Abstract
The aim of this study was to investigate the supposed vertical diel migration and the accompanying physiology of Beggiatoa bacteria from hypersaline microbial mats. We combined microsensor, stable-isotope, and molecular techniques to clarify the phylogeny and physiology of the most dominant species inhabiting mats of the natural hypersaline Lake Chiprana, Spain. The most dominant morphotype had a filament diameter of 6 to 8 μm and a length varying from 1 to >10 mm. Phylogenetic analysis by 16S rRNA gene comparison revealed that this type appeared to be most closely related (91% sequence identity) to the narrow (4-μm diameter) nonvacuolated marine strain MS-81-6. Stable-isotope analysis showed that the Lake Chiprana species could store nitrate intracellularly to 40 mM. The presence of large intracellular vacuoles was confirmed by fluorescein isothiocyanate staining and subsequent confocal microscopy. In illuminated mats, their highest abundance was found at a depth of 8 mm, where oxygen and sulfide co-occurred. However, in the dark, the highest Beggiatoa densities occurred at 7 mm, and the whole population was present in the anoxic zone of the mat. Our findings suggest that hypersaline Beggiatoa bacteria oxidize sulfide with oxygen under light conditions and with internally stored nitrate under dark conditions. It was concluded that nitrate storage by Beggiatoa is an optimal strategy to both occupy the suboxic zones in sulfidic sediments and survive the dark periods in phototrophic mats.
Beggiatoa spp. are large, filamentous, gliding, colorless sulfur bacteria and are known to occur worldwide in diverse habitats with a wide range of salinities. They form visible white mats on the surfaces of organic-rich freshwater sediments (39, 46, 63, 64) and in sulfur springs (36), marine caves (37), marine eutrophic coastal zones (22, 58, 59), upwelling regions (7, 15), whale falls (11), cold seeps (10, 43, 48), and gas seeps (4, 28). These filamentous bacteria migrate in daily cycles in microbial mats (16, 23, 67). Microbial mats are dense, cohesive communities with a visible laminated pattern due to the zonation of different metabolic groups with different pigmentations. Within a few millimeters of the upper mat, steep physicochemical gradients occur due to high metabolic activities of the densely packed microorganisms. The main functional groups are cyanobacteria, purple sulfur bacteria, colorless sulfur bacteria, and sulfate-reducing bacteria (67). Most mats are characterized by a microbiologically controlled rapid sulfur cycle. In phototrophic mats, oxygenic photosynthesis and dissimilatory sulfate reduction result in opposing gradients of oxygen and sulfide, which sometimes overlap in a narrow transition zone. The sulfide and oxygen distributions underlie diel cycles (54), which induce migratory behavior of phototrophic and nonphototrophic organisms (8, 55). Gliding motility and a tactile response to diverse parameters, e.g., light, oxygen, and sulfide (41, 44, 45), enable Beggiatoa spp. to follow the movement of the dynamic transition zone in phototrophic mats. Beggiatoa spp. have been reported to exist at the oxic-anoxic interface in hypersaline microbial mats (16, 23), but none of these organisms have been characterized for phylogeny and function. The preference for a niche where oxygen and sulfide co-occur has been shown for both marine and freshwater Beggiatoa species. For some filaments, the capability of anaerobic sulfide oxidation with nitrate has been demonstrated, and the larger relatives can store nitrate, presumably in intracellular vacuoles (30, 47, 64, 65). Although narrow nonvacuolated Beggiatoa spp. proliferate in zones of <1 millimeter thick where both oxygen and sulfide co-occur (26, 45), wider, vacuolated forms of Beggiatoa and Thioploca are found in broader (up to 10 cm wide) suboxic zones where neither oxygen nor sulfide occurs (1). Especially larger filamentous species (9 to 375 μm in diameter) and other representatives of big colorless sulfur bacteria, such as Thioploca spp. and Thiomargarita namibiensis, possess internal vacuoles (24, 32, 38, 42, 60), which are presumably the main location of nitrate storage. These organisms can use internally stored nitrate as an electron acceptor for anaerobic sulfide oxidation in deep suboxic sediment layers (1, 38, 58). Therefore, it is thought that sulfur bacteria play an important role in and form a link between the sulfur and nitrogen cycles (7, 33, 40). In a recent study, 21 Beggiatoa spp. from various freshwater and marine habitats were characterized phylogenetically by 16S rRNA gene sequencing (2). Fourteen of the 21 studied Beggiatoa relatives are known to be vacuolated and able to store nitrate. Species from freshwater (5 species) and marine (16 species) habitats form phylogenetically separate subclusters, with vacuolated forms being restricted to a subcluster within the marine cluster of larger (>9 μm) Beggiatoa spp. Species with smaller filament diameters (1 to 6 μm) have not been shown to be vacuolated, including some Beggiatoa members originating from phototrophic microbial mats.
The purpose of our study was to understand the behavior and metabolism of currently uncultured hypersaline Beggiatoa spp. in intact microbial mats and, furthermore, to clarify their phylogenetic relationship to other filamentous sulfur bacteria. We characterized for the first time a hypersaline Beggiatoa sp. which originates from the natural hypersaline Lake Chiprana, Spain. The vertical distribution of this morphotype and its physiology in relation to environmental parameters were studied in intact mat samples by using direct methods, i.e., microsensor, stable-isotope, and microscopy techniques. In addition, the 16S rRNA gene of this species was sequenced. Morphology was documented in a detailed microscopic study, with emphasis on the presence of vacuoles for nitrate storage.
MATERIALS AND METHODS
Sampling site and mesocosm description.
The permanent hypersaline natural inland Lake Chiprana in northeastern Spain (41°14′30″N, 0°10′50"W) has a total surface area of 31 ha and a maximum depth of 5.6 m. The lakebed is composed of tertiary paleochannels with fossilized sands and silt deposits (66). Through permanent groundwater inflow, the ionic composition of the lake water is dominated by magnesium sulfate (SO42−, 0.5 mol liter−1; Mg2+, 0.35 mol liter−1), and it has an average salinity of ∼80‰ (21). Intact microbial mat samples (15 by 15 by 4 cm) were collected in September 2004 approximately 1 m from the shoreline at a 50-cm water depth and were transferred to Bremen, Germany, for further analysis and laboratory experiments. The mat samples were kept in the laboratory in an open plastic basin (1.2 by 1.2 by 0.5 m) with a continuous flowthrough of aerated artificial lake water (seawater plus 80 g/liter MgSO4·7H2O). Illumination was set as close as possible to natural conditions, with a light intensity of 500 μmol photons m−2 s−1 and an illumination regimen of 14 h of light and 10 h of darkness. During the whole incubation period of 5 months, and specifically at the times that experiments with these laboratory-incubated mats were performed (after 3 months of mesocosm incubation), oxygen and sulfide profiles were measured for mat characterization to establish that major physiological processes, i.e., oxygen and sulfide production, persisted in the mat. Most Beggiatoa filaments were found in microbial mats inhabited by the charophyte Lamprothamium papulosum. These mats were subsampled by coring, with a diameter of 5 cm and a thickness of 2 cm, for further detailed microsensor measurements.
Microsensor measurements of oxygen, sulfide, pH, and light.
