Abstract
Bile salts are natural detergents that facilitate the digestion and absorption of the hydrophobic components of the diet. However, their amphiphilic nature makes them very inhibitory for bacteria and strongly influences bacterial survival in the gastrointestinal tract. Adaptation to and tolerance of bile stress is therefore crucial for the persistence of bacteria in the human colonic niche. Bifidobacterium animalis subsp. lactis, a probiotic bacterium with documented health benefits, is applied largely in fermented dairy products. In this study, the effect of bile salts on proteomes of B. animalis subsp. lactis IPLA 4549 and its bile-resistant derivative B. animalis subsp. lactis 4549dOx was analyzed, leading to the identification of proteins which may represent the targets of bile salt response and adaptation in B. animalis subsp. lactis. The comparison of the wild-type and the bile-resistant strain responses allowed us to hypothesize about the resistance mechanisms acquired by the derivative resistant strain and about the bile salt response in B. animalis subsp. lactis. In addition, significant differences in the levels of metabolic end products of the bifid shunt and in the redox status of the cells were also detected, which correlate with some differences observed between the proteomes. These results indicate that adaptation and response to bile in B. animalis subsp. lactis involve several physiological mechanisms that are jointly dedicated to reduce the deleterious impact of bile on the cell's physiology.
Probiotics are live microorganisms which, when administered in adequate amounts, confer a health benefit to the host (3). It is widely believed that a high survival rate through the gastrointestinal tract is required in order for these bacteria to exert their beneficial effects (10, 26). One of the widest uses of probiotics is to include them in dairy products which have been historically consumed by humans. In this context, Bifidobacterium animalis subsp. lactis is a gram-positive bacterium which has been used largely in fermented milks as adjunct cultures (31). Well documented, clinically established effects of this species are the treatment of diarrhea, the balancing of the intestinal microbiota, and the alleviation of atopic eczema in infants (45).
After ingestion, bifidobacteria meet several biological barriers, with the gastric acidity and the high bile salt concentrations in the intestine being the most important ones. Bile is composed mainly of bile salts, which are detergent-like biological compounds synthesized in the liver from cholesterol and stored as amino acid conjugates in the gall bladder. During digestion, bile is secreted into the intestine, where it plays a major role in the emulsification and absorption of fats. In addition, bile displays a strong antimicrobial activity by inducing membrane damage and by causing oxidative stress to the DNA (6). Therefore, tolerance to physiological bile salt concentrations is crucial in order for intestinal bacteria to colonize the gut.
Even though a great deal of effort has been dedicated in recent years to characterize probiotics, very little is known about the mechanisms of response and tolerance to bile in lactic acid bacteria and bifidobacteria (4). Several membrane proteins are thought to be involved in bile resistance mechanisms in Lactococcus (56), Lactobacillus (16, 17, 18), and Bifidobacterium (40), by mediating the efflux of the bile salts out of the cells. Some researchers have suggested a possible involvement of bile salt hydrolases, enzymes responsible for bile salt deconjugation, in a moderate increase of bile resistance in many intestinal bacteria brought about by decreasing the toxicity of conjugated bile salts (14, 22). In a previous study with Bifidobacterium longum, we showed that growth in the presence of bile salts affects mainly the expression of enzymes involved in the so-called bifid shunt (47). We also showed that the acquired bile salt resistance in B. animalis subsp. lactis widely affects the metabolism in strain 4549dOx (34, 35, 43, 46). In the present study, we used B. animalis subsp. lactis IPLA 4549 and its derivative 4549dOx, with acquired resistance to bile, to unravel the molecular mechanisms leading to adaptation and resistance to bile in this species. We compared the proteomes of both strains in the presence and in the absence of bile salts. This allowed us to draw conclusions about the specific response of B. animalis subsp. lactis to bile and about the mechanisms involved in the acquired bile salt resistance in the resistant mutant derivative.
MATERIALS AND METHODS
Strains and growth conditions.
The bacterial strains used in this study were B. animalis subsp. lactis IPLA 4549 and its oxgall-resistant derivative B. animalis subsp. lactis 4549dOx (35). The mutant has a stable resistance phenotype and probably harbors several mutations, since it displays remarkable physiological changes compared to the wild-type (WT) parent strain (34, 35, 43, 46). Strains were cultured on MRS agar (BD Diagnostic Systems, Sparks, MD) supplemented with 0.05% l-cysteine (wt/vol) (Merck KGaA, Darmstadt, Germany) (MRSC). Single colonies were picked and cultured in liquid MRSC medium in a chamber (Bactron anaerobic/environmental chamber; Sheldon Manufacturing Inc., Cornelius, OR) with an anaerobic atmosphere (5% CO2-5% H2-90% N2) at 37°C. Volumes of 250 ml of fresh MRSC were inoculated at 1% (vol/vol) with fresh overnight cultures and incubated anaerobically until the optical density at 600 nm (OD600) reached 0.6 (mid-log phase for all conditions). The time necessary to reach this OD varied between 6 and 8 h depending on the strain and the bile concentration tested. When necessary, bile salts (oxgall; Oxoid Limited, Hampshire, United Kingdom) were added to final concentrations of 1.2, 3.0, or 10.0 g liter−1. Oxgall shows higher bile salt and phospholipid concentrations and lower phospholipid/bile salt molar ratios than human bile (11), whereas the ratio of the glycine- to taurine-conjugated bile acids is similar (23). In spite of this and due to the practical difficulty in having human bile available, this study was performed with oxgall. Experiments were performed at least in triplicate.
DNA techniques.
Chromosomal DNA of B. animalis was isolated using a GenElute bacterial genomic DNA kit (Sigma-Aldrich, St. Louis, MO). For amplification of the region encompassing the gene coding for the putative protein BA46 and its 600 bp upstream region, we used the forward primer S54F (5′-GTGAAGGCATGGATGGCCATCCATC), the reverse primer S54R (5′-ACGGTGTTTGCAACAACACTCCTAC), and the two internal primers S54F1 (5′-AGGCGCCTTGTGACGAAACTGGTG) and S54R1 (5′-CGTGAGCTTGTCGAGCGCCAGATC). PCR fragments were purified with the GenElute PCR clean-up kit (Sigma) and sequenced in the Servicio de Secuenciación de ADN (SSAD, CIB, Cantoblanco, Madrid).
Two-dimensional gel electrophoresis, statistical analysis, and protein identification.
Preparation of cell extracts and two-dimensional (2D) gel electrophoresis were performed basically as we previously described for B. longum (47). Significant differences in protein expression were established with at least three independent gels per condition using an ImageScanner (Amersham Biosciences, Buckinghamshire, United Kingdom) and the ImageMaster 2D Platinum software (version 5.0; Amersham Biosciences and Geneva Bioinformatics S.A.). Spots were detected and defined by the software by means of an algorithm that takes into account several parameters related to the shape and surroundings of the spots. Volumes of each spot were normalized (percent by volume) by referring values to the sum of total spot volumes within each gel. Gels were matched automatically, and groups of spots (spots which share similar relative positions in gels) were generated. We considered a protein to be under- or overproduced when, after image analysis and subsequent computing of the normalized spot volumes, the means from at least four gels coming from independent cultures were 1.4-fold different among the conditions tested at a significance level of P < 0.05 (Student's t test for paired samples).
Selected spots were excised from gels and subjected to peptide mass fingerprinting by matrix-assisted laser desorption ionization-time-of-flight mass spectrometry (MS) analysis. MS-Fit (University of California at San Francisco Mass Spectrometry Facility; http://prospector.ucsf.edu) and Mascot (Matrix Science Inc., Boston, MA), installed locally, were used to identify proteins from peptide mass fingerprints, and annotations were made according to the cluster of orthologous genes (COG) functional groups. For Mascot analysis, the B. animalis BB-12 theoretical peptide database, constructed at Chr. Hansen from the hypothetical protein database of this bacterium, was used (20). Putative functions were assigned if the first entries returned in both search engines were the same, if it returned a high molecular weight search score (37), and if there was an agreement (±10%) on the observed and theoretical Mrs and pIs of the protein.
Estimation of redox balance by fluorescence spectroscopy.
The fluorescence properties of buffered cell suspensions (OD600, 0.6) in 50 mM Tris-HCl buffer, pH 7.0, were monitored in an Eclipse fluorescence spectophotometer (Varian, Inc., Palo Alto, CA). The intensity values corresponding to NADH were calculated from the 413-nm emission at a λex of 316 nm, whereas for flavin adenine dinucleotide (FAD) the intensity values were calculated from the 436-nm emission at a λex of 380 nm, as described by Ammor et al. (2). The redox ratio was deduced from the NADH- and FAD-related signals using the equation redox ratio = FAD intensity/(FAD intensity + NADH intensity) (27).
