Abstract
A phylogenetic analysis of three genomic regions revealed that Tomato yellow leaf curl virus (TYLCV) from western North America is distinct from TYLCV isolated in eastern North America and the Caribbean. This analysis supports a second introduction of this Old World begomovirus into the New World, most likely from Asia.
Tomato yellow leaf curl virus (TYLCV) is an emergent, monopartite begomovirus (of the family Geminiviridae) that causes perhaps the most devastating epidemics in tomato agriculture (32). TYLCV was first identified in Israel in 1939 (32) and was isolated and sequenced in 1988 (isolate TYLCV-IL [7, 29]). Molecular typing of outbreaks has shown that viruses closely related to TYLCV-IL have spread worldwide (19, 27, 32, 46).
TYLCV was first introduced into the New World, either through the Dominican Republic in the early 1990s (26, 33) or through Cuba (19, 36), possibly in a shipment of tomato seedlings from Israel (34). TYLCV subsequently spread to other Caribbean islands and to North America (24). Previously published phylogenies have supported the monophyly of New World TYLCV (8, 17, 23, 25, 28, 43, 48), suggesting that all New World TYLCV isolates were derived from a single introduction of a TYLCV-IL-like virus into the New World (13, 17, 23, 28, 48). However, recently obtained isolates of TYLCV from western North America (WNA; from Sinaloa, Mexico, and California) are not as closely related to other New World TYLCV isolates (5). Herein, we test whether these WNA isolates could have descended from a second, independent introduction of TYLCV into the New World.
New World sequences of TYLCV were downloaded from GenBank (Table 1 [names following current nomenclature {9}]). BLASTN (1) was used to identify sequences closely related to the WNA TYLCV, and two recent isolates of TYLCV from Asia that have 99% identity to TYLCV-US:TX and TYLCV-MX:Cul were included in our analysis (Table 1). TYLCV-IL, which is closely related to the New World TYLCV (13, 17, 23, 28, 48), was also included. Sequences were aligned by eye with SE-AL (http://evolve.zoo.ox.ac.uk). Most of the available sequences were partial genome sequences, some covering less than 10% of the TYLCV genome (Table 1). To include all of the extant sequences from the New World, we identified three adjacent, nonoverlapping regions of the TYLCV genome for phylogenetic analysis, for which different numbers of sequences were available: ∼255 nucleotides (nt) of the intergenic region, which is known to be the most variable portion of the genome (10, 11, 30); 61 noncoding nt from the intergenic region with two-thirds of the pre-coat protein-coding region (V2), ∼360 nt; and a portion of the coat protein (CP)-coding region, ∼538 nt. Together, these regions covered ∼40% of the TYLCV genome.
TABLE 1.
New World (and sister taxa found on other continents) isolates of TYLCV used in analysesa
| Origin | Name | Yr(s) of isolation | Source plant | GenBank accession no. | Sequence length (nt) | Sequence data for indicated region
|
Source or referencec | ||
|---|---|---|---|---|---|---|---|---|---|
| Int (255 nt) | V2 (360 nt) | CP (538 nt) | |||||||
| Antigua | TYLCV-AG:StJ | NA | Tomato | EF028239 | 545 | • | Direct submission | ||