Subsamples of the laboratory-incubated mat were taken for respective experimental procedures, and these were subsequently characterized for oxygen and sulfide dynamics by microsensor analysis. These microbial mat subsamples were exposed to a 14-h light (500 μmol photons m−2 s−1)-10-h dark diel cycle at room temperature (20°C) within a small aquarium (15 by 15 by 5 cm). The overlying water had a similar salinity and composition to those of the original Lake Chiprana water and was aerated by an air jet over the water surface. The mats were illuminated by a fiber-optic halogen light source (KL 1500; Schott, Germany), and the irradiance at the mat surface was determined with an underwater scalar irradiance meter (LI-250; LiCor). To ensure steady-state conditions, microsensors for oxygen (O2), sulfide (H2S), and pH were applied at the end of each experimental light or dark setting. Since repeated profiles measured at the same spot varied <2% (results not shown), we assumed steady-state conditions. The mat profiles were taken by mounting all microsensors on a motorized micromanipulator (Oriel) stabilized by a heavy stand. Sensors were operated and their signals were recorded via a computer acquisition system (LabView; National Instruments). The sediment surface relative to the sensor tips was determined with the aid of a dissection microscope (SV6; Zeiss, Germany). Profiles of O2, H2S, and pH were recorded in steps, with a vertical depth resolution of 200 μm. The Clark-type O2 microsensor (53) had a tip diameter of 15 μm and a response time of less than 3 s. It was two-point calibrated by using the signals in the overlying air-saturated brine (100% air saturation) and the anoxic zones of the mat (0%). Dissolved oxygen concentrations at a given salinity (80‰) and temperature (20°C) were determined by the method of Sherwood et al. (62). The pH glass-microelectrode (54) and the H2S microsensor (20) were calibrated before their application to microbial mats. The pH microsensor had a tip diameter of ∼15 μm and response times of less than 20 s and was calibrated with standard solutions (Mettler-Toledo, Switzerland). The H2S microsensor had a tip diameter of 20 μm, was coated with black enamel paint to avoid light interference (34), and had response times of less than 20 s. It was calibrated in anoxic phosphate buffer (0.2 M; pH 8.2) by adding Na2S from a stock solution, and exact concentrations were determined spectrophotometrically by the Pachmeyer method (52). Calculations of total sulfide derived from the local pH and H2S concentration (20) were done according to the procedure described by Wieland and Kühl (68). Where mentioned below, total sulfide denotes the sum of H2S, HS−, and S2−. Spectral light gradients in intact mat pieces illuminated with a light intensity of 500 μmol photons m−2 s−1 were measured using an optic fiber light microsensor (27) with a tip size of approximately 30 μm. The sensor was connected to a USB2000 Ocean Optics spectrometer. The measurements were started from below at a depth where no light penetration could be detected, and the sensor tip was moved upwards in steps of 500 μm until the mat surface was reached. At each depth, a light spectrum between 400 and 800 nm was recorded, and all spectra were normalized to the light spectrum taken at the mat surface.
Depth distribution of Beggiatoa filament densities and biomass determination.
The vertical distribution of Beggiatoa filament densities in the mat was determined under both light and dark conditions, directly after finishing the microsensor profiling. The microbial mat subsamples were cut perpendicular to the mat surface at former microsensor measurement spots, placed immediately within a petri dish while still wet, and kept in darkness. With the aid of a dissection microscope at a magnification of ×16, images of the cross section were taken with a Nikon Coolpix digital camera mounted on a camera adapter. To minimize the effect of retraction of Beggiatoa filaments back into the mat, less than 10 min passed between cross-sectioning and digital imaging. However, due to the motility of Beggiatoa filaments, typically up to 2.5 μm s−1, an unknown part of the population may have retracted into the microbial mat during the mat handling time. Although this may have led to an underestimation of the total population size, this potential effect did not likely affect the relative depth distribution of the Beggiatoa population. Several images of three different mat sections were analyzed. Images were processed using Adobe Photoshop 9.0 for Windows to enhance the contrast and the brightness of the white filaments. The depth distribution of filament densities was analyzed by counting the numbers of filaments in 1-mm-wide depth zones from the surface down to a depth of 10 mm. Percentages of filaments were determined as the number of filaments per depth zone against the total filament number for the mat cross section (average of three samples), and corresponding standard errors were calculated. Beggiatoa filament biovolume was estimated from the determined filament densities and a determined average filament volume value, which was obtained by measuring the lengths and widths of 100 individual filaments and calculating the value with the formula for a cylindrical geometrical shape. A Beggiatoa filament-specific weight density of 1 g cm−3 was assumed, which enabled calculation of biomass per 1-mm depth interval.
Total extractable nitrate and elemental sulfur.
For analysis of nitrate (NO3−) and elemental sulfur (S0), triplicate samples of the microbial mats were frozen at −20°C directly after determination of the Beggiatoa filament depth distribution. The frozen samples were sliced horizontally with a cryomicrotome (Microm, GmbH, Walldorf, Germany) into 1-mm-thick slices from the surface down to a depth of 10 mm. Each slice was transferred to a glass vial containing 0.5 ml of a 20% zinc acetate solution. After being freeze-dried for porosity and wet bulk density determinations, gastight vials (6 ml; Labco) were filled with demineralized water, spiked with a 10 μM 15NO3− solution, and subsequently flushed with helium gas to reduce the N2 background and to introduce a 1-ml headspace. In order to reduce the NO3− to NO gas, 0.5 ml of titanium(III) chloride was injected directly afterwards, and vials were immediately vigorously shaken (9, 56). Gas samples of the headspace were injected into a gas chromatograph-mass spectrometer which was equipped with a reduction oven (630°C), reducing the NO gas to N2. The stable-isotope composition of the N2 gas was analyzed by mass spectrometry. Due to the high 15NO3− background (10 μM), nearly all (>99%) of the 14NO3− formed an excess 14N15N (29N2)/15N15N (30N2) ratio compared to the 15NO3− medium (99% pure). Based on the known amount of 15NO3−, we determined the 14NO3− content by using the principle of the isotope pairing method (49).
For zero-valent sulfur analysis (S0), mat pieces fixed with 0.5 ml 20% zinc acetate in 10-ml glass vials were extracted with 10-ml 100% high-performance liquid chromatography (HPLC)-grade methanol aliquots for 2 days. Subsequently, samples were completely homogenized by sonication and vigorous shaking. The methanol phase was then separated from the mat slurry by centrifugation (1 min at 3,000 × g). Elemental sulfur was quantified by HPLC and UV detection at 265 nm as described by Zopfi et al. (69).
Measurements of Beggiatoa internal nitrate and elemental sulfur concentrations.
One Beggiatoa type, based on its morphological characteristics, was found to dominate the Lake Chiprana Beggiatoa community. For internal nitrate concentration determination, Beggiatoa filaments were picked out of intact mat samples with the aid of a glass needle and transferred to a droplet of artificial saline water (80 g/liter NaCl) on a microscope slide. Filament biovolume (V) was calculated from length (l) and diameter (d) determinations according to the equation V = π(0.5d)2 × l. The biovolume of the entire filament was taken for calculations of internal nitrate and elemental sulfur. Single filaments were then placed into 300-μl glass vials filled with 150-μl aliquots of deionized water and frozen at −20°C to disrupt the cells. After thawing, the samples were centrifuged (1 min at 3,000 × g), and 100-μl supernatant aliquots were taken and transferred into 6-ml glass vials filled with 10 μM 15NO3−. The 14NO3− content was determined by the isotope method as described above for the mat samples. The elemental sulfur concentration was determined on the same samples by 100% HPLC-grade methanol extraction of the remaining air-dried pellet.
In an incubation experiment with an enriched Beggiatoa population for the determination of Beggiatoa nitrate storage potential, filaments were picked out of intact mat pieces and placed in oxygen-free artificial lake water on top of 50 μM NO3−-containing agar. Filaments were then picked at two sampling times with a 24-h time interval. Samples were analyzed for internal nitrate and elemental sulfur concentrations as described above.
16S rRNA gene analysis of Lake Chiprana Beggiatoa spp.
General bacterial primers (Interativa, Ulm, Germany) were used to amplify almost-full-length 16S rRNA genes from single filaments. The master mix for the PCR was prepared as follows: 50 pmol each of primers 8f (19) and 1492r (31), 2.5 μmol of each deoxyribonucleoside triphosphate, 1× Super-Taq buffer (HT Biotechnology Ltd., Cambridge, United Kingdom), and 1 U of Taq DNA polymerase (Eppendorf, Germany) were amended with sterile water to a total volume of 100 μl. Live Beggiatoa filaments taken directly out of intact mat pieces were rinsed in artificial seawater before the addition of the universal primers. The PCR was started with an initial denaturation step of 4 min at 94°C, followed by 32 cycles of 0.5 min at 94°C, 0.5 min at 48°C, and 1.5 min at 72°C, and was terminated with a final step of 10 min at 72°C.
PCR products were purified (PCR purification kit; QIAGEN, Hilden, Germany), ligated using a TOPO-TA sequencing kit (Invitrogen, Carlsbad, CA), and then transformed according to the company's specifications. Clones with inserts were extracted from the clone library and sequenced by Taq cycle sequencing performed with vector primers and a model ABI sequencer (Applied Biosystems). The retrieved Beggiatoa sequences were analyzed with the ARB software package (35). The alignment was corrected manually, and the phylogenetic tree was constructed by maximum parsimony, neighbor joining, and maximum likelihood analyses with different filter sets. For tree reconstruction, only full-length sequences were taken into account. The tree was constructed by including subsets of data and an outgroup reference sequence. The tree structure was computed by these different approaches to build a consensus tree. All partial trees were checked for branching incongruities. Multifurcation in specific branching orders was used for different results by the applied methods. Analyses included closely related BLAST matches (3).
Confocal laser scanning microscopy.
