotoxin B (SEB) is a single-chain polypeptide (molecular mass, 28.4 kDa) consisting of 239 amino acid residues (14). In our study SEB was used as a model system to set up an MS-based assay capable of selectively detecting protein toxin in complex matrices because (i) SEB most commonly causes classic food poisoning, (ii) it is the best-characterized staphylococcal enterotoxin, (iii) it is an important cause of non-menstruation-associated toxic shock syndrome, and (vi) it has also been studied as a potential biological warfare and incapacitating agent, because it is easily aerosolized, stable, and can cause widespread sickness. Sabotage of food and/or water with SEB is therefore a possibility for terrorist attacks. SEB is the most heat-stable staphylococcal enterotoxin and is also resistant to the proteolytic enzymes of the gastrointestinal tract and to low pH. The toxic and lethal doses of SEB depend on the animal species and also on the route of exposure (35, 37). Between 100 and 200 ng of ingested SEB can cause symptoms of staphylococcal intoxication. In humans, currently the estimated 50% effective dose and 50% lethal dose are 0.4 and 20 ng/kg, respectively, for aerosol exposure (12, 35). Due to the high toxicity of SEB, detection methods must be sensitive enough to measure food contamination at low-nanogram amounts of the toxin per gram of food. An enzyme-linked immunosorbent assay (ELISA) is the most frequently used method to search for SEB in direct analyses of food (2, 4) and in biological samples (29). Traditional immunochemical methods, however, are time-consuming, and in some cases antibody cross-reactivity with food and biological matrices may prevent efficient and reliable identification of the toxin, resulting in false-positive results and/or a reduced detection limit (13). A number of alternative methods have also been developed for the detection of SEB, including a protein chip (18), surface plasmon resonance (24), biosensors (32), a time-resolved fluorescence assay (27), a piezoelectric crystal immunosensor (23), and an automated fiber optic biosensor (21). Commercial immunoassay kits (30) are also available for rapid analysis of SEB.
The aim of our study was to develop a new method based on selective immunoaffinity capture enrichment that can be performed in a simple single-tube experiment for selective isolation of the protein toxin of interest. Immunoaffinity separation provides a rapid and efficient way to increase the amount of the target molecule in a biological sample (1). In this work, we bound anti-SEB antibody to magnetic particles functionalized by a p-toluene-sulfonyl (tosyl) group in order to prepare an immunoaffinity molecular probe, which was then used for affinity capture of SEB. MALDI-TOF MS analysis was employed for selective detection of SEB directly on beads or after elution (off beads).
MATERIALS AND METHODS
Materials.
SEA, SEB, and polyclonal affinity-purified anti-SEB immunoglobulin G (IgG) (≥95% IgG) and anti-SEA IgG (≥95% IgG) were obtained from Toxin Technology, Inc. (Sarasota, FL). Fractionated antiserum, anti-SEB developed in rabbits, 1,2-dimethoxy-4-hydroxycinnamic acid (sinapinic acid), and all other chemicals were obtained from Sigma-Aldrich (St. Louis, MO). M-280 Dynabeads (tosyl-activated superparamagnetic polystyrene beads coated with a polyurethane layer) were obtained from Dynal (Norway). The physical characteristics of the beads were as follows: diameter, 2.8 ± 0.2 μm; surface area, 4 to 8 m2/g; active chemical functionality, 50 to 70 μmol/g; density, 1.3 g/cm3; and concentration, 2 × 109 beads/ml (approximately 30 mg/ml). Prior to use beads were washed twice in buffer A. A magnetic particle concentrator (Dynal, Norway) was used to facilitate supernatant removal and washing steps. High-performance liquid chromatography gradient-grade solvents were purchased from Merck (Darmstadt, Germany). ZipTip C18 and Millex-GX filters were obtained from Millipore (Vimodrone, Italy). A Vivaspin 20 molecular filter was obtained from Sartorius AG (Goettingen, Germany). A reversed passive latex agglutination (RPLA) SET-RPLA kit was obtained from Oxoid Limited (United Kingdom).
Preparation of anti-SEB- and anti-SEA-coated magnetic particles.
The following buffers were used for coating and elution. Buffer A consisted of 0.1 M sodium phosphate buffer (pH 7.4) (2.62 g NaH2PO4·H2O and 14.42 g Na2HPO4·2H2O were dissolved in water, and the volume was adjusted to 1,000 ml). Buffer B consisted of phosphate-buffered saline (pH 7.4) with 0.1% (wt/vol) bovine serum albumin (BSA) (0.88 g NaCl and 0.8 g BSA were dissolved in 0.01 M sodium phosphate [pH 7.4], and the volume was adjusted to 100 ml). Buffer C consisted of 0.2 M Tris (pH 8.5) with 0.1% (wt/vol) BSA (2.42 g Tris was dissolved in distilled water, the pH was adjusted to 8.5 with 1 M HCl, and the volume was adjusted to 100 ml). Buffer D consisted of 100 mM glycine (the pH was adjusted to 2.5 with 1 M HCl).