| TYLCV-AG:StP | NA | Tomato | EF028240 | 530 | • | Direct submission | |||
| Cuba | TYLCV-CU | 1995-1997 | NA | AJ223505 | 2,781 | • | • | • | Direct submission; E. Bejarano, personal communication |
| TYLCV-CU2 | NA | Tomato | U65089 | 777 | • | 37 | |||
| TYLCV-CU3 | 2001 | Pepper | AF414089 | 630 | • | 36 | |||
| TYLCV-CU4 | 2003 | Squash | DQ207810 | 449 | • | 20 | |||
| Dominican Republic | TYLCV-DO | 1994 | Tomato | AF024715 | 2,781 | • | • | • | 24, 39 |
| Guadeloupe | TYLCV-GP | 2001 | Tomato | AY319645 | 542 | • | 44 | ||
| TYLCV-GP2 | 2001 | Tomato | AY319646 | 966 | • | 44 | |||
| Jamaica | TYLCV-JM:2T | 1994-1996b | Tomato | U84146 | 315 | • | Direct submission | ||
| TYLCV-JM:10T | 1994-1996b | Tomato | U84147 | 312 | • | Direct submission | |||
| TYLCV-JM:52 | 1994-1996b | Pepper | U88889 | 252 | • | Direct submission | |||
| Mexico | TYLCV-MX:Yuc | 1996-1997 | Tomato | AF168709 | 753 | • | • | 2 | |
| TYLCV-MX:Cul | 2005 | Tomato | DQ631892 | 2,781 | • | • | • | 5 | |
| TYLCV-MX:Sin | NA | Tomato | DQ377367 | 742 | • | • | Direct submission | ||
| TYLCV-MX:Sin2 | NA | NA | EF523478 | 2,781 | • | • | • | Direct submission | |
| Puerto Rico | TYLCV-PR | 2001 | Tomato | AY134494 | 2,781 | • | • | • | 3 |
| United States | TYLCV-Flo | 1997 | Tomato | AY530931 | 2,781 | • | • | • | 34, 47 |
| TYLCV-US:Flo | 1997 | Tomato | AF260331 | 1,320 | • | • | • | 48 | |
| TYLCV-US:SC | 2005 | Tomato | DQ139329 | 806 | • | • | • | 18 | |
| TYLCV-US:TX | 2006 | Tomato | EF110890 | 2,752 | • | • | • | 16 | |
| TYLCV-US:CA | 2007 | Tomato | EF539831 | 2,781 | • | • | • | Direct submission | |
| Israel | TYLCV-IL | 1988 | Tomato | X15656 | 2,787 | • | • | • | 29 |
| China | TYLCV-CN:SH2 | 2006 | Tomato | AM282874 | 2,781 | • | • | • | 46 |
| Japan | TYLCV-JA:Tosa | 2004 | Tomato | AB192965 | 2,781 | • | • | • | 43 |
NA, not available; Int, intergenic region; •, sequence used in this alignment.
The TYLCV from Jamaica could not have been isolated before the virus was first observed on the island, in 1994 (21).
Direct submission, information in GenBank annotation (submitted by the researchers who sequenced the given isolate) not published elsewhere.
MODELTEST (35) was used to select the most appropriate model of nucleotide substitution for phylogenetic analysis (intergenic region, HKY85; V2 region, JC+Γ; CP region, Tamura-Nei), which was performed with PAUP*, using maximum likelihood (ML) and tree-bisection-reconnection branch-swapping (41). To assess the reliability of specific groups, a neighbor-joining bootstrap analysis was employed (1,000 replicates under the ML substitution model). The bootstrap values for all nodes with more than 50% support were reported (Fig. 1).
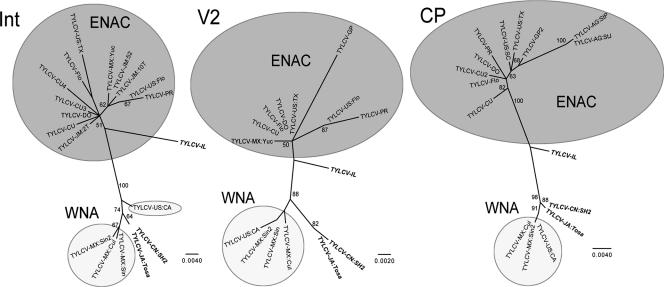
FIG. 1.
Evolutionary relationships among New World TYLCV isolates based on 255 nt of the intergenic region (Int), 360 nt of the intergenic/V2-coding region (V2), and 538 nt of the CP-coding region. An Israeli (TYLCV-IL) and two Asian (TYLCV-CN:SH2 and TYLCV-JA:Tosa) isolates were included and are shown italicized and bolded. The ENAC sequences are shown in the darker shading; the WNA sequences are shown in the lighter shading. All branch lengths are scaled according to the numbers of nucleotide substitutions per site.