For visualization of intracellular vacuoles and other morphological cell characteristics, several specific fluorochromes were applied to freshly picked Beggiatoa filaments. Fluorescein isothiocyanate (FITC; Research Organics, Cleveland, OH), Newport Green (Molecular Probes, Invitrogen), and a vacuole membrane marker (MDY-64; Molecular Probes, Invitrogen) were used to stain intracellular constituents (29). Nucleic acids were marked with SYBR green (Molecular Probes, Invitrogen). SYPRO orange (Molecular Probes, Invitrogen) was used for protein staining. Some filaments were double stained with SYBR green and SYPRO orange as described previously (70). Beggiatoa filaments were transferred to and stained on microscope slides. The stained filaments were examined by means of a TCS-SP1 confocal microscope controlled by Leica confocal software, version 2.61, build 1537 (Leica, Heidelberg, Germany). The TCS-SP1 microscope was attached to an upright microscope and equipped with three different lasers (488, 561, and 633 nm). For microscopy, a 63×, 1.2-numerical-aperture water immersion lens corrected for coverslips was used. Images were projected with the microscope software and Imaris (Bitplane, Switzerland). Images were printed from Photoshop without any corrections.
Determination of migration speed.
Freshly picked Beggiatoa bacteria from intact microbial mats were placed under different conditions (in medium or artificial seawater and in gradient tubes or microcapillary tubes, but always at 20°C) to visualize movement and determine gliding velocities. Movements were recorded with a charge-coupled-device camera under red light illumination, which presumably does not affect the movement of these bacteria (44). Image sequences were analyzed, and movement speeds were determined. All values for several measurements in the same data set were averaged.
Nucleotide sequence accession number.
The nucleotide sequence of the Beggiatoa morphotype described here (Chiprana, mesocosm mat [MM]) has been deposited in GenBank under accession number EF 428583.
RESULTS
Comparison of intact Lake Chiprana and mesocosm-incubated mats.
The composition of the microbial mats kept in the mesocosm was similar to that of mats obtained directly from Lake Chiprana. Vertical cuts of mats from the mesocosm system showed similarly colored depth layers, with each inhabited by similar species to those in the natural microbial mat (21). The surface layers of both mats were composed mainly of diatoms (Nitzschia spp.), Chloroflexus spp., and unicellular cyanobacteria. The underlying green and brown layers, with whitish embedded calcium carbonate crystals, included several types of filamentous cyanobacteria, dominated by species of the genera Oscillatoria and Microcoleus. Below a depth of 1 cm, a dark gray zone of iron sulfide (FeS) precipitates emerged. In the green and brown mat layers, several white filaments appeared, with enough to detect them easily by the naked eye but too few to form a distinct layer of Beggiatoa sp.-like organisms. Microscopic analysis of these filaments revealed a dominant morphotype of 6 to 8 μm in diameter, with filament lengths varying between 1 and 10 mm. Differential interference contrast microscopy (6, 50) revealed sulfur inclusions as well as internal vacuoles. Filament ends were always rounded. There was no visible difference in filament morphotype in distinct mat layers.
Beggiatoa distribution dynamics in intact mats.
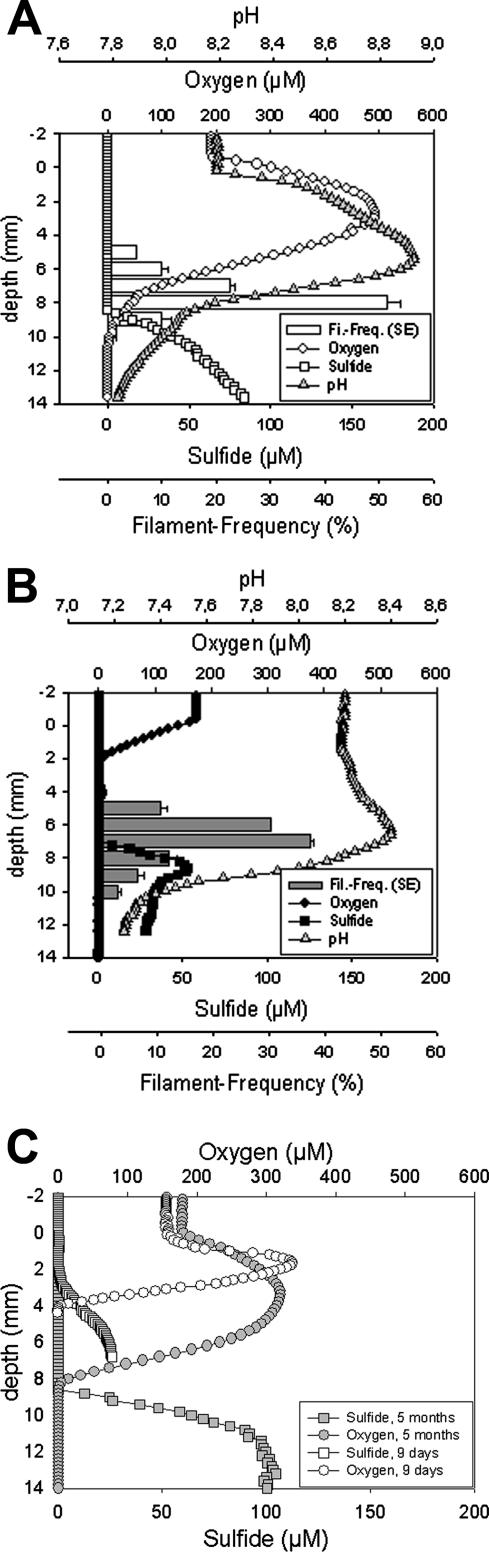
The total estimated Beggiatoa biomasses in the 0- to 10-mm depth zone for the illuminated and dark mats were 11.5 μg mm−2 and 10.1 μg mm−2 (wet weight), with total biovolumes of 0.012 and 0.010 μl mm−3 mat, respectively. The vertical distributions of Beggiatoa filaments were slightly but significantly different in dark- and light-incubated mats (Fig. 1). The maximum filament abundance in light-incubated mats was at a depth of 8 mm, and after 10 h of dark incubation, the maximum filament abundance was at a depth of 7 mm. In the light, the maximum abundance was found in the depth zone where oxygen and sulfide co-occurred (Fig. 2A). Upon darkening of the environment, oxygen retreated close to the surface, whereas the sulfide front hardly moved. In mats incubated in the dark, a gap of more than 5 mm existed between the oxic and sulfidic zones, and the Beggiatoa filaments remained in the anoxic layer close to the sulfidic zone (Fig. 2B). Thus, the vertical distribution of Beggiatoa seemed more related to the distribution of sulfide than to that of oxygen. Routine oxygen and sulfide profiling of the mesocosm-incubated intact Lake Chiprana microbial mat showed that although the mat expanded in thickness over the incubation period, oxygen and sulfide production in the mat persisted (Fig. 2C). The presence of sulfide in the mesocosm-incubated mats suggests that active sulfate reduction in the surface layer continued during the incubation period. Previous studies concerning Lake Chiprana microbial mats showed that the concentrations of dissolved organic carbon (21, 21a) as well as sulfate reduction rates (34a) were indeed highest in the photic zone of the microbial mat, indicating that excreted photosynthates may primarily fuel the process of sulfate reduction in these mats.
FIG. 1.
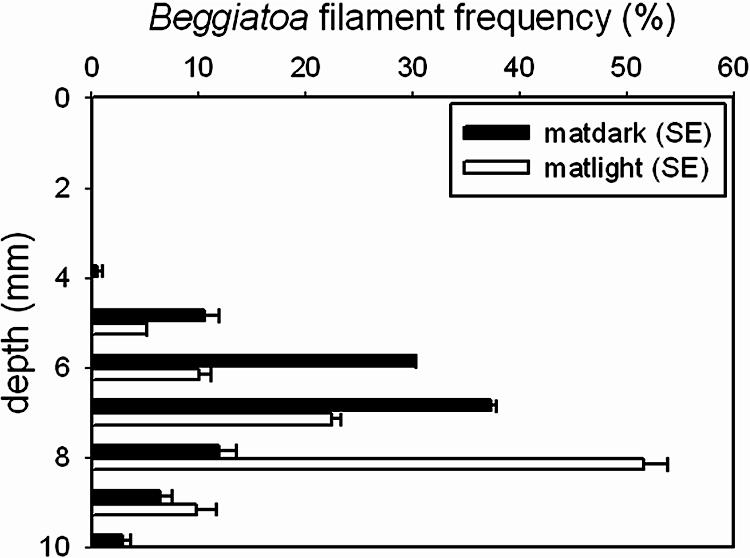
Depth distribution of Beggiatoa filaments in a microbial mat after 10 h of dark and 14 h of light incubation (500 μmol photons m−2 s−1). The data shown are frequencies (%) with standard errors (SE). Black bars, incubation in the dark; white bars, incubation in the light.