The final volume of affinity-purified anti-SEB IgG (60 μg, unless otherwise stated) or anti-SEB antiserum (11 μl) was adjusted to 100 μl with buffer A, and the preparation was added to a Dynabead suspension (100 μl), mixed for 1 min, and incubated with shaking at 37°C for 24 h. The supernatant was removed, and the particles were washed two times with buffer B (400 μl) at 4°C for 5 min. Free tosyl groups were blocked by buffer C (400 μl) at 37°C for 4 h. Finally, the particles were washed in buffer B (400 μl) at 4°C for 5 min. Anti-SEA IgG-coated magnetic beads were prepared in the same way.
Immunomagnetic isolation of SEB.
Anti-SEB-coated magnetic beads were washed twice in buffer B (400 μl) and resuspended in an SEB-containing solution (sample). During optimization of immobilization and affinity capture, unless otherwise stated, SEB (20 pmol) dissolved in buffer B (100 μl) was used as the sample. The suspension was incubated with shaking at 37°C for 1 h, and the supernatant was removed. The beads were washed five times in buffer B (400 μl). The supernatant was removed, and beads (1 μl) were loaded directly onto a MALDI target plate for online MALDI-TOF MS analysis. Elution of SEB from the surface-immobilized antibody-antigen complex and regeneration of anti-SEB-coated beads for reuse were performed using buffer D (50 μl). The solution was vortexed for 1 min, and the supernatant was removed immediately. The beads were washed five times in buffer B (400 μl) and finally resuspended in buffer A (400 μl) for the next immunomagnetic separation. The immunomagnetic isolation-elution procedure for purified SEB was repeated 11 times using the same affinity probe. Eluted samples were analyzed in triplicate by MALDI-TOF MS and ELISA.
MALDI-TOF MS.
MALDI-TOF MS experiments were performed in positive ion mode using a Voyager DE-Pro (Applied Biosystems, Framingham, MA) mass spectrometer with delayed extraction equipped with a 337-nm nitrogen laser in linear acceleration mode. Freshly prepared 1,2-dimethoxy-4-hydroxycinnamic acid (10 mg/ml) was used as the matrix in 50% acetonitrile-0.1% (vol/vol) trifluoroacetic acid. The following source parameters were used: accelerating voltage, 24 kV; grid voltage, 76% of the extraction field; and delay time, 180 ns. Spectra were acquired in positive ion mode in the mass range from m/z 1,000 to 35,000. The final mass spectrum was the sum of 10 mass spectra obtained by applying 50 laser shots for each. The analysis was performed in triplicate. The instrument parameters and laser energy were kept constant during a series of experiments performed on the same day for comparison of intensity values (cps). The SEB standard was desalted using ZipTip C18 prior MS analysis.
ELISA.
Antigen immunodetection was performed using an ELISA classified as an “indirect antibody capture assay” (16). The samples were diluted in 50 mM NH4HCO3, and each dilution was analyzed in triplicate. A standard solution of SEB was included in each experiment. Incubation, washing steps, the enzyme-linked antibody reaction, and detection were performed as previously described (28). Sample concentrations were calculated by interpolation using the standard curve and the Microplate Manager II data analysis software (Bio-Rad, Hercules, CA).
Bacterial culture.
Bacterial strains used in this study are described in Table 1. Bacteria were grown in 50 ml Luria-Bertani broth (Oxoid, Basingstoke, United Kingdom) at 37°C with constant shaking for 24 h (late exponential phase) unless otherwise stated. Growth was monitored every hour by measuring the optical density at 600 nm and by counting colonies on plates. Cells were removed by centrifugation at 12,500 × g for 15 min at 4°C and were filtered using a 0.45-μm filter (Millex-GX). Twenty-microliter portions of culture supernatants were used for the solid-phase immunoaffinity MALDI-TOF MS experiments. All samples were prepared and analyzed in triplicate. RPLA analyses were performed using a commercially available SET-RPLA toxin detection kit specific for detection and semiquantitative analysis of enterotoxins A, B, C, and D.
TABLE 1.