Growth in medium containing glucose or sodium oxalate and preparation and incubation of resting cells.
Ten milliliters of B. animalis subsp. lactis IPLA 4549 and B. animalis subsp. lactis 4549dOx cultures, previously grown in MRSC containing different concentrations of oxgall up to mid-log phase, were collected by centrifugation (10,000 × g for 15 min), washed twice with 50 mM Tris-HCl buffer (pH 7.0), and resuspended in 10 ml of 50 mM Tris-HCl buffer (pH 5.0) containing glucose (25 mM). The resting cell suspensions were incubated with constant gentle stirring for 4 h at 37°C. Additionally, MRSC containing 7 mM sodium oxalate was inoculated with Bifidobacterium cultures and incubated for 5 days.
Determination of organic acids and glucose by HPLC and of ethanol by dynamic headspace extraction and gas chromatography-MS analysis.
Cells were removed from cultures by centrifugation, and suspensions of resting cells and the supernatants were analyzed by high-performance liquid chromatography (HPLC). Determination of the amounts of glucose and oxalate consumed and acetic, lactic, and formic acids produced was carried out essentially as described by Ruas-Madiedo et al. (43). Briefly, a chromatographic system composed of an Alliance 2690 module injector, a PDA 996 photodiode array detector, a 410 differential refractometer detector, and Millenium 32 software (Waters Corporation, Milford, CA) was used. Samples (50 μl) were separated in isocratic conditions using a HPX-87H Aminex ion-exchange column (Hewlett Packard, Palo Alto, CA) with diluted sulfuric acid as the mobile phase. Standard solutions of organic acids and glucose were used as controls for both identification and quantification. The results presented are the means from at least three separate experiments. The acetic/lactic acid ratio was calculated, as well as the total carbon balance of the bifid shunt expressed as moles carbon formed/moles glucose consumed using the following formula: 2 × [acetic acid] + 3 × [lactic acid] + 2 × [ethanol] + 1 × [formic acid]/6 × [glucose].
A dynamic headspace extraction and gas chromatography-MS analysis was used to determine ethanol in supernatants of resting cells as previously described by Ruas-Madiedo et al. (43). A system composed of a 3100 Purge and Trap Concentrator (Tekmar and Dohrmann, Cincinnati, OH) fitted with a Vocarb 3000 trap (Supelco/Sigma-Aldrich Química S.A., Madrid, Spain) was used. The concentrator was coupled to a gas chromatograph (GC 6890N; Agilent Technologies Inc., Palo Alto, CA) connected to a mass spectrometer detector (MS 5973N; Agilent). Data were collected with the enhanced ChemStation G1701DA software (Agilent). Briefly, supernatants of resting cells were mixed with an internal standard solution of propyl acetate (Chem Service Inc., West Chester, PA), placed in a needle sparger tube, and purged with helium. The volatile compounds were trapped by absorption to the trap and were desorbed from it by heating directly into the injection port. Separation was carried out in a HP-Innowax capillary column (Agilent) with several temperature ramps. Ethanol was identified by comparison of its mass spectrum with that in the HP-Wiley 138 library (Agilent) and by comparison of its retention time with that of an ethanol standard (Sigma). The peak of ethanol was quantified as the relative abundance with respect to the internal standard. The concentration (mM) of ethanol was calculated using the linear regression equation (r2 ≥ 0.99) from the standard curve obtained with at least three replicate measurements.
Data on the consumption of glucose and the formation of organic acids and ethanol by resting cells were subjected to one-way analysis of variance with the SPSS 11.0 software (SPSS Inc., Chicago, IL) using the factor “concentration of bile salts” with two categories: 0 and 3 or 0 and 10 g liter−1 for the WT and mutant, respectively.
RESULTS AND DISCUSSION
Determination of the proteins involved in bile adaptation in B. animalis and in the bile resistance phenotype of the mutant.
To unravel the molecular mechanisms leading to bile adaptation, high-throughput molecular techniques are being widely used as a first approach to understand the physiological changes involved in this process (7, 8, 29, 30, 57). Here, we used proteomics to detect target proteins affected by growth in the presence of bile in B. animalis subsp lactis. Moreover, in order to gain insight into both the bile salt response and resistance mechanisms, we used a previously isolated and partially characterized strain which had acquired the capacity to resist high concentrations of bile. We compared the proteomes of the WT strain with and without bile in order to obtain the general response to bile. We also compared the proteomes of the WT strain and its derivative in the absence of bile to detect constitutive changes occurring in the mutant. Finally, we compared the responses to bile in both strains in order to discriminate the proteins affected by bile specifically in the mutant.
Since the mutant 4549dOx was more tolerant to bile than the WT IPLA4549 (46), we selected 3 g liter−1 as the highest concentration tested for IPLA 4549 and 10 g liter−1 for 4549dOx. The growth rate of IPLA4549 at 3 g liter−1 was similar to that of 4549dOx at 10 g liter−1 (data not shown). These two different bile concentrations should thus reflect the effect of bile under a similar cell status.
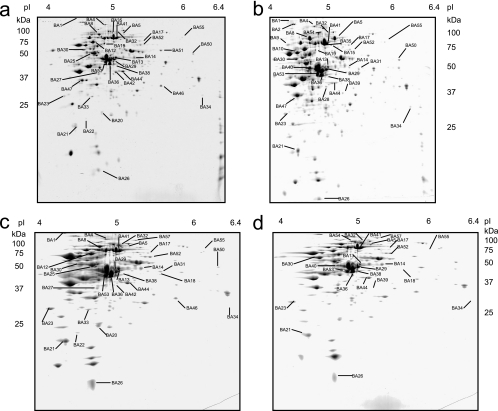
Figure 1 shows 2D gels of protein extracts of IPLA 4549 and 4549dOx grown either in the absence or in the presence of bile salts. In total 45 spots were detected, the amount of which was different in at least one of the conditions tested (Table 1). These results are consistent with a previous transcriptomic study which showed that the probiotic strain B. animalis subsp. lactis BB-12, when grown in the presence of bile, displays up-regulation of 86 genes and down-regulation of 123 (20). They also indicate the capacity of bile compounds to massively modulate protein expression in this species.
FIG. 1.
Proteomes of the B. animalis WT and bile salt-resistant derivative. 2D gels of cytosolic extracts from mid-exponential-phase cells of B. animalis IPLA 4549 (a and c) and B. animalis 4549dOx (b and d) grown without (a and b) and with (c and d) bile salts are shown. Spots identified by peptide mass fingerprinting are labeled, and the identifications of the 45 spots affected by bile salts are listed in Table 1.
TABLE 1.