Our data set of TYLCV contains four Mexican sequences (Table 1), one of which is from eastern Mexico (Yucatán), near the Caribbean (2), with the other three from the important tomato-growing region of Sinaloa, in western Mexico (6). By contrast, most of the sequences isolated from the United States have been found near the Caribbean (e.g., Florida [34], Louisiana [45], Georgia [22], and Mississippi [15]), even though the virus has recently been identified in western states, such as California and Arizona (14, 38). In all three trees, the sequences from eastern Mexico, the eastern United States, and the Caribbean grouped together, forming an eastern North America and Caribbean (ENAC) clade, albeit with strong support in the CP gene only (bootstrap values of 51% [Fig. 1, Int], 50% [Fig. 1, V2], and 100% [Fig. 1, CP]). The western Mexican (Sinaloa) and western United States (California) sequences grouped together in the two protein-coding region trees (Fig. 1, V2 [91% bootstrap support] and CP). In the intergenic region (Fig. 1, Int), the four WNA viruses did not form a monophyletic clade, and this was likely due to a single informative base that TYLCV-US:CA shares with TYLCV-IL and the ENAC clade (a cytosine at nt 176; nt 188 in TYLCV-IL). Despite this probable parallelism, all four WNA viruses were part of well-supported clades with the Asian TYLCV sequences in all three trees (bootstrap values of 100% [Fig. 1, Int], 88% [Fig. 1, V2], and 98% [Fig. 1, CP]). Crucially, this Asian/WNA clade was reciprocally monophyletic with the ENAC clade and thereby provided strong support for the assumption that the WNA viruses are more distantly related to ENAC TYLCV and likely came from a second introduction of TYLCV into the New World. Similarly, this analysis also indicates that this second introduction most likely came to the Pacific coast of Mexico from Asia.
To further test the hypothesis of multiple entry of TYLCV into the New World, we performed a Shimodaira-Hasegawa test (full optimization, with PAUP* [40]). Accordingly, the ML trees for each genomic region, which support two entries of TYLCV into the New World, were compared to “model” trees in which all New World isolates were forced to be monophyletic, as expected under a single entry into the New World. In all cases, the single entry hypothesis was rejected, though the V2 region offered only marginal support (intergenic region P = 0.017; V2 P = 0.105; CP P = 0.041). These results contradict the conventional wisdom that TYLCV has spread through the New World from a single introduction (14).
Previous studies have noted that there is a large (>1,500-mile) separation between the Yucatán, where TYLCV was first introduced into Mexico in 1996 (2), and Sinaloa, where TYLCV was observed in 2005 (5). Hence, a second introduction of TYLCV explains why TYLCV would be found on both coasts of Mexico without having migrated through the bulk of Mexico.
After the confirmation of TYLCV in Sinaloa, TYLCV was found farther north in Mexico (14), in Texas (16), in Arizona (14), and in southern California (38). Our analyses further confirm that TYLCV-US:CA is most closely related to the western Mexican viruses (38) and therefore likely spread over the border from Mexico (Fig. 1). However, others describe an Arizonian isolate (sequence currently unavailable) that is more closely related to TYLCV-US:TX, which is part of the ENAC clade (Fig. 1), than to viruses from Mexico (14). We therefore conclude that the ENAC strain of TYLCV, which had spread to Louisiana by 2000 (45), has spread through Texas to Arizona but that the first TYLCV to be isolated from California is more closely related to other WNA TYLCV strains from Mexico. This is not surprising, as two other geminiviruses infecting tomato, Tomato leaf curl Sinaloa virus (12) and Chino del Tomate virus (4, 42), have previously spread from Sinaloa into the United States.
Though our results shed some light on the passage of TYLCV through the New World, a combination of recent common ancestry and the limited availability of whole-genome sequences (only a third of the New World TYLCV sequences in GenBank are of whole genomes) (Table 1) meant that it was not possible to fully resolve the phylogenetic history of all New World isolates. Further, as recombination is an important aspect of geminivirus evolution (10, 31), sequencing only a portion of the genome could be revealing only part of the New World TYLCV evolutionary history.
Acknowledgments
This research was funded by National Science Foundation grant DBI-0603070.