S. aureus strains analyzed by immunomagnetic separation and MALDI-TOF MS
| Strain | Sourcea | Origin | Genotype and/or phenotypeb | RPLA assay results | Affinity MS resultsc |
|---|---|---|---|---|---|
| ATCC 14458 | ATCC | Feces of child with acute diarrhea | SEB+d | SEB positive | SEB positive |
| ATCC 25923 | ATCC | Clinical sample | egc+seg+sem+seu+e | NDf | ND |
| A900322 | DSAN | Clinical strain from a patient with toxic shock syndrome | egc+seg+sen+seo+sem+sei+e | ND | ND |
| RIMD 31092 | DSAN | Clinical methicillin-resistant S. aureus strain | egc+sem+seg+seb+sec+e | SEB positive, SEC positive | SEB positive |
| AB-8802 | DSAN | Raw poultry meat, food-derived strain | egc+segv+seiv+e | ND | ND |
| ROS | ISA | Patient | Nonenterotoxigenic | ND | ND |
ATCC, American Type Culture Collection, Rockville, MD; DSAN, Dipartimento di Scienza degli Alimenti, Università degli Studi di Napoli Federico II, Portici, Italy (kindly provided by G. Blaiotta); ISA, Istituto di Scienza dell'Alimentazione, CNR, Avellino, Italy.
egc+, enterotoxin gene cluster positive. The subscript v indicates a variant.
Results of immunocapture using the anti-SEB molecular probe, followed by MALDI-TOF MS detection.
Data from reference 14.
Data from reference 9.
ND, not detected.
Preparation of milk sample.
Untreated and heat-treated whole bovine milk (purchased locally) samples (10 ml) were inoculated with 1% (vol/vol) S. aureus ATCC 14458 grown as described previously. Milk samples then were incubated at room temperature and 37°C for 24 and 48 h. Caseins and fat were removed by isoelectric precipitation at pH 4.6 by adding 10% (vol/vol) acetic acid (1 ml) at 40°C for 5 min, 1 M sodium acetate (100 μl), and dichloromethane (10 ml). Each sample was mixed and centrifuged at 5,000 rpm for 10 min. After centrifugation three phases were observed (from bottom to top): dichloromethane containing fat, precipitated caseins, and the aqueous phase containing mainly whey proteins. The whey protein fraction was concentrated and purified using a molecular filter (VivaSpin 20; molecular mass cutoff, 10 kDa). Concentrated whey proteins containing SEB were used for subsequent immunoaffinity separation. Milk samples were prepared and analyzed in three independent experiments.
RESULTS
A novel method based on combined use of affinity capture on magnetic particles and MALDI-TOF MS for rapid detection of SEB was developed. The general scheme of the method is shown in Fig. 1. The first step of the procedure is preparation of the affinity probe by functionalization of monodisperse, hydrophobic magnetic particles with anti-SEB antibodies. The antibodies are immobilized through their primary amino groups on the surface of the magnetic beads, which are activated by p-toluene-sulfonyl (tosyl) reactive groups. In these experiments, we used both rabbit serum anti-SEB and affinity-purified polyclonal anti-SEB IgG for immobilization. The second step is immunomagnetic separation, in which the sample, containing the enterotoxin B protein, is reacted with the anti-SEB-coated magnetic particles. After incubation, the particles are washed in order to remove nonbinding components. The concentrated and purified enterotoxin B protein bound to the particles is then either directly analyzed by MALDI-TOF MS or eluted with appropriate solvent and subsequently detected by traditional immunochemical methods like ELISA and/or by MS.
FIG. 1.
Experimental scheme: detection of SEB by immunoaffinity capture and MALDI-TOF MS. Large circles, magnetic particles; Y shape, anti-SEB antibody; small circles, molecules other than SEB; triangles, SEB.
MALDI-TOF MS analysis of SEB.
Enterotoxin B protein was analyzed by MALDI-TOF MS at different concentrations to determine the detection limit. The protonated molecular ion peak (MH+) of SEB was easily detectable down to a level of 2 ng (corresponding to 70 fmol) with a signal-to-noise ratio of more than 50 (Fig. 2). This detection limit is comparable to that of other immunochemical assays. MALDI-TOF MS is generally known as a nonquantitative method; however, using rigorous sample preparation and the data acquisition method, the intensity of the MH+ peak was found to increase linearly with increasing quantities of SEB in the range from 70 to 1,700 fmol. Therefore, in this concentration range SEB could be analyzed in a semiquantitative way.
FIG. 2.
MALDI-TOF mass spectrum of SEB (85 fmol or 2.4 ng). Peaks corresponding to singly and doubly charged protonated molecular ions of the protein are indicated.
Optimization of immobilization and affinity capture.
Magnetic beads were coated using either affinity-purified anti-SEB polyclonal IgG or fractionated rabbit serum for immobilization. A large excess of antigen (20 pmol) was added during affinity capture in order to saturate all the possible binding sites of the molecular probes. The quantity of eluted SEB was measured by ELISA. A molecular probe prepared with fractionated serum bound significantly less SEB (approximately 1.8 pmol or 51 ng) than probes prepared with affinity-purified antibody (4.3 pmol or 122 ng). For this reason in all subsequent experiments affinity-purified anti-SEB polyclonal IgG was used to coat magnetic particles.