Proteins affected by bile salt exposure in B. animalis and proteins with altered expression in the bile-resistant B. animalis 4549dOx relative to the WT strain, B. animalis IPLA 4549
| COG functional group | Spot no.a | Putative functionb | Coverage (%)c | Normalized vold without oxgall
|
Variation factor, mutant/WTe (fold) | Normalized vold with oxgall
|
Variation factorf by oxgall
|
|||
|---|---|---|---|---|---|---|---|---|---|---|
| IPLA 4549 | 4549dOx | IPLA 4549 | 4549dOx | 3 g liter−1 in WT | 10 g liter−1 in mutant | |||||
| Carbohydrate transport and metabolism | BA31 | Xylulose-5-phosphate/fructose-6-phosphate phosphoketolase | 30 | — | 0.209 ± 0.065 | 0.321 ± 0.090 | ++ | |||
| BA41 | Xylulose-5-phosphate/fructose-6-phosphate phosphoketolase | 45 | 0.944 ± 0.101 | 0.823 ± 0.098 | 2.143 ± 0.230 | 1.308 ± 0.192 | 2.27 | 1.59 | ||
| BA35 | Xylulose-5-phosphate/fructose-6-phosphate phosphoketolase | 54 | 1.204 ± 0.272 | 2.402 ± 1.023 | 1.99 | |||||
| BA50 | Glucose-6-phosphate 1-dehydrogenase | 66 | 0.141 ± 0.024 | 0.297 ± 0.078 | 2.11 | 0.319 ± 0.076 | 2.26 | |||
| BA36 | Glyceraldehyde-3-phosphate dehydrogenase/erythrose-4-phosphate dehydrogenase | 70 | 1.632 ± 0.411 | 2.752 ± 0.121 | 1.68 | |||||
| BA38 | Glyceraldehyde-3-phosphate dehydrogenase/erythrose-4-phosphate dehydrogenase | 69 | 0.321 ± 0.052 | 0.652 ± 0.154 | 2.03 | 0.078 ± 0.011 | 0.288 ± 0.083 | −4.12 | −2.26 | |
| BA53 | Glyceraldehyde-3-phosphate dehydrogenase/erythrose-4-phosphate dehydrogenase | 69 | 0.856 ± 0.136 | 0.425 ± 0.040 | −2.02 | 2.714 ± 0.433 | 0.591 ± 0.126 | 3.17 | 1.39 | |
| BA10 | α-Glucosidase | 53 | 0.339 ± 0.141 | 0.675 ± 0.030 | 1.99 | |||||
| BA15 | α-Galactosidase | 46 | 0.147 ± 0.018 | 0.245 ± 0.069 | 1.66 | |||||
| BA14 | 6-Phosphogluconate dehydrogenase | 70 | 0.100 ± 0.072 | 0.089 ± 0.053 | 0.215 ± 0.068 | 0.486 ± 0.074 | 2.14 | 5.46 | ||
| BA18 | Sugar transport ATP binding protein | 51 | — | — | 0.213 ± 0.056 | — | ++ | |||
| BA25 | Glucose-6-phosphate isomerase | 62 | 0.256 ± 0.093 | 0.241 ± 0.061 | 0.686 ± 0.121 | 2.68 | ||||
| BA28 | UDP-glucose 4-epimerase | 40 | 0.260 ± 0.045 | 0.651 ± 0.229 | 2.50 | |||||
| Energy production and conversion | BA29 | Formyl-CoA transferase | 67 | 0.474 ± 0.056 | 0.850 ± 0.216 | 1.79 | 0.877 ± 0.203 | 1.649 ± 0.396 | 1.85 | 1.94 |
| BA13 | Formyl-CoA transferase | 74 | 0.213 ± 0.054 | 0.268 ± 0.048 | 0.614 ± 0.153 | 0.630 ± 0.177 | 2.88 | 2.35 | ||
| BA32 | Formate acetyltransferase | 64 | 0.177 ± 0.017 | 0.184 ± 0.063 | 0.301 ± 0.155 | 0.730 ± 0.381 | 1.7 | 3.97 | ||
| Amino acid transport and metabolism | BA5 | Methionine synthase | 64 | 0.104 ± 0.011 | 1.616 ± 0.230 | 15.54 | 0.033 ± 0.009 | 0.619 ± 0.136 | −3.11 | −2.61 |
| BA40 | O-Acetylhomoserine sulfhydrolase | 77 | 0.312 ± 0.093 | 2.441 ± 0.505 | 7.82 | 0.765 ± 0.285 | −3.19 | |||
| Nucleotide transport and metabolism | BA52 | CTP synthase | 50 | 0.210 ± 0.076 | 0.380 ± 0.136 | 1.81 | 0.670 ± 0.240 | 1.056 ± 0.153 | 3.19 | 2.78 |
| BA20 | Adenylate kinase | 66 | 0.222 ± 0.049 | 0.204 ± 0.117 | 0.827 ± 0.151 | 3.73 | ||||
| BA54 | Polyribonucleotide nucleotidyltransferase | 54 | 0.219 ± 0.034 | 0.211 ± 0.123 | 0.553 ± 0.160 | 2.62 | ||||
| BA4 | Carbamoylphosphate synthase large subunit | 48 | 0.050 ± 0.011 | 0.133 ± 0.070 | 2.66 | 0.102 ± 0.014 | 2.05 | |||
| BA17 | Ribonucleotide reductase, α subunit | 13 | — | 0.041 ± 0.033 | — | 0.014 ± 0.004 | −2.93 | |||
| BA42 | Inosine-uridine nucleoside N-Ribohydrolase | 43 | 0.495 ± 0.091 | 0.105 ± 0.008 | −4.72 | |||||
| Translation, ribosomal structure, and biogenesis | BA19 | Aspartyl tRNA synthetase | 58 | 0.507 ± 0.071 | 0.139 ± 0.062 | −3.65 | ||||
| BA34 | RNase PH | 47 | 0.070 ± 0.018 | 0.075 ± 0.022 | 0.012 ± 0.003 | 0.014 ± 0.005 | −5.9 | −5.33 | ||
| BA44 | 30S ribosomal protein S2 | 84 | 0.935 ± 0.084 | 0.127 ± 0.052 | −7.36 | 2.412 ± 0.336 | 0.325 ± 0.073 | 2.58 | 2.56 | |
| BA33 | Peptide deformylase | 49 | 0.162 ± 0.071 | 0.156 ± 0.065 | 0.365 ± 0.104 | 2.25 | ||||
| BA46 | Hypothetical protein in sigma 54 modulation protein/S30ea ribosomal protein family | 59 | 0.595 ± 0.092 | 0.048 ± 0.120 | −12.40 | 0.229 ± 0.053 | −2.6 | |||
| Transcription | BA57 | DNA-directed RNA polymerase beta prime chain | 60 | 0.119 ± 0.010 | 0.121 ± 0.032 | 0.345 ± 0.094 | 0.422 ± 0.082 | 2.9 | 3.49 | |
| Coenzyme metabolism | BA39 | 2-Dehydropantoate 2-reductase | 60 | — | 0.199 ± 0.065 | — | 0.036 ± 0.007 | −5.57 | ||
| BA27 | Ketol-acid reductoisomerase 1 | 55 | 0.279 ± 0.077 | 0.571 ± 0.193 | 2.05 | 0.661 ± 0.140 | 2.37 | |||
| Cell division and chromosome partitioning | BA9 | COG 3599, cell division initiation protein | 48 | 0.096 ± 0.052 | 0.287 ± 0.084 | 2.99 | ||||
| BA2 | COG 1196, chromosome segregation ATPases | 73 | 0.168 ± 0.128 | 0.298 ± 0.115 | 1.77 | |||||
| Lipid metabolism | BA55 | Long-chain-fatty-acid CoA ligase | 19 | 0.144 ± 0.042 | 0.026 ± 0.003 | −5.54 | 0.011 ± 0.005 | 0.006 ± 0.001 | −12.91 | −4.5 |
| Posttranslational modification, protein turnover, chaperones | BA8 | Heat shock protein ClpB | 58 | 0.898 ± 0.100 | 1.625 ± 0.386 | 1.81 | 2.407 ± 0.435 | 2.68 | ||
| BA1 | Trypsin-like serine protease | 23 | — | 0.021 ± 0.012 | 0.026 ± 0.012 | ++ | ||||
| BA22 | Thioredoxin-dependent thiol peroxidase | 81 | 0.017 ± 0.004 | 0.014 ± 0.006 | 0.067 ± 0.013 | 3.95 | ||||
| BA23 | GrpE | 62 | 0.114 ± 0.078 | 0.128 ± 0.051 | 0.529 ± 0.239 | 0.245 ± 0.065 | 4.64 | 1.92 | ||
| BA26 | Chaperone protein GroES | 77 | 0.283 ± 0.106 | 0.291 ± 0.123 | 0.738 ± 0.208 | 0.556 ± 0.079 | 2.61 | 1.91 | ||
| BA30 | Chaperone protein GroEL | 76 | 0.330 ± 0.114 | 0.357 ± 0.201 | 0.944 ± 0.114 | 0.700 ± 0.157 | 2.86 | 1.96 | ||
| BA12 | Chaperone protein DnaK | 65 | 0.661 ± 0.153 | 0.650 ± 0.186 | 1.295 ± 0.231 | 1.96 | ||||
| DNA replication, recombination, and repair | BA21 | DNA protection during starvation protein | 60 | 0.561 ± 0.031 | 1.140 ± 0.118 | 2.03 | 1.273 ± 0.145 | 2.27 | ||
| Cell envelope biogenesis, outer membrane | BA47 | Bile salt hydrolase | 60 | 0.962 ± 0.361 | 2.438 ± 0.632 | 2.53 | ||||
| Function unknown | BA51 | Hypothetical protein BL0016 | 54 | 0.423 ± 0.016 | — | |||||
Spot numbers refer to the proteins labeled in Fig. 1.
Putative functions were established according to the theoretical proteome of B. animalis subsp. lactis BB-12.
