Abstract
Phages are a main mortality factor for marine bacterioplankton and are thought to regulate bacterial community composition through host-specific infection and lysis. In the present study we demonstrate for a marine phage-host assemblage that interactions are complex and that specificity and efficiency of infection and lysis are highly variable among phages infectious to strains of the same bacterial species. Twenty-three Bacteroidetes strains and 46 phages from Swedish and Danish coastal waters were analyzed. Based on genotypic and phenotypic analyses, 21 of the isolates could be considered strains of Cellulophaga baltica (Flavobacteriaceae). Nevertheless, all bacterial strains showed unique phage susceptibility patterns and differed by up to 6 orders of magnitude in sensitivity to the same titer of phage. The isolated phages showed pronounced variations in genome size (8 to >242 kb) and host range (infecting 1 to 20 bacterial strains). Our data indicate that marine bacterioplankton are susceptible to multiple co-occurring phages and that sensitivity towards phage infection is strain specific and exists as a continuum between highly sensitive and resistant, implying an extremely complex web of phage-host interactions. Hence, effects of phages on bacterioplankton community composition and dynamics may go undetected in studies where strain identity is not resolvable, i.e., in studies based on the phylogenetic resolution provided by 16S rRNA gene or internal transcribed spacer sequences.
Marine viruses are the most abundant biological entity in the world's oceans (4) and are a main mortality factor for marine bacterioplankton. Most marine virus particles are believed to infect bacteria, and it has been estimated that 10 to 20% of planktonic marine bacteria are lysed by viruses (phages) per day (55). Thereby, up to a quarter of the photosynthetically fixed carbon in the ocean is recycled back to dissolved organic material (65). Phage infection affects bacterial evolution by transferring genetic material between bacteria (41, 62). Further, by selective infection and lysis of predominant bacterial populations, it is thought that lytic phages control bacterial community composition and sustain coexistence of bacterial species (“killing the winner” [57, 58]). This view is supported by predator-prey-like oscillations of phage-host systems in aquatic environments (19, 69) but is likely to simplify actual conditions, as it does not account for broad-host-range phages or bacterial acquisition of resistance (45).
Many marine phages appear to have a narrow host range, infecting only the bacterial strain with which they were isolated (24, 38, 40, 63). However, some findings indicate that the dogma of extreme host specificity among bacteriophages may be an artifact produced by commonly used methods for phage isolation. For instance, the single-host enrichment protocol may select for phages with a narrow host range (22, 64), and infection and lysis by low-virulence phages, which typically have a broad host range (22), may not be detected in plaque assays (10). However, some marine phages are known to infect different strains of the same species (12, 51, 64) or even different species (10-12). Hence, the specificity of phage-host interactions in marine waters needs to be better characterized and understood.
Development of resistance is a common bacterial response to the presence of lytic phages. Chemostat experiments have shown that when exposed to lytic phages, the composition of heterotrophic marine bacterial populations (34, 35) and Escherichia coli populations (13, 20, 30, 37) rapidly shifts from dominance by phage-sensitive clones to dominance by phage-resistant clones. Consequently, phage-resistant bacterial mutants seem to be constantly produced and tend to replace the sensitive strains when the population is exposed to a strong selective pressure from infectious phages. Further, for E. coli a strong coevolution of resistant bacteria and phages with an extended host range has been observed (27, 37). In a comprehensive study of hundreds of phage-host systems from several marine regions, a complex pattern of resistance and susceptibility was observed (40). Similarly, Synechococcus communities may consist of co-occurring resistant and sensitive strains (61). Hence, acquisition and maintenance of resistance against phage infection may be an integral part of marine bacterioplankton ecology, but the incidence of resistance in natural communities is difficult to evaluate directly (14).
In studies of bacterioplankton community composition, the delineation of bacterial species has been based on the 16S rRNA gene and, more recently, on the internal transcribed spacer (ITS) (8, 47), providing the phylogenetic resolution governed by these particular gene regions. The limitations associated with such species delineation has recently been discussed in the context of strain-specific niche occupation (21), where freshwater bacterial isolates with identical or almost-identical 16S rRNA gene and ITS sequences were suggested to represent distinct populations and to probably occupy different ecological niches (17, 21). These limitations may also apply in the context of strain-specific phage-induced bacterial mortality.
In the present study a collection of 21 Cellulophaga baltica (Bacteroidetes) strains, which were highly similar genotypically and phenotypically, allowed us to address phage-host interactions at the strain level. Our results demonstrate large strain-specific differences in phage susceptibility among hosts and a pronounced variability in specificity and efficiency of infection for the C. baltica phages.
MATERIALS AND METHODS
Isolation of bacteria and phages.
Strains NN014847 to NN016048 were isolated from various Danish coastal waters in 1994 (23). Strains MM#3 and #4 to #19 were isolated from Øresund surface water (56°2′N, 12°37′E) in February 2000, and strain OL12a was isolated from Baltic Sea surface water east off Öland (56°37′N, 16°45′E) in April 2005. The strains were selected as yellow colonies on CYT agar plates (1.0 g casein, 0.5 g yeast extract, 0.5 g CaCl2·H2O, 0.5 g MgSO4·H2O, 15 g agar, 1,000 ml deionized water, pH 7.3) with colony morphology (size, texture, and edge appearance) and restriction fragment length polymorphism (RFLP) profiles (16S rRNA gene, HaeIII, and NdeII; Roche) similar to those of the C. baltica type strain (strain NN015840) (23).