The immobilization procedure was optimized in order to obtain the maximal density of anti-SEB on the surface of the magnetic beads. Particles were reacted with increasing amounts of affinity-purified antibody (0 [i.e., blank], 12, 60, 100, and 300 μg) during coating and used to affinity capture the same amount of SEB. The quantities of SEB after elution were measured by ELISA (Fig. 3). Using 12 μg antibody for coating, approximately 0.8 pmol (23 ng) enterotoxin B was isolated. With a fivefold increase in the amount of antibody used for immobilization, approximately five times more SEB (∼4.4 pmol or 125 ng) was detected, showing a good linear correlation. A further increase in the amount of antibody did not result in a significant increase in the quantity of isolated SEB (e.g., using 300 μg antibody in the immobilization step, we detected ∼6 pmol or 170 ng SEB), showing that the surface was saturated. Therefore, in subsequent experiments we used 60 μg of antibody (for 100 μl of magnetic beads) as a good compromise between the consumption of anti-SEB and the capacity of the molecular probe.
FIG. 3.
Capacity of anti-SEB IgG (Toxin Technology)-coated magnetic beads as a function of the amount of antibody used for coating based on ELISA measurements.
The efficiency of the elution step was also monitored by quantifying SEB using ELISA before and after the immunomagnetic separation. The results showed that the level of recovery of the protein from the affinity beads was nearly 100% in a single elution step. The quantity of SEB eluted from the magnetic beads did not increase with increased elution time or with repeated elution steps. However, prolonged presence of SEB in the acidic glycine solution (pH 2.5) used for elution resulted in a significant loss of the ELISA response, probably due to denaturation of the antigen.
On- and off-bead detection of enterotoxin B by MALDI-TOF MS.
Two different MALDI-TOF MS detection methods were compared using the optimized affinity capture procedure. In the first method, SEB was detected by MALDI-TOF MS directly on the magnetic beads by mixing a portion of affinity-captured beads and matrix solution (Fig. 4a). In the second method, SEB was eluted and separated from the magnetic particles before MALDI analysis (Fig. 4b). The final quantities of SEB on the MALDI target plate were the same in the two experiments. Using 60 fmol of starting material, SEB could be successfully detected by both approaches. Off-bead detection yielded a slightly better signal-to-noise ratio.
FIG. 4.
MALDI-TOF mass spectra of affinity-captured SEB obtained (a) by applying 1 μl of affinity beads directly onto the MALDI target plate (on beads) and (b) after glycine elution (off beads).
Repeated use of anti-SEB-coated paramagnetic particles.
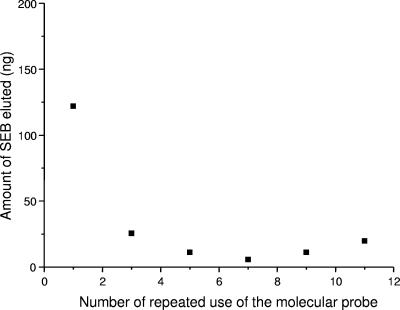
Antibody-coated magnetic particles can be used as affinity probes several times, if an appropriate buffer is used in the elution step. In our experiments, 100 mM glycine (pH 2.5) buffer was used for a short time (1 min) to elute SEB, but, at the same time, it did not denature the antibody. The reuse of anti-SEB-coated magnetic beads was controlled in an experiment in which the immunomagnetic isolation procedure was repeated 11 times and on-bead and off-bead samples were analyzed by MALDI-TOF MS. All samples yielded molecular ion peaks of SEB in the MALDI spectrum. During the first use the molecular probe captured a larger amount of SEB, resulting in higher-intensity ion peaks. In subsequent experiments similar intensities were observed. The amount of eluted SEB was measured by ELISA for samples 1, 3, 5, 7, and 9, as shown in Fig. 5. It was noted that the quantity of SEB detected in the first experiment was significantly greater than the quantities detected in the subsequent experiments. After the first use of the beads, the intensity of SEB became consistent. These results suggest that part of the antibody is bound to the surface of the magnetic beads by noncovalent interaction, and these molecules are removed in the elution step. The adsorbed antibody increases the capacity of the beads during the first use. However, after the physically adsorbed molecules are removed, the coated particles show constant activity.
FIG. 5.
Amounts of eluted SEB measured by ELISA during repeated use of the anti-SEB-coated affinity probe. The data are means of three wells. The experimental error was less than 10%.
Specificity of anti-SEB molecular probe.