FIG. 2.
Depth distribution of Beggiatoa filaments (frequency), oxygen and total sulfide concentrations, and pH in illuminated (A) and dark (B) Lake Chiprana microbial mats incubated for 3 months in a laboratory mesocosm system. Illuminated mats were incubated for 14 h at a light intensity of 500 μmol photons m−2 s−1, while dark mats were incubated in the dark for 10 h prior to measurements. Note the different scale for pH. Measured profiles of oxygen and sulfide concentrations in mats incubated in the mesocosm for 9 days and 5 months (C) show that although the mats expanded vertically, both oxygen and sulfide production in the mats persisted during the incubation period.
Some Beggiatoa species show a phobic response to wavelengths between 400 and 500 nm (41, 44). Microprobe light measurements revealed, however, that light of these wavelengths was already completely absorbed within the top 2 mm of illuminated mats (Fig. 3), which is much shallower than the main Beggiatoa biomass depth distribution (between 5 and 9 mm). However, in the latter depth zone, significant intensities of infrared light (800 nm) were still measurable (i.e., about 15% of the mat surface intensity at a depth of 6 mm). In additional laboratory tests, we could not find any indices for either a phobic or a philic response of Lake Chiprana Beggiatoa organisms subjected to infrared light (results not shown). These results show that light itself likely does not directly affect the Beggiatoa depth distribution.
FIG. 3.
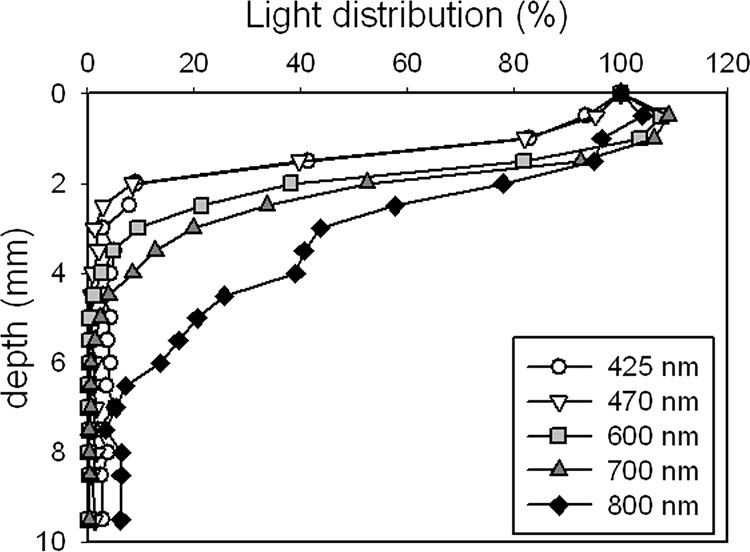
Light profiles in the studied mat, illuminated at 500 μmol photons m−2 s−1. The shorter wavelengths (<500 nm) were virtually completely absorbed in the first 2 mm, while the longer wavelengths penetrated deeper into the mat.
Microsensor pH measurements of light- and dark-incubated mats (Fig. 2) showed pH maxima. The pH maximum in illuminated mats was positioned close to the depth where the oxygen concentration reached its maximum, while in dark mats it occurred in the depth zone with the highest Beggiatoa filament abundance and increasing sulfide values. The pH maximum in the illuminated mat was likely mostly due to CO2 fixation by oxygenic phototrophs (cyanobacteria and diatoms) and other autotrophic bacteria (Beggiatoa), which resulted in a shift in the carbonate equilibrium. The maximum in the dark mat could be explained by the anaerobic metabolism of Beggiatoa, i.e., sulfide oxidation to elemental sulfur with concomitant nitrate reduction to ammonia, as this is a proton-consuming process, as also shown by Sayama et al. (59).
Total nitrate concentrations of between 1 and 15 nmol cm−3 were determined within the mat (Fig. 4) and included pore-water nitrate as well as intracellular nitrate that would be released from the Beggiatoa filaments during freezing and thawing. Due to the small sample size (∼10 μg mm−2), the lower concentrations (1 to 2 nmol cm−3) measured in the lower part of the mat (between 9 and 10 mm deep) might be caused by minimal contamination and therefore indistinguishable from zero. In the illuminated mat, nitrate concentrations were below 5 nmol cm−3 throughout the profile, and no clear trend was recognizable. The dark-incubated mat revealed a nitrate maximum at a depth of 4 mm, just below the disappearance of oxygen and at the same depth as the first Beggiatoa filaments. Below this maximum, nitrate values decreased toward the detection limit at a 10-mm depth, where only small numbers of Beggiatoa filaments were found.
FIG. 4.
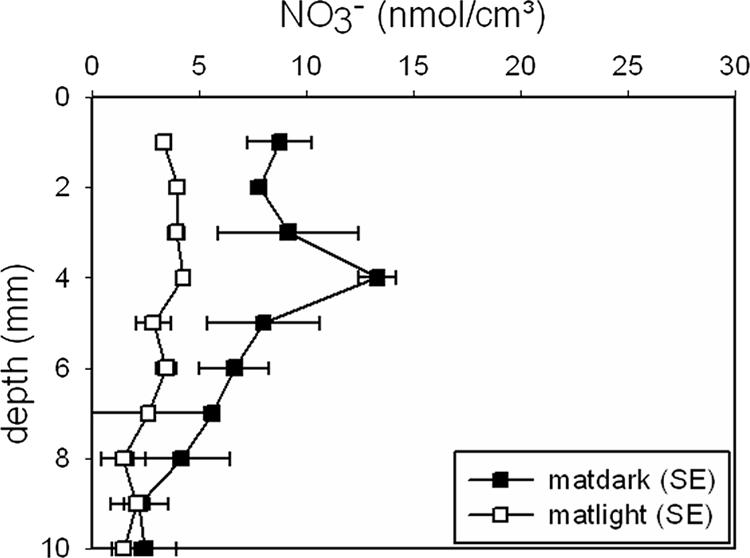
Microbial mat profiles of total extractable nitrate after light (14 h at 500 μmol photons m−2 s−1) and dark (10 h) incubation.
The elemental sulfur microbial mat profiles (Fig. 5) reflected the Beggiatoa filament depth distribution, with the highest values for depths below 4 mm and above 9 mm for both light and dark incubation. Similar to the nitrate results, the measured profiles of S0 in the dark mat showed higher values than the profiles for the illuminated mat. The dip in S0 concentration at the depths with the highest filament abundances may be explained by Beggiatoa S0 oxidation by oxygen and internally stored nitrate under light and dark incubation conditions, respectively, although external sulfur deposition or oxidation by other mat community members cannot be excluded.
FIG. 5.
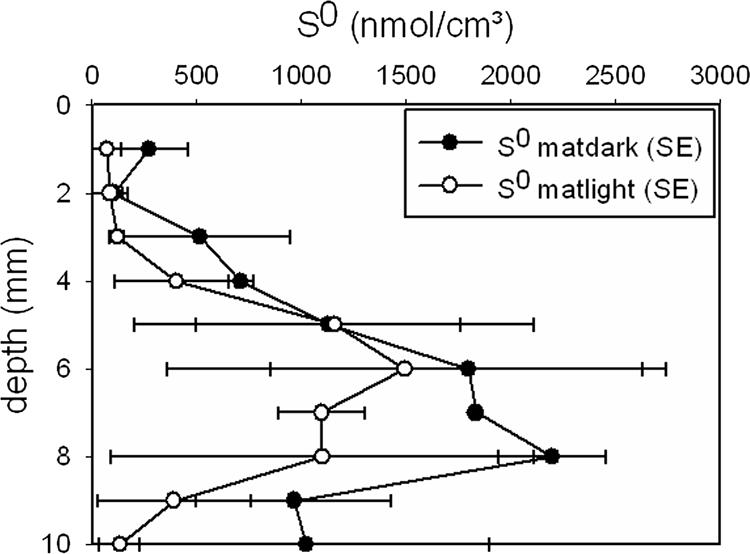
Microbial mat profiles of total extractable elemental sulfur after light (14 h of 500 μmol photons m−2 s−1) and dark (10 h) incubation.
Intracellular nitrate and sulfur storage.