Footnotes
Published ahead of print on 7 September 2007.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Ascencia-Ibáñez, J. T., R. Diaz-Plaza, J. Méndez-Lozano, Z. I. Monsalve-Fonnegra, G. R. Argüello-Astorga, and R. F. Rivera-Bustamante. 1999. First report of Tomato yellow leaf curl geminivirus in Yucatán, México. Plant Dis. 83:1178. [DOI] [PubMed] [Google Scholar]
- 3.Bird, J., A. M. Idris, D. Rogan, and J. K. Brown. 2001. Introduction of the exotic Tomato yellow leaf curl virus-Israel in tomato to Puerto Rico. Plant Dis. 85:1028. [DOI] [PubMed] [Google Scholar]
- 4.Brown, J. K., A. M. Idris, I. Torres-Jerez, G. K. Banks, and S. D. Wyatt. 2001. The core region of the coat protein gene is highly useful for establishing the provisional identification and classification of begomoviruses. Arch. Virol. 146:1581-1598. [DOI] [PubMed] [Google Scholar]
- 5.Brown, J. K., and A. M. Idris. 2006. Introduction of the exotic monopartite Tomato yellow leaf curl virus into west coast Mexico. Plant Dis. 90:1360. [DOI] [PubMed] [Google Scholar]
- 6.Cook, R., and L. Calvin. 2005. Greenhouse tomatoes change the dynamics of the North American fresh tomato industry. Economic Research Report no. ERR2. http://www.ers.usda.gov/publications/err2/.
- 7.Czosnek, H., and H. Laterrot. 1997. A worldwide survey of Tomato yellow leaf curl viruses. Arch. Virol. 142:1391-1406. [DOI] [PubMed] [Google Scholar]
- 8.Fauquet, C. M., S. Sawyer, A. M. Idris, and J. K. Brown. 2005. Sequence analysis and classification of apparent recombinant begomoviruses infecting tomato in the Nile and Mediterranean basins. Phytopathology 95:549-555. [DOI] [PubMed] [Google Scholar]
- 9.Fauquet, C. M., and J. Stanley. 2005. Revising the way we conceive and name viruses below the species level: a review of geminivirus taxonomy calls for new standardized isolate descriptors. Arch. Virol. 150:2151-2179. [DOI] [PubMed] [Google Scholar]
- 10.García-Andrés, S., G. P. Accotto, J. Navas-Castillo, and E. Moriones. 2007. Founder effect, plant host, and recombination shape the emergent population of begomoviruses that cause the tomato yellow leaf curl disease in the Mediterranean basin. Virology 359:302-312. [DOI] [PubMed] [Google Scholar]
- 11.Ge, L., J. Zhang, X. Zhou, and H. Li. 2007. Genetic structure and population variability of Tomato yellow leaf curl China virus. J. Virol. 81:5902-5907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Idris, A. M., and J. K. Brown. 1998. Sinaloa tomato leaf curl geminivirus: biological and molecular evidence for a new subgroup II virus. Phytopathology 88:648-657. [DOI] [PubMed] [Google Scholar]
- 13.Idris, A. M., and J. K. Brown. 2005. Evidence for interspecific-recombination for three monopartite begomoviral genomes associated with the tomato leaf curl disease from central Sudan. Arch. Virol. 150:1003-1012. [DOI] [PubMed] [Google Scholar]
- 14.Idris, A. M., J. C. Guerrero, and J. K. Brown. 2007. Two distinct isolates of Tomato yellow leaf curl virus threaten tomato production in Arizona and Sonora, Mexico. Plant Dis. 91:910. [DOI] [PubMed] [Google Scholar]
- 15.Ingram, D. M., and A. Henn. 2001. First report of Tomato yellow leaf curl virus in Mississippi. Plant Dis. 85:1287. [DOI] [PubMed] [Google Scholar]
- 16.Isakeit, T., A. M. Idris, G. Sunter, M. C. Black, and J. K. Brown. 2007. Tomato yellow leaf curl virus in tomato in Texas, originating from transplant facilities. Plant Dis. 91:466. [DOI] [PubMed] [Google Scholar]