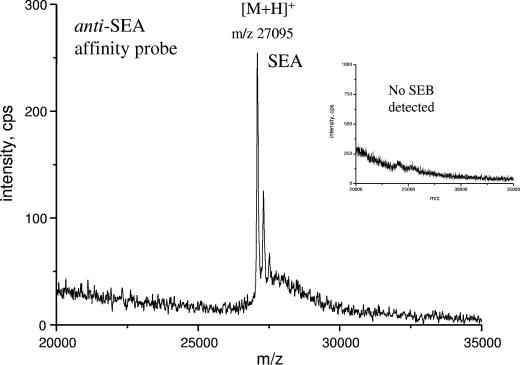
Specific detection of staphylococcal enterotoxins by traditional immunoassays is often troublesome because of the high level of sequence homology of the enterotoxins. In the MS-based method selective isolation of the toxin is followed by a selective detection step. Detection by MALDI-TOF MS is based on measurement of an intrinsic physical property, the molecular mass of the toxin. This is one of the main advantages of the combined method over classical immunoassays which lack such a control. To demonstrate specificity, the following tests were performed. SEA was used as a negative control to test the specificity of the anti-SEB affinity probe for SEB. In this experiment no SEA could be detected by MALDI-TOF MS, which demonstrated that the magnetic affinity probe is specific for SEB and not for SEA. In a reverse experiment, anti-SEA affinity probe was prepared, which proved to be specific for SEA and not for SEB (Fig. 6).
FIG. 6.
MALDI-TOF MS spectrum of SEA after immunocapture using the anti-SEA affinity probe. (Inset) Immunocapture of SEB using anti-SEA affinity probe (no binding observed).
Use of immunomagnetic separation and MALDI-TOF MS for detection of enterotoxin B in complex biological samples.
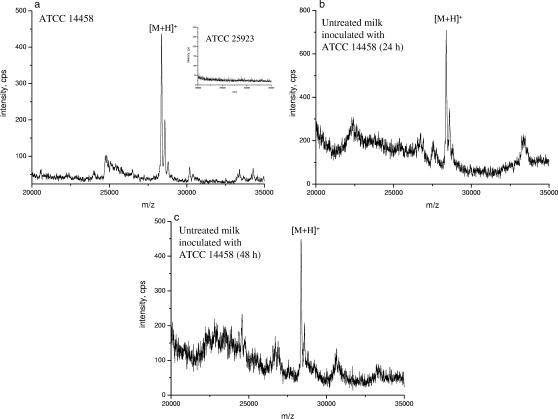
The optimized method based on the combination of solid-phase immunocapture with MALDI-TOF MS was tested using complex biological samples (i.e., culture supernatants) and milk samples (Fig. 7). Culture media of six S. aureus strains, including five enterotoxigenic strains (ATCC 14458, ATCC 25923, A900322, RIMD 31092, and AB-8802) and one nonenterotoxigenic strain (ROS) (Table 1) were analyzed by the RPLA test and immunomagnetic separation combined with MALDI-TOF MS. Bacterial culture broth was applied directly onto the immunoaffinity probe without further purification. ATCC 14458 is known to produce SEB, and the level of SEB in the culture supernatant of this strain was 280 ng/ml as measured by ELISA and the RPLA test. Figure 7a shows an abundant peak corresponding to the molecular ion of SEB in the MALDI-TOF mass spectrum obtained after immunomagnetic separation of the culture broth of ATCC 14458. On the other hand, there was no detectable peak in the mass range from m/z 20,000 to 30,000 in the mass spectra of the other non-SEB-producing strains (Fig. 6a, inset).
FIG. 7.
MALDI-TOF mass spectra of affinity-captured SEB obtained after glycine elution for (a) the culture supernatant of S. aureus SEB-positive strain ATCC 14458 (the inset shows the mass spectrum of SEB-negative strain ATCC 25923) and a milk sample (b) 24 h and (c) 48 h after inoculation with SEB-producing strain ATCC 14458.
In order to demonstrate the applicability of the solid-phase immunocapture method in food analysis, milk samples (untreated and heat treated) were contaminated with SEB-producing S. aureus strain ATCC 14458. Samples were incubated at different temperatures for different time intervals. After incubation, the SEB in the water-soluble whey protein fraction was enriched by the immunoaffinity method and the fraction was analyzed by MALDI-TOF MS. SEB was detected in all milk samples. Figures 7b and c show the MALDI-TOF MS spectra obtained for untreated milk after 24 and 48 h of incubation at room temperature. Based on an ELISA measurement, the amount of SEB in the sample at 24 h was 6 ng/ml. After 48 h the intensity of SEB in the MALDI spectra was almost the same. This can be explained by the biological competition between the pathogenic bacteria and the natural bacterial flora (like lactic acid bacteria) present in milk, which alters foods to preclude the growth of pathogens.