Stable-isotope gas chromatography-mass spectrometry and elemental sulfur HPLC analysis revealed that intracellular concentrations of both nitrate and elemental sulfur were variable (4 to 42 mM for nitrate and 26 to 252 mM for S0) in Beggiatoa organisms picked directly out of intact mats (Table 1). In an incubation experiment with an enriched Beggiatoa population in water containing 50 μM nitrate, filaments were able to increase internal nitrate concentrations about fivefold during a 24-hour period. The longer that filaments were exposed to nitrate, the more internal nitrate was detectable. After an initial 24-hour incubation period, Beggiatoa internal nitrate values amounted to 8 mM. An additional 24-hour incubation period with excess nitrate resulted in an increased internal nitrate concentration of 44 mM. The values for internally stored sulfur remained relatively unchanged (26 to 34 mM) during this experiment.
TABLE 1.
Beggiatoa sp. intracellular nitrate and sulfur storage
| Filament sample group | Internal concn (mM)a
|
|
|---|---|---|
| Nitrate | Sulfur | |
| Filaments from fresh, intact mats | 4 ± 0.8 | 252 ± 0.5 |
| Filaments from dark-incubated mats | 42 ± 1.9 | 26 ± 0.4 |
| Filaments in incubation expt | ||
| 1 day | 8 ± 0.7 | 26 ± 2.4 |
| 2 days | 44 ± 3.4 | 34 ± 5.3 |
Data are means ± standard deviations.
Morphology and phylogeny.
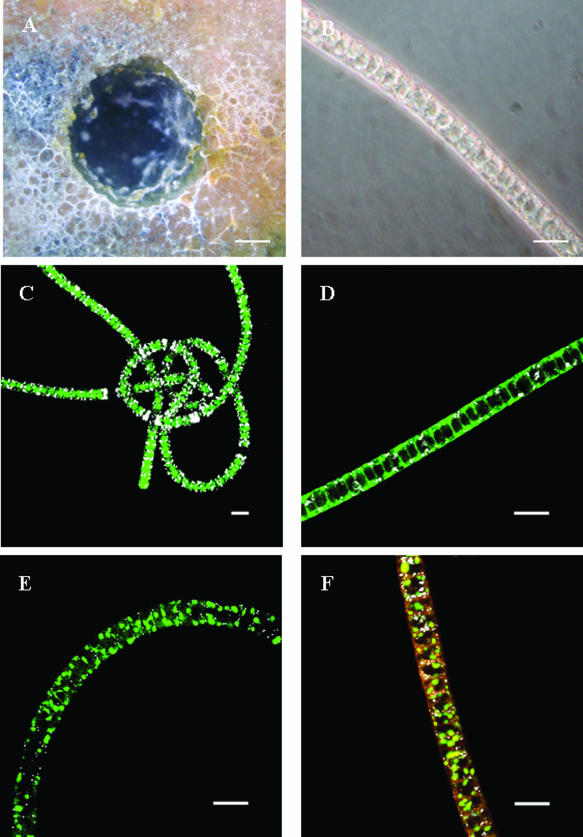
Microscopic analysis revealed a dominant morphotype with a mean diameter of 6 to 8 μm, while the length varied from <1 mm to >10 mm. Differential interference contrast microscopy showed the presence of vacuoles (Fig. 6B), which was further verified by specific staining procedures with diverse fluorochromes and confocal laser scanning microscopy (Fig. 6C to F). Images of Beggiatoa filaments stained with FITC, an amine-reactive dye, clearly showed the presence of vacuolar structures (Fig. 6C) by positively staining the central vacuole. Intracellular sulfur granules were also recorded in white due to their reflection signals (Fig. 6C, D, E, and F). In addition, the black gaps which appeared within filaments could be interpreted as lysed (empty) cells, which may indicate potential filament breaking points (Fig. 6C). Some cells seemed to be packed with sulfur inclusions only (Fig. 6C). MDY-64, a known yeast vacuole membrane marker, was employed here to test its applicability for Beggiatoa vacuole and other membrane-coated cell constituents. This fluorochrome is known to distribute throughout the cell but becomes enriched within membrane structures (57). It showed a negative imprint of the vacuole lumen (Fig. 6D) but apparently also stained other small membrane-coated particles within the cytoplasm. With SYBR green, multiple areas of highly enriched nucleic acid accumulations could be demonstrated within single cells (Fig. 6E and F). SYPRO orange, a protein-specific fluorochrome, stained the cell surface as well as a thin cytoplasmic layer surrounding the vacuole (Fig. 6F).
FIG. 6.
Images of hypersaline Beggiatoa sp. originating from Lake Chiprana. Bar, 10 mm (A) or 10 μm (B to E). (A) Top view of a cored anoxic microbial mat covered by Beggiatoa bacteria. (B) Differential interference contrast micrograph of a vacuolated and sulfur globule-containing Beggiatoa filament. (C) FITC-stained filament. White areas, reflection of sulfur globules. (D) Staining with yeast vacuole membrane marker MDY-64. White areas, reflection of sulfur globules. (E) SYBR green nucleic acid stain. White areas, reflection of sulfur globules. (F) Red, protein stain with SYPRO orange; green, SYBR green stain for nucleic acids; white, reflection of sulfur globules.
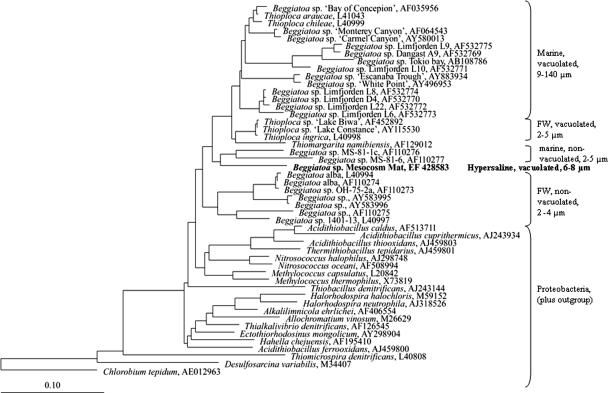
Several single filaments of the dominant Beggiatoa morphotype were picked from intact mat pieces for the construction of a 16S rRNA gene clone library. One of 80 clones contained a nearly full-length 16S rRNA gene sequence related to the Beggiatoa sequence, while the other sequences were related to contaminant, i.e., nontarget, bacteria. The Beggiatoa-related sequence was assumed to correspond to the dominant morphotype observed in the mat, as only one single filament was applied in the PCR. To avoid Beggiatoa DNA autolysis, the picked filaments were immediately transferred into the PCR vial without a previous rigorous cleaning step. This procedure, however, apparently caused the dominance of sequences of contaminant bacteria in the clone library. The obtained target sequence grouped with a cluster of marine Beggiatoa isolates (Fig. 7). So far, all characterized larger Beggiatoa species (>9 μm) in this group contain vacuoles. The hypersaline morphotype from Lake Chiprana appeared to be most closely related (91% sequence similarity) to the marine, narrow (4 μm), and reportedly nonvacuolated Beggiatoa sp. strain MS-81-6 (AF 110277), which originated from the Sippewissett salt marshes (Massachusetts) (47).
FIG. 7.
16S rRNA gene-based phylogenetic reconstruction showing the positions of the hypersaline nitrate-storing Beggiatoa strain originating from Lake Chiprana and other representatives of the gamma subdivision of the Proteobacteria. Bar, 10% estimated sequence divergence.
Gliding motility.
The gliding movement of filamentous, multicellular Beggiatoa organisms proceeds with a reported average speed of 3 μm s−1 (41). The measured gliding speed of the hypersaline Beggiatoa strain from Lake Chiprana incubated under diverse conditions was between 1 and 2 μm s−1. The slowest gliding movement, 0.94 μm s−1 (±0.44 μm s−1 [standard deviation]), was detected in microcapillaries containing artificial anoxic seawater and a nitrate concentration of 50 μM. Filaments moving in medium in gradient tubes with 20 mM H2S added from one side were slightly faster (1.49 ± 0.11 μm s−1). The highest gliding speed, 2.00 ± 0.29 μm s−1, was recorded for microcapillaries filled with anoxic medium only. With an average speed of 2 μm s−1, filaments could have covered a distance of 72 mm in 10 h, the time of the dark incubation period, if they would have followed a straight line.