- 17.Köklü, G., A. Rojas, and A. Kvarnheden. 2006. Molecular identification and the complete nucleotide sequence of a Tomato yellow leaf curl virus isolate from Turkey. J. Plant Pathol. 88:61-66. [Google Scholar]
- 18.Ling, K. S., A. M. Simmons, R. L. Hassell, A. P. Keinath, and J. E. Polston. 2006. First report of Tomato yellow leaf curl virus in South Carolina. Plant Dis. 90:379. [DOI] [PubMed] [Google Scholar]
- 19.Martinez Zubiaur, Y., I. Zabalgogeazcoa, C. De Blas, F. Sanchez, E. L. Peralta, J. Romero, and F. Ponz. 1996. Geminiviruses associated with diseased tomatoes in Cuba. J. Phytopath. 144:277-279. [Google Scholar]
- 20.Martinez Zubiaur, Y., D. Fonseca, M. Quiones, and I. Palenzuela. 2004. Presence of Tomato yellow leaf curl virus infecting squash (Curcubita pepo) in Cuba. Plant Dis. 88:572. [DOI] [PubMed] [Google Scholar]
- 21.McGlashan, D., J. E. Polston, and D. Bois. 1994. Tomato yellow leaf curl geminivirus in Jamaica. Plant Dis. 78:1219. [Google Scholar]
- 22.Momol, M. T., G. W. Simone, W. Dankers, R. K. Sprenkel, S. M. Olson, E. A. Momol, J. E. Polston, and E. Hiebert. 1999. First report of Tomato yellow leaf curl virus in tomato in south Georgia. Plant Dis. 83:487. [DOI] [PubMed] [Google Scholar]
- 23.Monci, F., S. Sánchez-Campos, J. Navas-Castillo, and E. Moriones. 2002. A natural recombinant between the geminiviruses Tomato yellow leaf curl Sardinia virus and Tomato yellow leaf curl virus exhibits a novel pathogenic phenotype and is becoming prevalent in Spanish populations. Virology 303:317-326. [DOI] [PubMed] [Google Scholar]
- 24.Morales, F. J. 2006. History and current distribution of begomoviruses in Latin America. Adv. Virus Res. 67:127-162. [DOI] [PubMed] [Google Scholar]
- 25.Moriones, E., and J. Navas-Castillo. 2000. Tomato yellow leaf curl virus, an emerging virus complex causing epidemics worldwide. Virus Res. 71:123-134. [DOI] [PubMed] [Google Scholar]
- 26.Nakhla, M. K., D. P. Maxwell, R. T. Martinez, M. G. Carvalho, and R. L. Gilbertson. 1994. Widespread occurrence of the eastern Mediterranean strain of Tomato yellow leaf curl geminivirus in tomatoes in the Dominican Republic. Plant Dis. 78:926. [Google Scholar]
- 27.Navas-Castillo, J., S. Sánchez-Campos, and J. A. Díaz. 1999. Tomato yellow leaf curl virus-Is causes a novel disease of common bean and severe epidemics in tomato in Spain. Plant Dis. 83:29-32. [DOI] [PubMed] [Google Scholar]
- 28.Navas-Castillo, J., S. Sánchez-Campos, E. Noris, D. Louro, G. P. Accotto, and E. Moriones. 2000. Natural recombination between Tomato yellow leaf curl virus-Is and Tomato leaf curl virus. J. Gen. Virol. 81:2797-2801. [DOI] [PubMed] [Google Scholar]
- 29.Navot, N., E. Pichersky, M. Zeidan, D. Zamir, and H. Czosnek. 1991. Tomato yellow leaf curl virus: a whitefly-transmitted geminivirus with a single genomic component. Virology 185:151-161. [DOI] [PubMed] [Google Scholar]
- 30.Padidam, M., R. N. Beachy, and C. M. Fauquet. 1995. Classification and identification of geminiviruses using sequence comparisons. J. Gen. Virol. 76:249-263. [DOI] [PubMed] [Google Scholar]
- 31.Padidam, M., S. Sawyer, and C. M. Fauquet. 1999. Possible emergence of new geminiviruses by frequent recombination. Virology 265:218-225. [DOI] [PubMed] [Google Scholar]