DISCUSSION
MS-based methods possess great potential for application in detection and structural characterization of bacterial protein toxins. Sample complexity, however, often limits their use and thus their efficacy with biological samples, foods, etc. There are two ways to improve the toxin detection limit of traditional electrospray ionization and MALDI MS methods. One is based on the highly specific enzymatic activity of some protein toxins. In this case, instead of detection of the toxin (i.e., botulinum or tetanus), the products resulting from hydrolysis can be analyzed (3, 19, 40). On the other hand, nonenzymatic toxins, like staphylococcal enterotoxins, could be successfully detected after selective enrichment of the target toxin in the sample. In this work, to obtain the highest recovery, we prepared and optimized a molecular probe, anti-SEB-coated magnetic beads, for solid-phase immunoaffinity capture of SEB from complex mixtures. Staphylococcal food intoxication is one of the leading causes of bacterial food-borne diseases due to the ingestion of foods containing staphylococcal enterotoxins. Staphylococci can readily multiply in many foods, but dairy products are probably the most frequently contaminated foods. Raw milk and cheeses prepared from raw milk have been repeatedly involved in staphylococcal outbreaks. Also, the presence of enterotoxin B is commonly associated with this food. In this work we demonstrated that immunoaffinity capture can be used for such a complex matrix online or offline with MALDI-TOF MS detection. This method determines the molecular mass of the target compound after immunochemical separation and therefore offers greater specificity than conventional immunoassays, and thus it minimizes the possibility of false-positive results. Other advantages of the method include minimal sample manipulation, thus avoiding loss of biological material, high sensitivity (detection limit, ∼2 ng), and rapidity, which allows us to obtain results in a short period of time (in minutes). In addition, reuse of anti-SEB-coated magnetic beads is possible with such a two-step approach that combines immunomagnetic separation with MS methods. Multiplex detection of different toxins is also plausible if molecular probes specific for all the given toxins are mixed. These advantages suggest that the two-step processes are ideal for automated applications and assays.
Acknowledgments
We are grateful to project SIFORTI (MIUR-CNR, Legge 449/97) for financial support, to the “Rete di Spettrometria di Massa” (Contract Fondo Europeo Regionale Sviluppo 94.05.09.103, ARINCO 94.IT.16.028), to CNR-NATO for a senior fellowship for Petr Kačer, and to Waters SPA (Milan, Italy) for a fellowship for Livia Malorni. G. Schlosser acknowledges financial support from the Magyary Zoltan Higher Educational Public Foundation (Hungary).
Footnotes
Published ahead of print on 7 September 2007.
REFERENCES
- 1.Alefantisa, T., P. Grewala, J. Ashtona, A. S. Khanb, J. J. Valdesb, and V. G. D. Vecchio. 2004. A rapid and sensitive magnetic bead-based immunoassay for the detection of staphylococcal enterotoxin B for high-throughput screening. Mol. Cell. Probes 18:379-382. [DOI] [PubMed] [Google Scholar]
- 2.Balaban, N., and A. Rasooly. 2001. Analytical chromatography for recovery of small amounts of staphylococcal enterotoxins from food. Int. J. Food Microbiol. 64:33-40. [DOI] [PubMed] [Google Scholar]
- 3.Barr, J. R., H. Moura, A. E. Boyer, A. R. Woolfitt, S. R. Kalb, A. Pavlopoulos, L. McWilliams, J. G. Schmidt, R. A. Martinez, and D. L. Ashley. 2005. Botulinum neurotoxin detection and differentiation by mass spectrometry. Emerg. Infect. Dis. 11:1578-1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bennett, R. W. 2005. Staphylococcal enterotoxin and its rapid identification in foods by enzyme-linked immunosorbent assay-based methodology. J. Food Prot. 68:1264-1270. [DOI] [PubMed] [Google Scholar]
- 5.Bernardo, K., S. Fleer, N. Pakulat, O. Krut, F. Hünger, and M. Krönke. 2002. Identification of Staphylococcus aureus exotoxins by combined sodium dodecyl sulfate gel electrophoresis and matrix-assisted laser desorption/ionization-time of flight mass spectrometry. Proteomics 2:740-746. [DOI] [PubMed] [Google Scholar]
- 6.Blaiotta, G., V. Fusco, C. von Eiff, F. Villani, and K. Becker. 2006. Biotyping of enterotoxigenic Staphylococcus aureus by enterotoxin gene cluster (egc) polymorphism and spa typing analyses. Appl. Environ. Microbiol. 72:6117-6123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boyer, A. E., H. Moura, A. R. Woolfitt, S. R. Kalb, L. G. McWilliams, A. Pavlopoulos, J. G. Schmidt, D. L. Ashley, and J. R. Barr. 2005. From the mouse to the mass spectrometer: detection and differentiation of the endoproteinase activities of botulinum neurotoxins A-G by mass spectrometry. Anal. Chem. 77:3916-3924. [DOI] [PubMed] [Google Scholar]
- 8.Boyle, M. D. P., T. G. Romer, A. K. Meeker, and D. D. Sledjeski. 2001. Use of surface-enhanced laser desorption ionization protein chip system to analyze streptococcal exotoxin B activity secreted by Streptococcus pyogenes. J. Microbiol. Methods 46:87-97. [DOI] [PubMed] [Google Scholar]