DISCUSSION
The retrieved sequence of the hypersaline nitrate-storing Beggiatoa sp. obtained in this study clearly showed a phylogenetic relationship to three (Thioploca, Thiomargarita, and Beggiatoa) of the four genera of the large sulfur bacteria, which group within the γ-subclass of the Proteobacteria. Only representatives of Beggiatoa and Thiothrix, the fourth genus of the large sulfur bacteria, are maintained in pure culture to date. The classification of uncultured morphotypes of Beggiatoa sp.-like bacteria has been based on filament diameter, the presence of intracellular vacuoles, nitrate storage capacity, and some metabolic properties. The dominant Beggiatoa sp.-like morphotype obtained from the hypersaline Lake Chiprana had an average filament width of 6 μm and clearly recognizable intracellular vacuoles. The closely affiliated marine strain MS-81-6 has been grown in pure culture and was studied by Nelson et al. (46, 47) and Hagen and Nelson (17, 18). This strain is characterized by a narrow filament width (4 μm) and the apparent absence of intracellular vacuoles. Although the morphological characteristics of these two morphotypes differ in at least two aspects (filament width and vacuolation), they both appear to be tolerant to salinities above those found in marine water: Lake Chiprana has an average salinity of 8%, and strain MS-81-6 was isolated from a salt marsh where salt concentrations during low tide can also increase strongly due to evaporation. Ahmad et al. (2) defined strain MS-81-6, together with another cultivated, nonvacuolated, but narrower (2 μm) strain (MS-81-1c) from the same salt marsh (47), as the root of the large vacuolated sulfur bacterium clade. Interestingly, the Lake Chiprana isolate seems to be an intermediate between both groups, as its cells are narrow but vacuolated. The Beggiatoa morphotype from Lake Chiprana appears to be more closely related phylogenetically to representatives of the marine narrow nonvacuolated Beggiatoa than to those of larger marine and vacuolated sulfur bacteria. Therefore, vacuolation and nitrate accumulation capability appear not to be restricted to wide-diameter Beggiatoa spp., as previously assumed (29).
The vertical diurnal migration behavior of hypersaline but further uncharacterized Beggiatoa organisms from photosynthetic mats from Guerrero Negro, Mexico, was studied before (16). In that study, it was observed that during the day, Beggiatoa bacteria were concentrated in the zone where oxygen and sulfide co-occurred. At night, however, this population was split into two subpopulations. One part rose to the surface of the mat, while another part of the population remained in the deeper, now anoxic, part of the mat. A similar behavior to that of the latter group was found for Lake Chiprana Beggiatoa. During the light (daytime) period, filaments were most abundant in the narrow oxygen and sulfide transition zone. Here they most likely oxidized sulfide by using oxygen as an electron acceptor, as the observed pH minimum in this zone reflects this acidic process. Simultaneously, the filaments could concentrate nitrate, presumably in the intracellular vacuoles, while at the oxic-anoxic transition zone other microbial mat community members produce nitrate during aerobic ammonium oxidation (nitrification). This scenario seems possible, as the ability for nitrate storage of the studied Lake Chiprana strain was confirmed by stable-isotope techniques. Although measured total extractable nitrate values were low, nitrate can be concentrated strongly, 1,000- to 10,000-fold, by Beggiatoa, as shown by the incubation experiment, similar to data reported for other strains (13, 38). The fact that the determined total (internal plus pore-water) extractable nitrate profiles did not reflect the Beggiatoa depth distribution can be explained by the ratio of relatively low Beggiatoa volume to total microbial mat volume, as this amounted to less than 1%. The analyzed microbial mat elemental sulfur concentration profiles, however, appeared similar to the Beggiatoa depth distribution, but this might be coincidental. Although internal sulfur concentrations were found to be significantly higher than internal nitrate concentrations, the actual impact of the rather low total Beggiatoa biomass (3.1 to 5.9 g m−2) and the related intracellular stored sulfur contribution to total (internal plus external) sulfur concentrations in the mat remain to be investigated. The relatively low areal biomass values actually suggest that the impact of Lake Chiprana Beggiatoa on the system's sulfur cycle may be rather low. Analogous examinations have been conducted in different habitats and have shown very low intracellular elemental sulfur concentrations in comparison to a large amount of extracellular elemental sulfur in the bulk sediment (25). Various authors made widely different estimations of the importance of nitrate-storing bacteria for S cycling. With generally 1 order of magnitude higher cell densities, estimations ranged from 3 to 91% of all produced sulfide being oxidized by these bacteria (12, 42, 51). While the importance of Beggiatoa bacteria for mat ecology is debatable, their physiological adaptation to the diel cycles typical for microbial mats is, without a doubt, highly interesting. Their nitrate storage capacity allows Beggiatoa bacteria to remain active during the night, when most mats are anoxic and thus exclude the metabolic activity of aerobic sulfide oxidizers. Beggiatoa bacteria are most likely repelled by steep oxygen and sulfide gradients, allowing them to find the overlapping zone. Nevertheless, the Beggiatoa filaments seemingly do not use their tactic responses to follow the retracting oxygen front during evening darkening. Nitrate storage also allows them at night to remain close to the position they found during the day. A similar strategy was proposed for the immotile large sulfur bacterium Thiomargarita namibiensis, which must overcome long periods (weeks to months) of anoxic conditions, as in its natural habitat dark marine sediments, oxygen, and nitrate are only incidentally introduced during turbulent mixing events (60, 61). Beggiatoa organisms from cold seeps and other sulfidic marine sediments preferably inhabit the suboxic zone (59), the zone that separates the oxic and sulfidic zones, where neither sulfide nor oxygen is present in detectable amounts (5, 14). Whereas marine filamentous strains use a characteristic suite of tools (gliding motility, negative tactic responses to sulfide and oxygen, and nitrate storage) to migrate through a permanent anoxic sediment, the hypersaline Beggiatoa organisms of this study apparently use the same tools to position themselves optimally in a dynamic environment. How far salinity itself, in terms of both quantity and quality, influences the Beggiatoa survival strategy remains to be investigated. We can only hypothesize at this stage that not salinity but, rather, oxygen and nitrate concentration dynamics of a specific sulfidic ecosystem are the determining factors influencing Beggiatoa migration behavior. The internally stored nitrate can thus be used to overcome spatially or temporally separated zones of sulfide and nitrate availability. This unique example shows that one set of characteristics can be applied in two different strategies and thereby allow a competitive advantage in two entirely different habitats.
Acknowledgments
This work was supported by the Max Planck Society (MPG), the Deutsche Forschungsgemeinschaft (DFG), and the Forschergruppe Geobiology of Organo- and Biofilms (DFG-FOR 571).
We gratefully acknowledge Rutger de Wit for assisting with the field work and the Lake Chiprana authorities for granting permission to access the lake and take microbial mat samples. Gaby Eickert, Ines Schröder, and Cäcilia Weigand provided excellent technical support. We highly acknowledge Ute Kuhlike for technical support during laser microscopy. We thank two anonymous reviewers for their constructive comments and helpful suggestions that improved the manuscript.
Footnotes
Published ahead of print on 31 August 2007.
REFERENCES
- 1.Ahmad, A., J. P. Barry, and D. C. Nelson. 1999. Phylogenetic affinity of a wide, vacuolate, nitrate-accumulating Beggiatoa sp. from Monterey Canyon, California, with Thioploca spp. Appl. Environ. Microbiol. 65:270-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ahmad, A., K. M. Kalanetra, and D. C. Nelson. 2006. Cultivated Beggiatoa spp. define the phylogenetic root of morphologically diverse, noncultured, vacuolate sulfur bacteria. Can. J. Microbiol. 52:591-598. [DOI] [PubMed] [Google Scholar]
- 3.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 4.Barry, J. P., H. G. Greene, D. L. Orange, C. H. Baxter, B. H. Robison, R. E. Kochevar, J. W. Nybakken, D. L. Reed, and C. M. McHugh. 1996. Biologic and geologic characteristics of cold seeps in Monterey Bay, California. Deep-Sea Res. I 43:1739-1762. [Google Scholar]
- 5.Berner, R. A. 1981. A new geochemical classification of sedimentary environments. J. Sediment Petrol. 51:359-365. [Google Scholar]
- 6.Beyer, H. 1974. Theorie und Praxis der Interferenzmikroskopie. Geest & Portig K.-G., Leipzig, Germany.