Percentage of amino acid coverage (peptides observed/theoretical value from sequence data).
Normalized relative volumes (percent volumes) for matching spots between wild-type and mutant strains. Only statistically significant results are presented. Values are means ± standard deviations; n ≥ 3 for each strain. —, not detected.
r = percent volume in the mutant/percent volume in the WT. When r > 1, variation factor = r. When r < 1, variation factor = −1/r. Only statistically significant results are presented.
r = percent volume with bile salt/percent volume without bile salt. When r > 1, variation factor = r. When r < 1, variation factor = −1/r. ++, spot present only after bile salt exposure.
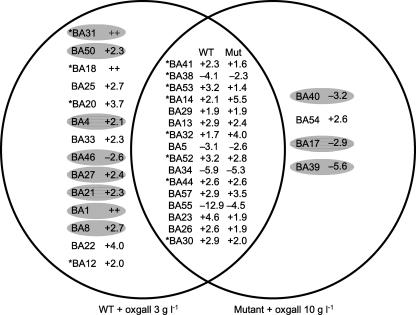
Among the 45 spots mentioned above, 34 were affected by oxgall in either the mutant, the WT, or both strains. Sixteen spots were affected by oxgall in both the WT and the mutant, 14 spots appeared to be affected by bile specifically in the WT IPLA 4549, and 4 were affected specifically in the derivative (Fig. 2). Since these last four proteins (BA40, BA54, BA17, and BA39) are affected only in the resistant mutant, they can be considered to belong to the functions responsible for its resistant phenotype. The 16 spots affected by bile in both strains could represent the global response to the presence of oxgall in B. animalis. This is strongly supported by previous results that we obtained with other Bifidobacterium species. Indeed, eight (BA41, BA38, BA53, BA14, BA32, BA52, BA44, and BA30) out of these 16 proteins have already been shown to be affected in a similar way by the presence of oxgall in B. longum (47). Among the 14 proteins affected by the presence of oxgall only in the WT, there could be either proteins that are not directly linked to the oxgall resistance in the mutant or proteins that are involved in the resistance but constitutively expressed in the mutant and thus escape the oxgall effect. Indeed, seven such proteins (BA31, BA50, BA4, BA27, BA21, BA1, and BA8) were up-regulated by oxgall in the WT and constitutively overexpressed in the resistant mutant whether oxgall was added or not. In contrast, spot BA46 was down-regulated by oxgall in the WT and constitutively underexpressed in the mutant. We thus considered that these 8 spots, in addition to the 16 spots whose expression varied depending on oxgall addition in the two strains, represent the bile salt response. The remaining spots that are bile induced in the WT but not in the mutant (BA18, BA25, BA20, BA33, BA22, and BA12) were considered not to be directly involved in the bile salt response in B. animalis but rather to be secondary targets. Furthermore, the comparison between the WT and the derivative in the absence of oxgall showed that 11 spots (BA35, BA36, BA10, BA28, BA42, BA19, BA9, BA2, BA15, BA47, and BA51) varied and were not sensitive to the addition of oxgall. These could thus be considered to not be directly involved in bile salt adaptation but could be partially responsible for the stable bile salt resistance phenotype of the mutant.
FIG. 2.
Proteins involved in the bile salt response of B. animalis. Left circle, proteins induced or repressed by oxgall in the WT. Right circle, proteins induced or repressed by oxgall only in the mutant. Intersection, proteins repressed or induced by oxgall in both strains. Shading indicates proteins whose expression is also constitutively modified in the mutant in the absence of oxgall. Asterisks indicate proteins whose synthesis is also affected by bile in B. longum (47). ++ indicates proteins that were detected only in the presence of bile salts.
Proteins responsible for the bile salt adaptation of B. animalis are involved in carbohydrate metabolism.
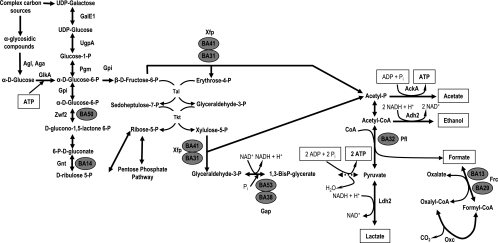
As mentioned above, we consider that the general response to bile in B. animalis is due to the proteins whose synthesis is affected by bile in both strains together with proteins affected by bile in the WT only and which are constitutively affected in the mutant (Fig. 2). Among those, BA31, BA41, BA38, BA53, BA14, BA32, BA29, BA13, and BA50 are involved in carbohydrate metabolism (Fig. 3). Some of them have already been shown to be implicated in the bile salt response in B. longum (47).
FIG. 3.
Schematic representation of the carbon catabolic pathway, the bifid shunt, and oxalic acid degradation. Enzymes that are involved in the general bile salt response in B. animalis are marked with gray circles. Abbreviations: AckA, acetate kinase; Adh2, aldehyde-alcohol dehydrogenase 2; Aga, α-galactosidase; Agl, α-glucosidase; Frc, formyl-CoA transferase; GalE1, UDP-glucose 4-epimerase; Gap, glyceraldehyde-3-phosphate dehydrogenase C; GlkA, glucokinase; Gnt, 6-phosphogluconate dehydrogenase; Gpi, glucose 6-phosphate isomerase; Ldh2, lactate dehydrogenase; Oxc, oxalyl-CoA decarboxylase; Pgm, phosphoglucomutase; Pfl, formate acetyltransferase; Tal, transaldolase; Tkt, transketolase; UgpA, UTP-glucose-1-phosphate uridylyltransferase; Xfp, xylulose-5-phosphate/fructose-6-phosphate phosphoketolase; Zwf2, glucose-6-phosphate 1-dehydrogenase; Pi, phosphate.
Spots BA31 and BA41 were identified as xylulose-5-phosphate/fructose-6-phosphate phosphoketolase (Xfp), the key enzyme of the bifid shunt (32). BA38 and BA53 were also identified as the same protein: glyceraldehyde 3-phosphate dehydrogenase (Gap), which is responsible for the interconversion of glyceraldehyde-3-phosphate and glyceraldehyde-1,3-bisphosphate, a major controlling point of carbon flux in Lactococcus lactis (55). This suggests the presence of different Gap isoforms, as already shown in Corynebacterium glutamicum (36) and L. lactis (53). Gap and Xfp were the sole enzymes of the bifid shunt whose expression changed in response to bile salts (Fig. 3). In addition, BA13 and BA29, which corresponded to the formyl coenzyme A (formyl-CoA) transferase, which catalyzes the reaction formyl-CoA + oxalate = formate + oxalyl-CoA, were also overproduced in the presence of bile. Another enzyme of carbohydrate catabolism, formate acetyltransferase (or pyruvate formate-lyase), which is responsible for the transfer of an acetyl group from acetyl-CoA to formate, leading to CoA and pyruvate, was also induced by oxgall (spot BA32).
Finally, BA50 and BA14, which are also involved in carbohydrate metabolism, were detected as overexpressed by oxgall. BA50 was identified as glucose-6-phosphate 1-dehydrogenase, an enzyme acting at the early stages of the pentose phosphate pathway by catalyzing the formation of d-glucono-1,5-lactone 6-phosphate from d-glucose 6-phosphate. Spot BA14 was identified as 6-phosphogluconate dehydrogenase. d-Glucono-1,5-lactone 6-phosphate can be oxidized to 6-phospho-d-gluconate by 6-phosphogluconolactonase and subsequently transformed into d-ribulose 5-phosphate by 6-phosphogluconate dehydrogenase. This pentose phosphate can be directly transformed into d-xylulose 5-phosphate by epimerization or can be isomerized to d-ribose 5-phosphate and then transformed by the transketolase.
Taken together, these findings suggest that, as we showed in B. longum (47), bile challenge leads to a modification of carbon metabolism. However, the enzymes affected by bile are different in the two species, suggesting a different effect of bile on sugar metabolism. Indeed, in B. animalis our results suggest an additional impact of bile salts on formate/oxalate metabolism. As shown in Fig. 3, this may lead to a possible enhancement of the degradation of oxalic acid with the subsequent formation of formic acid under bile challenge. To test this hypothesis, several metabolites were measured.