- 9.Carol, J., M. C. J. K. Gorseling, C. F. de Jong, H. Lingeman, C. E. Kientz, B. L. van Baar, and H. Irth. 2005. Determination of denaturated proteins and biotoxins by on-line size-exclusion chromatography-digestion-liquid chromatography-electrospray mass spectrometry. Anal. Biochem. 346:150-157. [DOI] [PubMed] [Google Scholar]
- 10.Clements, M. O., and S. Foster. 1999. Stress resistance in Staphylococcus aureus. Trends Microbiol. 7:458-462. [DOI] [PubMed] [Google Scholar]
- 11.De Boer, M. L., and A. W. Chow. 1994. Toxic shock syndrome toxin 1-producing Staphylococcus aureus isolates contain the staphylococcal enterotoxin B genetic element but do not express staphylococcal enterotoxin B. J. Infect. Dis. 170:818-827. [DOI] [PubMed] [Google Scholar]
- 12.Dinges, M. M., P. M. Orwin, and P. M. Schlievert. 2000. Enterotoxins of Staphylococcus aureus. Clin. Microbiol. Rev. 13:16-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Edwin, C., S. R. Tatini, and S. K. Maheswaran. 1986. Specificity and cross-reactivity of staphylococcal enterotoxin A monoclonal antibodies with enterotoxins B, C1, D, and E. Appl. Environ. Microbiol. 52:1253-1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fueyo, J. M., M. C. Martin, M. A. Gonzalez-Hevia, and M. C. Mendoza. 2001. Enterotoxin production and DNA fingerprinting in Staphylococcus aureus isolated from human and food samples. Relations between genetic types and enterotoxins. Int. J. Food Microbiol. 67:139-145. [DOI] [PubMed] [Google Scholar]
- 15.Gill, D. M. 1982. Bacterial toxins: a table of lethal amounts. Microbiol. Rev. 46:86-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harlow, E., and D. Lane. 1988. Antibodies. A laboratory manual, p. 553-612. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 17.Hines, H. B., F. Lebeda, M. Hale, and E. E. Brueggemann. 2005. Characterization of botulinum progenitor toxins by mass spectrometry. Appl. Environ. Microbiol. 71:4478-4486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huelseweh, B., R. Ehricht, and H.-J. Marschall. 2006. A simple and rapid protein array based method for the simultaneous detection of biowarfare agents. Proteomics 6:2972-2981. [DOI] [PubMed] [Google Scholar]
- 19.Kalb, S. R., H. Moura, A. E. Boyer, L. G. McWilliams, J. L. Pirkle, and J. R. Barr. 2006. The use of Endopep-MS for the detection of botulinum toxins A, B, E, and F in serum and stool samples. Anal. Biochem. 351:84-92. [DOI] [PubMed] [Google Scholar]
- 20.Kawano, Y., Y. Ito, Y. Yamakawa, T. Yamashino, T. Horii, T. Hasegawa, and M. Ohta. 2000. Rapid isolation and identification of staphylococcal exoproteins by reverse phase capillary high performance liquid chromatography-electrospray ionization mass spectrometry. FEMS Microbiol. Lett. 189:103-108. [DOI] [PubMed] [Google Scholar]
- 21.King, K. D., G. P. Anderson, K. E. Bullock, M. J. Regina, E. W. Saaski, and F. S. Ligler. 1999. Detecting staphylococcal enterotoxin B using an automated fiber optic biosensor. Biosens. Bioelectron. 14:163-170. [DOI] [PubMed] [Google Scholar]
- 22.Krakauer, T. 1999. Immune response to staphylococcal superantigens. Immunol. Res. 20:163-173. [DOI] [PubMed] [Google Scholar]
- 23.Lin, H.-C., and W.-C. Tsai. 2003. Piezoelectric crystal immunosensor for the detection of staphylococcal enterotoxin B. Biosens. Bioelectron. 18:1479-1483. [DOI] [PubMed] [Google Scholar]
- 24.Naimushin, A. N., S. D. Soelberg, D. K. Ngueyen, L. Dunlap, D. Bartholomew, J. Elkind, J. Melendez, and C. E. Furlong. 2002. Detection of Staphylococcus aureus enterotoxin B at femtomolar levels with a miniature integrated two-channel surface plasmon resonance (SPR) sensor. Biosens. Bioelectron. 17:573-584. [DOI] [PubMed] [Google Scholar]
- 25.Nakano, M., Y. Kawano, M. Kawagishi, T. Hasegawa, Y. Iinuma, and M. Ohta. 2002. Two-dimensional analysis of exoproteins of methicillin-resistant Staphylococcus aureus (MRSA) for possible epidemiological applications. Microbiol. Immunol. 46:11-22. [DOI] [PubMed] [Google Scholar]
- 26.Nedelkov, D., and R. W. Nelson. 2003. Detection of staphylococcal enterotoxin B via biomolecular interaction analysis mass spectrometry. Appl. Environ. Microbiol. 69:5212-5215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peruski, A. H., L. H. R. Johnson, and L. F. J. Peruski. 2002. Rapid and sensitive detection of biological warfare agents using time-resolved fluorescence assays. J. Immun. Methods 263:35-41. [DOI] [PubMed] [Google Scholar]
- 28.Pizzano, R., M. A. Nicolai, and F. Addeo. 1997. Antigenicity of the 139-149 αs1-casein region in different species revealed by ELISA and immunoblotting using antipeptide antibodies. J. Agric. Food Chem. 45:2807-2813. [Google Scholar]
- 29.Poli, M. A., V. R. Rivera, and D. Neal. 2002. Sensitive and specific colorimetric ELISAs for Staphylococcus aureus enterotoxins A and B in urine and buffer. Toxicon 40:1723-1726. [DOI] [PubMed] [Google Scholar]
- 30.Portocarrero, S. M., M. Newman, and B. Mikel. 2002. Staphylococcus aureus survival, staphylococcal enterotoxin production and shelf stability of country-cured hams manufactured under different processing procedures. Meat Sci. 62:267-273. [DOI] [PubMed] [Google Scholar]
- 31.Rappuoli, R., and C. Montecucc. 1997. Guidebook to protein toxins and their use in cell biology. Oxford University Press, Oxford, United Kingdom.