- 7.Bruchert, V., B. B. Jorgensen, K. Neumann, D. Riechmann, M. Schlosser, and H. Schulz. 2003. Regulation of bacterial sulfate reduction and hydrogen sulfide fluxes in the central Namibian coastal upwelling zone. Geochim. Cosmochim. Acta 67:4505-4518. [Google Scholar]
- 8.Castenholz, R. W., B. B. Jorgensen, E. Damelio, and J. Bauld. 1991. Photosynthetic and behavioral versatility of the cyanobacterium Oscillatoria-boryana in a sulfide-rich microbial mat. FEMS Microbiol. Ecol. 86:43-58. [Google Scholar]
- 9.Cox, R. D., and R. F. Earp. 1982. Determination of trace level organics in ambient air by high-resolution gas-chromatography with simultaneous photo-ionization and flame ionization detection. Anal. Chem. 54:2265-2270. [Google Scholar]
- 10.de Beer, D., E. Sauter, H. Niemann, N. Kaul, J. P. Foucher, U. Witte, M. Schlüter, and A. Boetius. 2006. In situ fluxes and zonation of microbial activity in surface sediments of the Hakon Mosby mud volcano. Limnol. Oceanogr. 51:1315-1331. [Google Scholar]
- 11.Deming, J. W., A. L. Reysenbach, S. A. Macko, and C. R. Smith. 1997. Evidence for the microbial basis of a chemoautotrophic invertebrate community at a whale fall on the deep seafloor: bone-colonizing bacteria and invertebrate endosymbionts. Microsc. Res. Tech. 37:162-170. [DOI] [PubMed] [Google Scholar]
- 12.Ferdelman, T. G., C. Lee, S. Pantoja, J. Harder, B. Bebout, and H. Fossing. 1997. Sulfate reduction and methanogenesis in a Thioploca-dominated sediment off the coast of Chile. Geochim. Cosmochim. Acta 61:3065-3079. [Google Scholar]
- 13.Fossing, H., V. A. Gallardo, B. B. Jorgensen, M. Huttel, L. P. Nielsen, H. Schulz, D. E. Canfield, S. Forster, R. N. Glud, J. K. Gundersen, J. Kuver, N. B. Ramsing, A. Teske, B. Thamdrup, and O. Ulloa. 1995. Concentration and transport of nitrate by the mat-forming sulphur bacterium Thioploca. Nature 374:713-715. [Google Scholar]
- 14.Froelich, P. N., G. P. Klinkhammer, M. L. Bender, N. A. Luedtke, G. R. Heath, D. Cullen, P. Dauphin, D. E. Hammond, B. Hartman, and V. Maynard. 1979. Early oxidation of organic matter in pelagic sediments of the eastern equatorial Atlantic: suboxic diagenesis. Geochim. Cosmochim. Acta 43:1075-1090. [Google Scholar]
- 15.Gallardo, V. A. 1977. Large benthic microbial communities in sulphide biota under Peru-Chile subsurface countercurrent. Nature 268:331-332. [Google Scholar]
- 16.Garcia-Pichel, F. 1994. Diel migrations of microorganisms within a benthic, hypersaline mat community. Appl. Environ. Microbiol. 60:1500-1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hagen, K. D., and D. C. Nelson. 1996. Organic carbon utilization by obligately and facultatively autotrophic Beggiatoa strains in homogeneous and gradient cultures. Appl. Environ. Microbiol. 62:947-953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hagen, K. D., and D. C. Nelson. 1997. Use of reduced sulfur compounds by Beggiatoa spp.: enzymology and physiology of marine and freshwater strains in homogeneous and gradient cultures. Appl. Environ. Microbiol. 63:3957-3964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hicks, R. E., R. I. Amann, and D. A. Stahl. 1992. Dual staining of natural bacterioplankton with 4′,6′-diamidino-2-phenylindole and fluorescent oligonucleotide probes targeting kingdom-level 16S ribosomal RNA sequences. Appl. Environ. Microbiol. 58:2158-2163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jeroschewski, P., C. Steuckart, and M. Kuhl. 1996. An amperometric microsensor for the determination of H2S in aquatic environments. Anal. Chem. 68:4351-4357. [Google Scholar]
- 21.Jonkers, H. M., R. Ludwig, R. De Wit, O. Pringault, G. Muyzer, H. Niemann, N. Finke, and D. De Beer. 2003. Structural and functional analysis of a microbial mat ecosystem from a unique permanent hypersaline inland lake: ‘La Salada de Chiprana’ (NE Spain). FEMS Microbiol. Ecol. 44:175-189. [DOI] [PubMed] [Google Scholar]
- 21a.Jonkers, H. M., I.-O. Koh, P. Behrend, G. Muyzer, and D. De Beer. 2005. Aerobic organic carbon mineralization by sulfate-reducing bacteria in the oxygen-saturated photic zone of a hypersaline microbial mat. Microb. Ecol. 49:291-300. [DOI] [PubMed] [Google Scholar]
- 22.Jorgensen, B. B. 1977. Distribution of colorless sulfur bacteria (Beggiatoa-spp) in a coastal marine sediment. Mar. Biol. 41:19-28. [Google Scholar]
- 23.Jorgensen, B. B., and D. J. Des Marais. 1986. Competition for sulfide among colorless and purple sulfur bacteria in cyanobacterial mats. FEMS Microbiol. Ecol. 38:179-186. [DOI] [PubMed] [Google Scholar]
- 24.Jorgensen, B. B., and V. A. Gallardo. 1999. Thioploca spp.: filamentous sulfur bacteria with nitrate vacuoles. FEMS Microbiol. Ecol. 28:301-313. [Google Scholar]
- 25.Jorgensen, B. B., and D. C. Nelson. 2004. Sulfide oxidation in marine sediments: geochemistry meets microbiology. Geol. Soc. Am. 379:63-81. [Google Scholar]
- 26.Jorgensen, B. B., and N. P. Revsbech. 1983. Colorless sulfur bacteria, Beggiatoa spp. and Thiovulum spp. in O2 and H2S microgradients. Appl. Environ. Microbiol. 45:1261-1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jorgensen, B. B., and D. J. Des Marais. 1986. A simple fiber optic microprobe for high resolution light measurements: application in marine sediments. Limnol. Oceanogr. 31:1376-1383. [DOI] [PubMed] [Google Scholar]
- 28.Joye, S., A. Boetius, K. Kalanetra, I. MacDonald, J. Montoya, B. Orcutt, and V. Samarkin. 2005. Biogeochemical signatures of cold seeps in benthic and pelagic environments along the Gulf of Mexico continental margin. Geophys. Res. Abstr. 2005:7. [Google Scholar]
- 29.Kalanetra, K. M., S. L. Huston, and D. C. Nelson. 2004. Novel, attached, sulfur-oxidizing bacteria at shallow hydrothermal vents possess vacuoles not involved in respiratory nitrate accumulation. Appl. Environ. Microbiol. 70:7487-7496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kamp, A., P. Stief, and H. N. Schulz-Vogt. 2006. Anaerobic sulfide oxidation with nitrate by a freshwater Beggiatoa enrichment culture. Appl. Environ. Microbiol. 72:4755-4760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kane, M. D., L. K. Poulsen, and D. A. Stahl. 1993. Monitoring the enrichment and isolation of sulfate-reducing bacteria by using oligonucleotide hybridization probes designed from environmentally derived 16S ribosomal RNA sequences. Appl. Environ. Microbiol. 59:682-686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kojima, H., and M. Fukui. 2003. Phylogenetic analysis of Beggiatoa spp. from organic rich sediment of Tokyo Bay, Japan. Water Res. 37:3216-3223. [DOI] [PubMed] [Google Scholar]
- 33.Kojima, H., A. Teske, and M. Fukui. 2003. Morphological and phylogenetic characterizations of freshwater Thioploca species from Lake Biwa, Japan, and Lake Constance, Germany. Appl. Environ. Microbiol. 69:390-398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kühl, M., C. Steuckart, G. Eickert, and P. Jeroschewski. 1998. A H2S microsensor for profiling biofilms and sediments—application in an acidic lake sediment. Aquat. Microb. Ecol. 15:201-209. [Google Scholar]
- 34a.Ludwig, R., F. A. Al-Horani, D. De Beer, and H. M. Jonkers. 2005. Photosynthesis-controlled calcification in a hypersaline microbial mat. Limnol. Oceanogr. 50:1836-1843. [Google Scholar]