HPLC analysis showed that when resting cells of both IPLA 4549 and 4549dOx were incubated in a glucose-containing buffer, glucose consumption as well as lactic and acetic acid formation decreased when bile was added (Table 2). In contrast, the amount of formic acid produced increased. Moreover, the acetic acid/lactic acid ratio increased, indicating that in the presence of bile the bifid shunt is displaced towards the acetic branch, which could allow B. animalis to obtain one extra molecule of ATP (Fig. 3). On the other hand, the total carbon balance of the bifid shunt decreased in the presence of bile, and this reduction was less pronounced in 4549dOx. This suggests that under bile challenging conditions, B. animalis 4549dOx may maintain its normal physiological metabolism better than IPLA 4549. In this respect, we recently demonstrated that the ability to survive acidic pH and the intracellular ATP content were significantly higher in 4549dOx than in IPLA 4549 (46). In addition, HPLC analysis showed a higher production of formic acid when B. animalis IPLA 4549 and B. animalis 4549dOx were grown in the presence of bile (Table 2).
TABLE 2.
Glucose consumed, formation of end metabolic products of the bifid shunt, and acetic acid/lactic acid ratios and carbon balances of buffered resting cells of B. animalis IPLA 4549 and its bile-resistant derivative B. animalis 4549dOx grown in the absence or presence of different bile salt concentrations
| Strain | Bile salt concn (g liter−1) | Mean ± SDa
|
||||||
|---|---|---|---|---|---|---|---|---|
| Glucose consumed (mM) | Organic acid formation (mM)
|
Ethanol formation (mM) | Acetic acid/lactic acid ratio | Carbon balance | ||||
| Lactic acid | Formic acid | Acetic acid | ||||||
| IPLA 4549 | 0 | 2.716 ± 0.379 | 1.506 ± 0.096 | 0.000 ± 0.000 | 3.829 ± 0.108 | 0.011 ± 0.002 | 2.500 ± 0.116 | 0.740 ± 0.080 |
| 3.0 | 0.848 ± 0.018 | 0.083 ± 0.021 | 0.128 ± 0.032 | 0.360 ± 0.093 | 0.020 ± 0.000 | 4.674 ± 0.510 | 0.268 ± 0.049 | |
| 4549dOx | 0 | 4.467 ± 0.221 | 2.461 ± 0.264 | 0.000 ± 0.000 | 6.594 ± 0.298 | 0.000 ± 0.000 | 2.621 ± 0.175 | 0.804 ± 0.080 |
| 10.0 | 1.355 ± 0.158 | 0.222 ± 0.038 | 0.692 ± 0.271 | 1.236 ± 0.180 | 0.007 ± 0.003 | 5.633 ± 0.207 | 0.469 ± 0.042 | |
Results are from at least three independent experiments. Means were all significantly different (P < 0.05).
It is remarkable that this increase in formate concentration is correlated with an overproduction of formyl-CoA transferase. This enzyme is one of the two coupled activities required for oxalate cleavage in Oxalobacter formigenes, an obligate anaerobe also colonizing the gastrointestinal tract, which employs oxalate breakdown to generate ATP (25). This metabolic pathway seems to be important in the human gastrointestinal tract, since the absence of O. formigenes has been related to kidney stone formation due to the high levels of oxalate in blood. As Fig. 3 shows, in B. animalis the formyl-CoA transferase could act together with the oxalyl-CoA decarboxylase, thus providing a way to detoxify oxalate, one of the strongest organic acids (pKa = 1.25).
Since previous studies reported that B. animalis was able to degrade oxalate when cultured in MRSC for 5 days (19), we determined the capacities of B. animalis IPLA 4549 and its bile-resistant derivative B. animalis 4549dOx to consume oxalate. Both strains were able to consume around 60% of the oxalate present in the medium after 5 days of incubation. In the absence of oxgall, this consumption was 0.77 ± 0.30 mM/OD600 unit and 0.91 ± 0.04 mM/OD600 unit for IPLA 4549 and 4549dOx, respectively. In the presence of oxgall and oxalate, the WT strain IPLA 4549 was not able to grow, and thus oxalate consumption could not be measured, whereas at 10 g liter−1 oxgall, IPLA 4549dOx increased its consumption of oxalate by around 50% (to 1.32 ± 0.46 mM/OD600).
Thus, changes affecting carbohydrate metabolism in B. animalis as a response to bile challenge seem to occur in three main ways: (i) activation of metabolic steps directed to the conversion of carbon sources into fructose-6-phosphate and d-xylulose 5-phosphate, the two sugars that feed the bifid shunt; (ii) an increase of the acetic acid/lactic ratio, focused to obtain extra ATP; and (iii) a shunt to formate instead of lactate/acetate production linked to enhanced oxalate consumption. Remarkably, the strategy of bile response in B. animalis is different from that in B. longum, in which we have found a general activation of the glycolytic pathway aimed at a faster conversion of glucose into lactic acid (47).
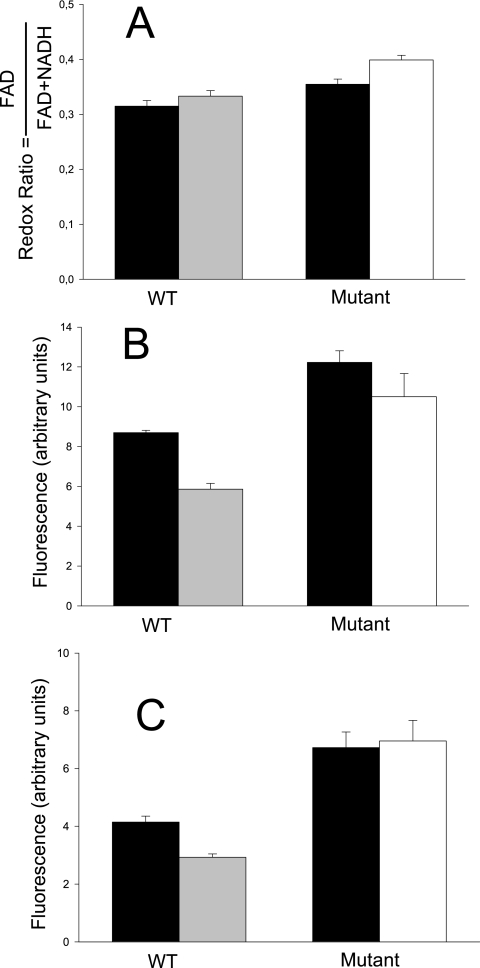
Bile affects the redox status of B. animalis cells.
As a consequence of the modification of carbon metabolism after bile challenge, one can expect multiple changes in the intracellular metabolites and thus an impact on the redox balance. In addition, several proteins involved in the redox status of the cells were involved in the bile salt response. This prompted us to determine fluctuations of intracellular pools of NADH and FAD+, whose fluorescence could be monitored under in vivo conditions (2). Changes in the redox ratio, calculated from FAD and NADH fluorescence-related signals (58), reflect changes in the balance between oxidized and reduced forms and hence fluctuation in the redox status of cells, which is dependent on the metabolic reactions occurring at a given time. As seen in Fig. 4, the redox ratio was affected when cells were grown in the presence of bile salts. However, this difference was observed only in the mutant. In the WT, the redox ratios were similar in the presence and absence of bile salts, as the relative amounts of both FAD and NADH were similarly lowered by bile.
FIG. 4.
Redox ratios (A), NADH-associated fluorescence (B), and FAD-associated fluorescence (C) of IPLA 4549 and 4549dOx grown in the absence of bile (solid bars) or in the presence of 3 g liter−1 (gray bars) or 10 g liter−1 (white bars) bile. Error bars represent standard deviations.
Oxidative stress is one of the major deleterious effects of bile on living cells. Prokaryotic and eukaryotic cells have bile-inducible genes whose products help the cell to cope with oxidative stress or DNA damage (6, 38) or are involved in redox reactions (7). Our data show that in B. animalis bile salts also lead to the overproduction of oxidative stress target proteins. It is known that oxidative stress causes the inhibition of methionine synthase in Escherichia coli (24). Methionine contains a sulfur group that can be oxidized and then reduced in the presence of thioredoxin, whose generation is dependent on the levels of NADH and NADPH (21, 49). In B. animalis, methionine synthase (BA5) is highly overproduced in the mutant (Table 1). Methionine synthase is responsible for the synthesis of l-methionine from l-homocysteine by transfer of a methyl group. Homocysteine is redox active and causes direct and indirect perturbation of redox homeostasis (58). The ketol-acid reductoisomerase corresponding to spot BA27 catalyzes the conversion of (R)-2,3-dihydroxy-3-methylbutanoate into (S)-2-hydroxy-2-methyl-3-oxobutanoate, with the formation of one molecule of NADPH. The overproduction of this enzyme under bile challenge could also contribute to modify the cell redox status. In addition, O-acetylhomoserine sulfhydrolase (BA40), catalyzing one of the last steps of the biosynthesis of methionine, was highly overproduced in 4549dOx. This enzyme, which is involved in sulfur metabolism, might thus have an indirect impact on the redox balance under bile challenge. Finally, BA21 was identified as a protein involved in DNA protection during starvation, which acts by protecting DNA from oxidative damage (1).