- 32.Rasooly, A., and K. E. Herold. 2006. Biosensors for the analysis of food- and waterborne pathogens and their toxins. J. AOAC Int. 89:873-883. [PubMed] [Google Scholar]
- 33.Ratts, R., C. Trujillo, A. Bharti, J. vander Spek, R. Harrison, and J. R. Murphy. 2005. A conserved motif in transmembrane helix 1 of diphtheria toxin mediates catalytic domain delivery to the cytosol. Proc. Natl. Acad. Sci. USA 102:15635-15640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rotun, S. S., V. McMath, D. J. Schoonmaker, P. S. Maupin, F. C. Tenover, B. C. Hill, and D. M. Ackman. 1999. Staphylococcus aureus with reduced susceptibility to vancomycin isolated from patent with fatal bacteremia. Emerg. Infect. Dis. 5:147-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rusnak, J. M., M. Kortepeter, R. Ulrich, M. Poli, and E. Boudreau. 2004. Laboratory exposures to staphylococcal enterotoxin B. Emerg. Infect. Dis. 10:1544-1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Seto, Y., and M. Kanamori-Kataoka. 2005. Mass spectrometric strategy for the determination of natural and synthetic organic toxins. J. Health Sci. 51:519-525. [Google Scholar]
- 37.Ulrich, R. G., S. Sidell, T. J. Taylor, C. L. Wilhelmsen, and D. R. Franz. 1997. Staphylococcal enterotoxin B and related pyrogenic toxins, p. 621-630. In F. R. Sidell, E. T. Takafugi, and D. R. Franz DR (ed.), Medical aspects of chemical and biological warfare. TMM Publications, Washington, DC.
- 38.van Baar, B. L., A. G. Hulst, C. F. de Jong, and E. R. Wils. 2002. Characterisation of botolinium toxins type A and B by matrix-assisted laser desorption ionisation and electrospray mass spectrometry. J. Chromatogr. A 970:95-115. [DOI] [PubMed] [Google Scholar]
- 39.van Baar, B. L., A. G. Hulst, C. F. de Jong, and E. R. Wils. 2004. Characterisation of botulinum toxins type C, D, E, and F by matrix-assisted laser desorption ionisation and electrospray mass spectrometry. J. Chromatogr. A 1035:97-114. [DOI] [PubMed] [Google Scholar]
- 40.van Baar, B. L., A. G. Hulst, B. Roberts, and E. R. Wils. 2002. Characterization of tetanus toxin, neat and in culture supernatant, by electrospray mass spectrometry. Anal. Biochem. 301:278-289. [DOI] [PubMed] [Google Scholar]
- 41.Whiting, R. C., S. Sackitey, S. Calderone, K. Morely, and J. G. Phillips. 1996. Model for the survival of Staphylococcus aureus in nongrowth environments. Int. J. Food Microbiol. 31:231-243. [DOI] [PubMed] [Google Scholar]
- 42.Williams, J. P., B. N. Green, D. C. Smith, K. R. Jennings, K. A. H. Moore, S. E. Slade, L. M. Roberts, and J. H. Scrivens. 2005. Noncovalent Shiga-like toxin assemblies: characterization by means of mass spectrometry and tandem mass spectrometry. Biochemistry 44:8282-8290. [DOI] [PubMed] [Google Scholar]
- 43.Yu, H. B., R. Kaur, S. Lim, X. H. Wang, and K. Y. Leung. 2007. Characterization of extracellular proteins produced by Aeromonas hydrophilia AH-1. Proteomics 7:436-449. [DOI] [PubMed] [Google Scholar]