- 35.Ludwig, W., O. Strunk, R. Westram, L. Richter, H. Meier, Yadhukumar, A. Buchner, T. Lai, S. Steppi, G. Jobb, W. Forster, I. Brettske, S. Gerber, A. W. Ginhart, O. Gross, S. Grumann, S. Hermann, R. Jost, A. Konig, T. Liss, R. Lussmann, M. May, B. Nonhoff, B. Reichel, R. Strehlow, A. Stamatakis, N. Stuckmann, A. Vilbig, M. Lenke, T. Ludwig, A. Bode, and K. H. Schleifer. 2004. ARB: a software environment for sequence data. Nucleic Acids Res. 32:1363-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Macalady, J. L., E. H. Lyon, B. Koffman, L. K. Albertson, K. Meyer, S. Galdenzi, and S. Mariani. 2006. Dominant microbial populations in limestone-corroding stream biofilms, Frasassi cave system, Italy. Appl. Environ. Microbiol. 72:5596-5609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mattison, R. G., M. Abbiati, P. R. Dando, M. F. Fitzsimons, S. M. Pratt, A. J. Southward, and E. C. Southward. 1998. Chemoautotrophic microbial mats in submarine caves with hydrothermal sulphidic springs at Cape Palinuro, Italy. Microb. Ecol. 35:58-71. [DOI] [PubMed] [Google Scholar]
- 38.McHatton, S. C., J. P. Barry, H. W. Jannasch, and D. C. Nelson. 1996. High nitrate concentrations in vacuolate, autotrophic marine Beggiatoa spp. Appl. Environ. Microbiol. 62:954-958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mezzino, M. J., W. R. Strohl, and J. M. Larkin. 1984. Characterization of Beggiatoa alba. Arch. Microbiol. 137:139-144. [Google Scholar]
- 40.Mills, H. J., R. J. Martinez, S. Story, and P. A. Sobecky. 2004. Identification of members of the metabolically active microbial populations associated with Beggiatoa species mat communities form Gulf of Mexico cold-seep sediments. Appl. Environ. Microbiol. 70:5447-5458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Moeller, M. M., L. P. Nielsen, and B. B. Joergensen. 1985. Oxygen responses and mat formation by Beggiatoa spp. Appl. Environ. Microbiol. 50:373-382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mussmann, M., H. N. Schulz, B. Strotmann, T. Kjaer, L. P. Nielsen, R. A. Rossello-Mora, R. I. Amann, and B. B. Jorgensen. 2003. Phylogeny and distribution of nitrate-storing Beggiatoa spp. in coastal marine sediments. Environ. Microbiol. 5:523-533. [DOI] [PubMed] [Google Scholar]
- 43.Nelson, D. C. 1992. The genus Beggiatoa, p. 3171-3180. In A. Balow, H. G. Trüper, M. Dworkin, W. Harder, and K. H. Schleifer (ed.), The prokaryotes, 2nd ed., vol. 166. Springer-Verlag, New York, NY. [Google Scholar]
- 44.Nelson, D. C., and R. W. Castenholz. 1982. Light responses of Beggiatoa. Arch. Microbiol. 131:146-155. [Google Scholar]
- 45.Nelson, D. C., B. B. Jorgensen, and N. P. Revsbech. 1986. Growth pattern and yield of a chemoautotrophic Beggiatoa sp. in oxygen sulfide microgradients. Appl. Environ. Microbiol. 52:225-233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nelson, D. C., N. P. Revsbech, and B. B. Jorgensen. 1986. Microoxic-anoxic niche of Beggiatoa spp.—microelectrode survey of marine and freshwater strains. Appl. Environ. Microbiol. 52:161-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nelson, D. C., J. B. Waterbury, and H. W. Jannasch. 1982. Nitrogen fixation and nitrate utilization by marine and fresh-water Beggiatoa. Arch. Microbiol. 133:172-177. [Google Scholar]
- 48.Nelson, D. C., C. O. Wirsen, and H. W. Jannasch. 1989. Characterization of large, autotrophic Beggiatoa spp. abundant at hydrothermal vents of the Guaymas Basin. Appl. Environ. Microbiol. 55:2909-2917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nielsen, L. P. 1992. Denitrification in sediment determined from nitrogen isotope pairing. FEMS Microbiol. Ecol. 86:357-362. [Google Scholar]
- 50.Nomarski, G. 1952. Interféromètre à polarization, Franz. Patent 1.059.123.
- 51.Otte, S., J. G. Kuenen, L. P. Nielsen, H. W. Paerl, J. Zopfi, H. N. Schulz, A. Teske, B. Strotmann, V. A. Gallardo, and B. B. Jørgensen. 1999. Nitrogen, carbon, and sulfur metabolism in natural Thioploca samples. Appl. Environ. Microbiol. 65:3148-3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pachmeyer, F. 1960. Vorkommen und bestimmung von Schwefelverbindungen in Mineralwasser. Ph.D. thesis. University of Munich, Munich, Germany.
- 53.Revsbech, N. P. 1989. An oxygen microsensor with a guard cathode. Limnol. Oceanogr. 34:474-478. [Google Scholar]
- 54.Revsbech, N. P., B. B. Jorgensen, T. H. Blackburn, and Y. Cohen. 1983. Microelectrode studies of the photosynthesis and O2, H2S, and pH profiles of a microbial mat. Limnol. Oceanogr. 28:1062-1074. [Google Scholar]
- 55.Richardson, L. L., and R. W. Castenholz. 1987. Diel vertical movements of the cyanobacterium Oscillatoria terebriformis in a sulfide-rich hot-spring microbial mat. Appl. Environ. Microbiol. 53:2142-2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Russow, R. 1999. Determination of N-15 in N-15-enriched nitrite and nitrate in aqueous samples by reaction continuous-flow quadrupole mass spectrometry. Rapid Commun. Mass Spectrom. 13:1334-1338. [DOI] [PubMed] [Google Scholar]
- 57.Sato, H., D. W. Frank, C. J. Hillard, J. B. Feix, R. R. Pankhaniya, K. Moriyama, V. Finck-Barbancon, A. Buchaklian, M. Lei, R. M. Long, J. Wiener-Kronish, and T. Sawa. 2003. The mechanism of action of the Pseudomonas aeruginosa-encoded type III cytotoxin, ExoU. EMBO J. 22:2959-2969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sayama, M. 2001. Presence of nitrate-accumulating sulfur bacteria and their influence on nitrogen cycling in a shallow coastal marine sediment. Appl. Environ. Microbiol. 67:3481-3487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sayama, M., N. Risgaard-Petersen, L. P. Nielsen, H. Fossing, and P. B. Christensen. 2005. Impact of bacterial NO3− transport on sediment biogeochemistry. Appl. Environ. Microbiol. 71:7575-7577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schulz, H. N., T. Brinkhoff, T. G. Ferdelman, M. H. Marine, A. Teske, and B. B. Jorgensen. 1999. Dense populations of a giant sulfur bacterium in Namibian shelf sediments. Science 284:493-495. [DOI] [PubMed] [Google Scholar]
- 61.Schulz, H. N., and B. B. Jorgensen. 2001. Big bacteria. Annu. Rev. Microbiol. 55:105-137. [DOI] [PubMed] [Google Scholar]
- 62.Sherwood, J. E., F. Stagnitti, M. J. Kokkinn, and W. D. Williams. 1991. Dissolved-oxygen concentrations in hypersaline waters. Limnol. Oceanogr. 36:235-250. [Google Scholar]
- 63.Strohl, W. R., and J. M. Larkin. 1978. Enumeration, isolation, and characterization of Beggiatoa from freshwater sediments. Appl. Environ. Microbiol. 36:755-770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sweerts, J., D. DeBeer, L. P. Nielsen, H. Verdouw, J. C. Vandenheuvel, Y. Cohen, and T. E. Cappenberg. 1990. Denitrification by sulfur oxidizing Beggiatoa spp. mats on fresh-water sediments. Nature 344:762-763. [Google Scholar]
- 65.Teske, A., and D. C. Nelson. August 2004, posting date. The genera Beggiatoa and Thioploca. In M. Dworkin et al. (ed.), The prokaryotes: an evolving electronic resource for the microbiological community, 3rd ed., release 3.17. Springer, New York, NY.
- 66.Valero-Garces, B. L., A. Navas, J. Machin, T. Stevenson, and B. Davis. 2000. Responses of a saline lake ecosystem in a semiarid region to irrigation and climate variability—the history of Salada Chiprana, central Ebro Basin, Spain. Ambio 29:344-350. [Google Scholar]
- 67.van Gemerden, H. 1993. Microbial mats—a joint venture. Mar. Geol. 113:3-25. [Google Scholar]
- 68.Wieland, A., and M. Kühl. 2000. Short-term temperature effects on oxygen and sulfide cycling in a hypersaline cyanobacterial mat (Solar Lake, Egypt). Mar. Ecol. Prog. Ser. 196:87-102. [Google Scholar]
- 69.Zopfi, J., T. G. Ferdelman, B. B. Jorgensen, A. Teske, and B. Thamdrup. 2001. Influence of water column dynamics on sulfide oxidation and other major biogeochemical processes in the chemocline of Mariager Fjord (Denmark). Mar. Chem. 74:29-51. [Google Scholar]
- 70.Zubkov, M. V., B. M. Fuchs, H. Eilers, P. H. Burkill, and R. Amann. 1999. Determination of total protein content of bacterial cells by SYPRO staining and flow cytometry. Appl. Environ. Microbiol. 65:3251-3257. [DOI] [PMC free article] [PubMed] [Google Scholar]