The general bile salt response in B. animalis has an impact on general stress response proteins.
We also observed general stress response mechanisms in the bile adaptation process in B. animalis. Five chaperones were found to be overproduced in the presence of bile, as previously found in other gram-positive bacteria (15, 29, 48): ClpB (BA8), a trypsin-like serine protease homolog to HtrA (BA1), GrpE (BA23), GroES (BA26), and GroEL (BA30). In a previous study, GroEL was shown to be overproduced in B. longum in the presence of bile salts (47), and GrpE and HtrA were also shown to be induced in the same species by heat shock (48). In addition, genes coding for GroES, GroEL, and ClpB were induced by osmotic shock, heat shock, or both in Bifidobacterium breve (50, 51, 52). HtrA-like proteins play a central role in protein secretion in L. lactis (39). In Bacillus subtilis two HtrA-like proteases, HtrA and HtrB, play a critical role in the cellular response against secretion and heat stress (13). This suggests that the patterns of protein secretion could be different in the presence of bile.
Some expression factors are also affected by the presence of bile.
Changes in three enzymes acting at the transcription/translation level (30S ribosomal protein S2 [BA44], DNA-directed RNA polymerase beta prime chain [BA57] [overproduced in both strains], and RNase PH [BA34] [underproduced in both strains]) suggest a modulation of protein synthesis as a response to bile. This is a common phenomenon reported for other stress factors in bacteria (41, 54).
Spot BA46 was underexpressed in the presence of bile in the WT, and its amount was dramatically, constitutively diminished in the derivative strain. It was identified as a hypothetical sigma 54 modulator. Sigma factors are bacterial transcription initiation factors that promote the attachment of the core RNA polymerase to specific initiation sites. They thus alter the specificity of promoter recognition. Sigma 54 is related to certain bacterial regulatory systems, particularly those associated with nitrogen metabolism (9). BA46 could thus be involved in bile salt adaptation, as a pleiotropic regulator. To determine if these changes in expression were related to mutations in the BA46 structural gene, its sequence and that of the 600-bp upstream region were determined in both strains. However, no mutation could be detected (accession number EF453721).
Regarding pyrimidine metabolism, the carbamoylphosphate synthase large subunit (BA4) and CTP synthase (BA52) were detected at higher levels in the presence of bile. This could be linked to the repair of oxidative DNA injuries caused by bile (6, 38).
Finally, long-chain-fatty-acid-CoA ligase (BA55) was found to be highly underproduced in the mutant, and in both strains in the presence of bile salts. This result is in agreement with previous transcriptomic studies with B. animalis subsp. lactis BB-12, indicating that several genes involved in the fatty acid biosynthesis were down-regulated in the presence of bile salts (20) and suggesting that bile salts could induce changes in the cytoplasmic membrane composition. Related to that, Kociubinski et al. (28) showed that growth of bifidobacteria in the presence of bile induced changes in lipid metabolism, which also affected the lipid composition of cell membranes. Our analysis of the cellular fatty acid composition also reflected drastic differences between B. animalis IPLA4549 and B. animalis 4549dOx grown in the absence of bile (44).
Proteins involved in the bile salt resistance phenotype of the 4549dOx derivative.
The above results give an overview of the global response of B. animalis during bile challenge. Among these oxgall target proteins, some are constitutively expressed in the mutant and escape the oxgall effect. Eleven additional spots were observed to vary between the WT and the mutant in the absence of bile and were thus not linked to the global response but may be responsible for the phenotype of the mutant. As well, four spots that respond to bile only in the mutant could also contribute to its bile-resistant phenotype. We thus considered that the mutant resistance phenotype was partially due to these 15 spots.
Several enzymes involved in sugar metabolism showed changes in amount when IPLA 4549 and 4549dOx cell extracts were compared (Fig. 1; Table 1). BA35 and BA36 were identified as isoforms of Xfp and Gap, respectively, two enzymes of the bifid shunt involved in the bile response. They thus should contribute to the bile resistance of the mutant. Interestingly, several spots corresponding to isoforms of these two proteins have been detected as varying in this study. However, no evidence of the activity of different isoforms is yet available. Additional MS/MS data could elucidate the nature of posttranslational modifications leading to these various isoforms. Overproduction of α-glucosidase (BA10) and α-galactosidase (BA15) occurred in 4549dOx, which releases α-d-glucose or α-d-galactose residues from 1,4-α-glucosidic or α-galactosidic compounds, respectively. In a previous work, we have shown that acquired bile salt resistance favored growth in the presence of maltose (43), which is in accordance with the higher production of α-glucosidase found in B. animalis 4549dOx. In addition, another enzyme was found to be overproduced in 4549dOx: UDP-glucose 4-epimerase (BA28), which catalyzes the interconversion of UDP-glucose to UDP-galactose. This protein is involved in fructose-6-phosphate formation from complex carbohydrates. From a physiological point of view, this could indicate that bile adaptation in the mutant triggers the preferential use of oligo- and polysaccharides, which are abundant in the colon, instead of glucose, promoting a metabolic adaptation to the efficient use of carbon sources in the B. animalis natural niche.
Several enzymes involved in nucleotide metabolism were identified as involved in the bile salt adaptation of the mutant: BA42, which is underproduced in 4549dOx, was identified as inosine-uridine nucleoside N-ribohydrolase, an enzyme that hydrolyzes ribosyl-purine compounds, releasing d-ribose and a purine, and BA17, identified as ribonucleoside-diphosphate reductase alpha chain, which was only found in 4549dOx.
Interestingly, BA47, identified as bile salt hydrolase, was found at higher levels in B. animalis 4549dOx. This result agrees with the higher levels of bile salt hydrolase activity of this strain described previously (34). Although suggested, a clear link between bile salt resistance and bile salt hydrolase activity could never be established (5, 12, 14, 22, 33). In our case, higher production of the enzyme and higher bile salt hydrolase activity (34) are related to the acquisition of bile salt tolerance in the mutant, which suggests a possible relationship between the resistance to bile salts and the presence of higher levels of bile salt hydrolase and deconjugating activity.
Two enzymes involved in the process of cell division, identified as cell division initiation protein (BA2) and the chromosome segregation ATPase (BA9), were also found to be overproduced in 4549dOx, which is consistent with previous studies indicating that bile strongly influences cell division and cell shape in bacteria (8, 42).
The last three proteins that may be involved in the mutant phenotype were BA51 (hypothetical protein), BA54 (polyribonucleotide nucleotidyltransferase), and BA19 (aspartyl-tRNA synthetase). For these no clear-cut conclusion could be drawn about their link to bile salt resistance.
Conclusion.
The proteomic and physiological data presented in this work reveal that bile salts induce a deep metabolic reorganization in B. animalis either during the process of acquisition of resistance or during growth in the presence of such compounds. The good correlation found between the changes detected after adaptation or exposure to bile in B. animalis suggests that both mechanisms go through five general lines in this species: (i) changes in the preferential branch through the glycolytic pathway and hence in the manner of energy production; (ii) changes associated with nitrogen metabolism, where a sigma factor could be involved; (iii) changes in fatty acid biosynthesis; (iv) an increase in the amount of molecular chaperones; and (v) changes in the redox balance of the cell. Remarkably, the effect of bile salts on the physiology of B. animalis indicates that in vitro screening of the potential probiotic properties of these bacteria could be dependent on the presence of these compounds, and bile could also affect the manner of interaction between bifidobacteria and the host. Our physiological proteomic approach establishes the basis for understanding bile adaptation in B. animalis, and constitutes a good starting point for further studies on metabolic processes modulated by bile in this probiotic bacterium.
Acknowledgments
This work was financed by European Union FEDER funds and the Spanish Plan Nacional de I + D (project AGL2004-06727-C02-01/ALI). B. Sánchez was the recipient of an FPI predoctoral fellowship from the Spanish Ministerio de Ciencia y Tecnología and from the MICA Department of the National Institute for Research in Agronomy (INRA). Matrix-assisted laser desorption ionization-time-of-flight/MS experiments were performed on the PAPPS platform of the INRA Center at Jouy en Josas.