- 32.Picó, B., M. J. Díez, and F. Nuez. 1996. Viral diseases causing the greatest economic losses to the tomato crop. II. The Tomato yellow leaf curl virus—a review. Sci. Hortic. 67:151-196. [Google Scholar]
- 33.Polston, J. E., D. Bois, C.-A. Serra, and S. Concepcion. 1994. First report of a tomato yellow leaf curl-like geminivirus in the Western Hemisphere. Plant Dis. 78:831. [Google Scholar]
- 34.Polston, J. E., R. J. McGovern, and L. G. Brown. 1999. Introductions of Tomato yellow leaf curl virus in Florida and implications for the spread of this and other geminiviruses of tomato. Plant Dis. 83:984-988. [DOI] [PubMed] [Google Scholar]
- 35.Posada, D., and K. A. Crandall. 1998. MODELTEST: testing the model of DNA substitution. Bioinformatics 14:817-818. [DOI] [PubMed] [Google Scholar]
- 36.Quinones, M., D. Fonseca, Y. Martinez, and G. P. Accotto. 2001. First report of Tomato yellow leaf curl virus infecting pepper plants in Cuba. Plant Dis. 86:73. [DOI] [PubMed] [Google Scholar]
- 37.Ramos, P. L., O. Guerra, V. Dorestes, N. Ramirez, R. Rivera-Bustamante, and P. Orama. 1996. Detection of TYLCV in Cuba. Plant Dis. 80:1208. [Google Scholar]
- 38.Rojas, M. R., T. Kon, E. T. Natwick, J. E. Polston, F. Akad, and R. L. Gilbertson. 2007. First report of Tomato yellow leaf curl virus associated with tomato yellow leaf curl disease in California. Plant Dis. 91:1056. [DOI] [PubMed] [Google Scholar]
- 39.Salati, R., M. K. Nahkla, M. R. Rojas, P. Guzman, J. Jaquez, D. P. Maxwell, and R. L. Gilbertson. 2002. Tomato yellow leaf curl virus in the Dominican Republic: characterization of an infectious clone, virus monitoring in whiteflies and identification of reservoir hosts. Phytopathology 92:487-496. [DOI] [PubMed] [Google Scholar]
- 40.Shimodaira, H., and M. Hasegawa. 1999. Multiple comparisons of log-likelihoods with applications to phylogenetic inference. Mol. Biol. Evol. 16:1114-1116. [Google Scholar]
- 41.Swofford, D. L. 2003. PAUP*. Phylogenetic analysis using parsimony (*and other methods). Version 4.0b8. Sinauer Associates. Sunderland, MA.
- 42.Torres-Pacheco, I., J. A. Garzón-Tiznado, J. K. Brown, A. Becerra-Flora, and R. F. Rivera-Bustamante. 1996. Detection and distribution of geminiviruses in Mexico and the southern United States. Phytopathology 86:1186-1192. [Google Scholar]
- 43.Ueda, S., S. Takeuchi, M. Okabayashi, K. Hanada, K. Tomimura, and T. Iwanami. 2005. Evidence of a new Tomato yellow leaf curl virus in Japan and its detection using PCR. J. Gen. Plant Pathol. 71:319-325. [Google Scholar]
- 44.Urbino, C., and K. Tassius. 2003. First report of Tomato yellow leaf curl virus in tomato in Guadeloupe. Plant Dis. 87:1397. [DOI] [PubMed] [Google Scholar]
- 45.Valverde, R. A., P. Lotrakul, A. D. Landry, and J. E. Boudreaux. 2001. First report of Tomato yellow leaf curl virus in Louisiana. Plant Dis. 85:230. [DOI] [PubMed] [Google Scholar]
- 46.Wu, J. B., F. M. Dai, and X. P. Zhou. 2006. First report of Tomato yellow leaf curl virus in China. Plant Dis. 90:1359. [DOI] [PubMed] [Google Scholar]
- 47.Yang, Y., T. A. Sherwood, C. P. Patte, E. Hiebert, and J. E. Polston. 2004. Use of Tomato yellow leaf curl virus (TYLCV) Rep gene sequences to engineer TYLCV resistance in tomato. Phytopathology 94:490-496. [DOI] [PubMed] [Google Scholar]
- 48.Ying, Z., and M. J. Davis. 2000. Partial characterization and host range of Tomato yellow leaf curl virus in south Florida. Annu. Meet. Fla. State Hortic. Soc. 113:185-190. [Google Scholar]