We thank Ana Hernández Barranco (IPLA-CSIC) for excellent technical assistance, Martin B. Pedersen (Chr. Hansen) for preparation of the peptide sequence database, and Christel Garrigues (Chr. Hansen) for critical reading of the manuscript.
Footnotes
Published ahead of print on 7 September 2007.
REFERENCES
- 1.Almiron, M., A. J. Link, D. Furlong, and R. Kolter. 1992. A novel DNA-binding protein with regulatory and protective roles in starved Escherichia coli. Genes Dev. 6:2646-2654. [DOI] [PubMed] [Google Scholar]
- 2.Ammor, S., K. Yaakoubi, I. Chevallier, and E. Dufour. 2004. Identification by fluorescence spectroscopy of lactic acid bacteria isolated from a small-scale facility producing traditional dry sausages. J. Microbiol. Methods 59:271-281. [DOI] [PubMed] [Google Scholar]
- 3.Araya, M., L. Morelli, G. Reid, M. E. Sanders, and C. Stanton. 2002. Guidelines for the evaluation of probiotics in foods. FAO/WHO report. http://www.who.int/foodsafety/fs_management/en/probiotic_guidelines.pdf.
- 4.Begley, M., C. G. Gahan, and C. Hill. 2005. The interaction between bacteria and bile. FEMS Microbiol. Rev. 29:625-651. [DOI] [PubMed] [Google Scholar]
- 5.Begley, M., C. Hill, and C. G. Gahan. 2006. Bile salt hydrolase activity in probiotics. Appl. Environ. Microbiol. 72:1729-1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bernstein, H., C. M. Payne, C. Bernstein, J. Schneider, S. E. Beard, and C. L. Crowley. 1999. Activation of the promoters of genes associated with DNA damage, oxidative stress, ER stress and protein malfolding by the bile salt, deoxycholate. Toxicol. Lett. 108:37-46. [DOI] [PubMed] [Google Scholar]
- 7.Bron, P. A., D. Molenaar, W. M. de Vos, and M. Kleerebezem. 2006. DNA micro-array-based identification of bile-responsive genes in Lactobacillus plantarum. J. Appl. Microbiol. 100:728-738. [DOI] [PubMed] [Google Scholar]
- 8.Bron, P. A., M. Marco, S. M. Hoffer, E. van Mullekom, W. M. de Vos, and M. Kleerebezem. 2004. Genetic characterization of the bile salt response in Lactobacillus plantarum and analysis of responsive promoters in vitro and in situ in the gastrointestinal tract. J. Bacteriol. 186:7829-7835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Buck, M., M. T. Gallegos, D. J. Studholme, Y. Guo, and J. D. Gralla. 2000. The bacterial enhancer-dependent σ54 (σN) transcription factor. J. Bacteriol. 182:4129-4136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chin, J., B. Turner, I. Barchia, and A. Mullbacher. 2000. Immune response to orally consumed antigens and probiotic bacteria. Immunol. Cell Biol. 78:55-66. [DOI] [PubMed] [Google Scholar]
- 11.Coleman, R., S. Iqbal, P. P. Godfrey, and D. Billington. 1979. Membranes and bile formation. Composition of several mammalian biles and their membrane-damaging properties. Biochem. J. 178:201-208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Corzo, G., and S. E. Gilliland. 1999. Bile salt hydrolase activity of three strains of Lactobacillus acidophilus. J. Dairy Sci. 82:472-480. [DOI] [PubMed] [Google Scholar]
- 13.Darmon, E., D. Noone, A. Masson, S. Bron, O. P. Kuipers, K. M. Devine, and J. M. van Dijl. 2002. A novel class of heat and secretion stress-responsive genes is controlled by the autoregulated CssRS two-component system of Bacillus subtilis. J. Bacteriol. 184:5661-5671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.De Smet, I., L. van Hoorde, M. Vande Woestyne, H. Christiaens, and W. Verstraete. 1995. Significance of bile salt hydrolytic activities of lactobacilli. J. Appl. Bacteriol. 79:292-301. [DOI] [PubMed] [Google Scholar]
- 15.Duché, O., F. Tremoulet, A. Namane, and J. Labadie. 2002. A proteomic analysis of the salt stress response of Listeria monocytogenes. FEMS Microbiol. Lett. 215:183-188. [DOI] [PubMed] [Google Scholar]
- 16.Elkins, C. A., and D. C. Savage. 1998. Identification of genes encoding conjugated bile salt hydrolase and transport in Lactobacillus johnsonii 100-100. J. Bacteriol. 180:4344-4349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Elkins, C. A., and D. C. Savage. 2003. CbsT2 from Lactobacillus johnsonii 100-100 is a transport protein of the major facilitator superfamily that facilitates bile acid antiport. J. Mol. Microbiol. Biotechnol. 6:76-87. [DOI] [PubMed] [Google Scholar]
- 18.Elkins, C. A., S. A. Moser, and D. C. Savage. 2001. Genes encoding bile salt hydrolases and conjugated bile salt transporters in Lactobacillus johnsonii 100-100 and other Lactobacillus species. Microbiology 147:3403-3412. [DOI] [PubMed] [Google Scholar]
- 19.Federici, F., B. Vitali, R. Gotti, M. R. Pasca, S. Gobbi, A. B. Peck, and P. Brigidi. 2004. Characterization and heterologous expression of the oxalyl coenzyme A decarboxylase gene from Bifidobacterium lactis. Appl. Environ. Microbiol. 70:5066-5073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Garrigues, C., B. Stuer-Lauridsen, and E. Johansen. 2005. Characterisation of Bifidobacterium animalis subsp, lactis BB-12 and other probiotic bacteria using genomics, transcriptomics and proteomics. Aust. J. Dairy Technol. 60:84-92. [Google Scholar]
- 21.Gleason, F. K., and A. Holmgren. 1988. Thioredoxin and related proteins in procaryotes. FEMS Microbiol. Rev. 4:271-297. [DOI] [PubMed] [Google Scholar]
- 22.Grill, J. P., S. Perrin, and F. Schneider. 2000. Bile salt toxicity to some bifidobacteria strains: role of conjugated bile salt hydrolase and pH. Can. J. Microbiol. 46:878-884. [DOI] [PubMed] [Google Scholar]
- 23.Hafkenscheid, J. C., and M. P. Hectors. 1975. An enzymatic method for the determination of the glycine/taurine ratio of conjugated bile acids in bile. Clin. Chim. Acta 65:67-74. [DOI] [PubMed] [Google Scholar]
- 24.Hondorp, E. R., and R. G. Matthews. 2004. Oxidative stress inactivates cobalamin-independent methionine synthase (MetE) in Escherichia coli. PLoS Biol. 2:1738-1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jonsson, S., S. Ricagno, Y. Lindqvist, and N. G. Richards. 2004. Kinetic and mechanistic characterization of the formyl-CoA transferase from Oxalobacter formigenes. J. Biol. Chem. 279:36003-36012. [DOI] [PubMed] [Google Scholar]
- 26.Kailasapathy, K., and J. Chin. 2000. Survival and therapeutic potential of probiotic organisms with reference to Lactobacillus acidophilus and Bifidobacterium spp. Immunol. Cell Biol. 78:80-88. [DOI] [PubMed] [Google Scholar]
- 27.Kirkpatrick, N. D., C. Zou, M. A. Brewer, W. R. Brands, R. A. Drezek, and U. Utzinger. 2005. Endogenous fluorescence spectroscopy of cell suspensions for chemopreventive drug monitoring. Photochem. Photobiol. 81:125-134. [DOI] [PubMed] [Google Scholar]
- 28.Kociubinski, G., A. G. Zavaglia, P. F. Perez, E. A. Disalvo, and G. L. De Antoni. 2002. Effect of bile components on the surface properties of bifidobacteria. J. Dairy Res. 69:293-302. [DOI] [PubMed] [Google Scholar]
- 29.Leverrier, P., D. Dimova, V. Pichereau, Y. Auffray, P. Boyaval, and G. Jan. 2003. Susceptibility and adaptive response to bile salts in Propionibacterium freudenreichii: physiological and proteomic analysis. Appl. Environ. Microbiol. 69:3809-3818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Leverrier, P., J. P. Vissers, A. Rouault, P. Boyaval, and G. Jan. 2004. Mass spectrometry proteomic analysis of stress adaptation reveals both common and distinct response pathways in Propionibacterium freudenreichii. Arch. Microbiol. 181:215-230. [DOI] [PubMed] [Google Scholar]
- 31.Masco, L., G. Huys, E. De Brandt, R. Temmerman, and J. Swings. 2005. Culture-dependent and culture-independent qualitative analysis of probiotic products claimed to contain bifidobacteria. Int. J. Food Microbiol. 102:221-230. [DOI] [PubMed] [Google Scholar]
- 32.Meile, L., L. M. Rohr, T. A. Geissman, M. Herensperger, and M. Teuber. 2001. Characterization of the d-xylulose 5-phosphate/d-Fructose 6-phosphate phosphoketolase gene (xfp) from Bifidobacterium lactis. J. Bacteriol. 183:2929-2936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moser, S. A., and D. C. Savage. 2001. Bile salt hydrolase activity and resistance to toxicity of conjugated bile salts are unrelated properties in lactobacilli. Appl. Environ. Microbiol. 67:3476-3480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Noriega, L., I. Cuevas, A. Margolles, and C. G. de los Reyes-Gavilán. 2006. Deconjugation and bile salts hydrolase activity by Bifidobacterium strains with acquired resistance to bile. Int. Dairy J. 16:850-855. [Google Scholar]
- 35.Noriega, L., M. Gueimonde, B. Sánchez, A. Margolles, and C. G. de los Reyes-Gavilán. 2004. Effect of the adaptation to high bile salts concentrations on glycosidic activity, survival at low PH and cross-resistance to bile salts in Bifidobacterium. Int. J. Food Microbiol. 94:79-86. [DOI] [PubMed] [Google Scholar]
- 36.Omumasaba, C. A., N. Okai, M. Inui, and H. Yukawa. 2004. Corynebacterium glutamicum glyceraldehyde-3-phosphate dehydrogenase isoforms with opposite, ATP-dependent regulation. J. Mol. Microbiol. Biotechnol. 8:91-103. [DOI] [PubMed] [Google Scholar]
- 37.Pappin, D. J., P. Hojrup, and A. J. Bleasby. 1993. Rapid identification of proteins by peptide-mass fingerprinting. Curr. Biol. 3:327-332. [DOI] [PubMed] [Google Scholar]
- 38.Payne, C. M., C. Crowley, D. Washo-Stultz, M. Briehl, H. Bernstein, C. Bernstein, S. Beard, H. Holubec, and J. Warneke. 1998. The stress-response proteins poly(ADP-ribose) polymerase and NF-kappaB protect against bile salt-induced apoptosis. Cell Death Differ. 5:623-636. [DOI] [PubMed] [Google Scholar]
- 39.Poquet, I., V. Saint, E. Seznec, N. Simoes, A. Bolotin, and A. Gruss. 2000. HtrA is the unique surface housekeeping protease in Lactococcus lactis and is required for natural protein processing. Mol. Microbiol. 35:1042-1051. [DOI] [PubMed] [Google Scholar]
- 40.Price, C. E., S. J. Reid, A. J. Driessen, and V. R. Abratt. 2006. The Bifidobacterium longum NCIMB 702259T ctr gene codes for a novel cholate transporter. Appl. Environ. Microbiol. 72:923-926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rallu, F., A. Gruss, and E. Maguin. 1996. Lactococcus lactis and stress. Antonie Leeuwenhoek 70:243-251. [DOI] [PubMed] [Google Scholar]
- 42.Rincé, A., Y. Le Breton, N. Verneuil, J. C. Giard, A. Hartke, and Y. Auffray. 2003. Physiological and molecular aspects of bile salt response in Enterococcus faecalis. Int. J. Food Microbiol. 88:207-213. [DOI] [PubMed] [Google Scholar]
- 43.Ruas-Madiedo, P., A. Hernández-Barranco, A. Margolles, and C. G. de los Reyes-Gavilán. 2005. A bile salt-resistant derivative of Bifidobacterium animalis has an altered fermentation pattern when grown on glucose and maltose. Appl. Environ. Microbiol. 71:6564-6570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ruiz, L., B. Sánchez, P. Ruas-Madiedo, C. G. de Los Reyes-Gavilán, and A. Margolles. 2007. Cell envelope changes in Bifidobacterium animalis ssp. lactis as a response to bile. FEMS Microbiol. Lett. 274:316-322. [DOI] [PubMed] [Google Scholar]
- 45.Salminen, S. J., M. Gueimonde, and E. Isolauri. 2005. Probiotics that modify disease risk. J. Nutr. 135:1294-1298. [DOI] [PubMed] [Google Scholar]
- 46.Sánchez, B., C. G. de los Reyes-Gavilán, and A. Margolles. 2006. The F1F0-ATPase of Bifidobacterium animalis is involved in bile tolerance. Environ. Microbiol. 8:1825-1833. [DOI] [PubMed] [Google Scholar]
- 47.Sánchez, B., M. C. Champomier-Vergès, P. Anglade, F. Baraige, C. G. de los Reyes-Gavilán, A. Margolles, and M. Zagorec. 2005. Proteomic analysis of global changes in protein expression during bile salt exposure of Bifidobacterium longum NCIMB 8809. J. Bacteriol. 187:5799-5808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Savijoki, K., A. Suokko, A. Palva, L. Valmu, N. Kalkkinen, and P. Varmanen. 2005. Effect of heat-shock and bile salts on protein synthesis of Bifidobacterium longum revealed by [35S]methionine labelling and two-dimensional gel electrophoresis. FEMS Microbiol. Lett. 248:207-215. [DOI] [PubMed] [Google Scholar]
- 49.Stadtman, E. R., H. van Remmen, A. Richardson, N. B. Wehr, and R. L. Levine. 2005. Methionine oxidation and aging. Biochim. Biophys. Acta 1703:135-140. [DOI] [PubMed] [Google Scholar]
- 50.Ventura, M., C. Canchaya, R. Zink, G. F. Fitzgerald, and D. van Sinderen. 2004. Characterization of the groEL and groES loci in Bifidobacterium breve UCC 2003: genetic, transcriptional, and phylogenetic analyses. Appl. Environ. Microbiol. 70:6197-6209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ventura, M., J. G. Kenny, Z. Zhang, G. F. Fitzgerald, and D. van Sinderen. 2005. The clpB gene of Bifidobacterium breve UCC 2003: transcriptional analysis and first insights into stress induction. Microbiology 151:2861-2872. [DOI] [PubMed] [Google Scholar]
- 52.Ventura, M., R. Zink, G. F. Fitzgerald, and D. van Sinderen. 2005. Gene structure and transcriptional organization of the dnaK operon of Bifidobacterium breve UCC 2003 and application of the operon in bifidobacterial tracing. Appl. Environ. Microbiol. 71:487-500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vido, K., H. Diemer, A. van Dorsselaer, E. Leize, V. Juillard, A. Gruss, and P. Gaudu. 2005. Roles of thioredoxin reductase during the aerobic life of Lactococcus lactis. J. Bacteriol. 187:601-610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Visick, J. E., and S. Clarke. 1995. Repair, refold, recycle: how bacteria can deal with spontaneous and environmental damage to proteins. Mol. Microbiol. 16:835-845. [DOI] [PubMed] [Google Scholar]
- 55.Wouters, J. A., H. H. Kamphuis, J. Hugenholtz, O. P. Kuipers, W. M. de Vos, and T. Abee. 2000. Changes in glycolytic activity of Lactococcus lactis induced by low temperature. Appl. Environ. Microbiol. 66:3686-3691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yokota, A., M. Veenstra, P. Kurdi, H. W. van Veen, and W. N. Konings. 2000. Cholate resistance in Lactococcus lactis is mediated by an ATP-dependent multispecific organic anion transporter. J. Bacteriol. 182:5196-5201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yuan, J., L. Zhu, X. Liu, T. Li, Y. Zhang, T. Ying, B. Wang, J. Wang, H. Dong, E. Feng, Q. Li, J. Wang, H. Wang, K. Wei, X. Zhang, C. Huang, P. Huang, L. Huang, Z. Ming, and H. Wang. 2006. A proteome reference map and proteomics analysis of Bifidobacterium longum NCC2705. Mol. Cell. Proteomics 5:1105-1118. [DOI] [PubMed] [Google Scholar]
- 58.Zou, C. G., and R. Banerjee. 2005. Homocysteine and redox signaling. Antioxid. Redox Signal. 7:547-559. [DOI] [PubMed] [Google Scholar]