Phages were obtained from Øresund surface water in 2000 and 2005. After a 0.45-μm prefiltration (Millipak; Millipore), phages were concentrated 250-fold by tangential flow filtration (Millipore) followed by Amicon ultracentrifugation (Millipore). Phages were isolated using the top-agar plating technique (plaque assay) (48), with all of the isolated bacterial strains used as hosts. Plaques with different morphologies were selected from each host and purified for three rounds of infection and lysis. Thereafter, 5 ml phage buffer (0.1 M NaCl, 0.008 M MgSO4, 0.05 M Tris-HCl, 2 mM CaCl2, 0.1% gelatin, 0.5 M tryptophan, 5% glycerol, pH 7.5) was added to a fully lysed plate, and the agar overlay was shredded with a sterile inoculation loop. The plate was incubated on a shaker for 20 min, and the mixture was then transferred to a sterile tube and centrifuged (5,000 × g, 10 min). The supernatant was passed through a 0.22-μm filter (Millex GS; Millipore), and the phage stock was stored at 4°C.
In order to examine potential effects of resistance acquisition on host range, strains resistant to two of the isolated phages, φSM and φST, were developed. These strains, #3 r φSM and #3 r φST, were obtained after exposure of strain MM#3 to the phages in laboratory experiments and isolated from fully lysed plates.
Sequencing of the 16S rRNA gene and the ITS.
Bacterial DNA was extracted using the DNeasy tissue kit (QIAGEN), and the 16S rRNA gene was PCR amplified using PuReTaq Ready-To-Go Beads (Amersham Biosciences) and primers 27f and 1492r (15). The thermal program consisted of 30 cycles of 30 s of denaturation at 95°C, 30 s of annealing at 50°C, and 45 s of extension at 72°C, followed by 7 min at 72°C. The ITS was amplified using primers G1 and 23Sr (21) and a thermal program consisting of 35 cycles of 30 s of denaturation at 95°C, 30 s of annealing at 54°C, and 45 s of extension at 72°C, followed by 72°C for 7 min. DNA was sequenced bidirectionally (commercially by Macrogen, Korea) using primers 27f, 530f, 519r, and 907r (26) for the 16S rRNA gene and primers G1 and 23Sr for the ITS. In some cases, the PCR-amplified ITS region could not be sequenced directly. The PCR products were therefore cloned prior to sequencing (TOPO TA cloning kit; Invitrogen). Sequences were aligned in Seqman (DNASTAR), and phylogenetic trees were constructed using the neighbor-joining algorithm in Clustal X (59).
Genotyping by UP-PCR.
Universally primed PCR (UP-PCR) is a whole-genome fingerprinting technique originally developed for fungi (see reference 31 for a review) and recently used to discriminate between genetically similar bacteria (7, 70). The UP-PCR method uses a single primer to prime genomes arbitrarily, and the primer is designed to primarily target less-conserved intergenic areas (31). Crude DNA templates for PCR (cell lysates) were prepared from each bacterial isolate as described by Brandt et al. (7) and stored at −20°C until use. UP-PCR was performed using the universal L15/AS19 primer 5′-GAG GGT GGC GGC TAG-3′ (32). PCR mixtures (20 μl) consisted of molecular biology reagent-grade water (Sigma), buffer (supplied with DNA polymerase), 2 mM MgCl2 (supplied with DNA polymerase), 100 μM of each deoxynucleoside triphosphate (AH Diagnostics), 1 ng μl−1 of PCR primer (DNA Technology), 1 unit of Dynazyme II DNA polymerase (Finnzymes, Finland), and 1 μl template DNA. PCR was carried out using the following program: initial denaturation at 94°C for 3 min and then 32 cycles consisting of denaturation at 94°C for 60 s, primer annealing 53°C for 60 s, and elongation at 72°C for 60 s. In the first and the last cycles, the elongation steps were extended to 90 s and 3 min, respectively. PCR products were subjected to agarose gel electrophoresis (1.5%, 100 V, 40 min) and stained with ethidium bromide. A 100-bp DNA ladder (Fermenta) was used as a molecular size marker. UP-PCR analysis was performed at least twice for each isolate to verify that reproducible band patterns were obtained. Subsequently, the isolates were grouped based on identical UP-PCR patterns.
Analysis of lipopolysaccharide (LPS) profiles.
Bacterial strains were grown in MLB medium (0.5 g Casamino Acids, 0.5 g peptone, 0.5 g yeast extract, 3 ml glycerol, 500 ml filtered seawater [Øresund], and 500 ml demineralized water) at room temperature for 72 h. Pseudomonas fluorescens strain Ag1, which was used as a reference, was grown to early stationary phase in LB medium (10 g tryptone, 5 g yeast extract, 1 g glucose, 10 g NaCl, 1 liter Milli-Q) at 28°C.
For preparation of proteinase K-digested cell lysates, cells were centrifuged (10,000 × g, 5 min), washed once in phosphate-buffered saline, and resuspended in lysis buffer (0.5 M Tris-HCl [pH 6.8], 2% sodium dodecyl sulfate (SDS), 10% glycerol). Cell concentrations were normalized during resuspension. Proteinase K (75 μg ml−1) was added and the lysates incubated at 56°C for 1 h, boiled for 5 min, centrifuged (14,000 × g, 30 min), and stored at −20°C. Lysates were analyzed by SDS-polyacrylamide gel electrophoresis using 14% acrylamide-0.4% bisacrylamide and 4 M urea as described by Sørensen et al. (52), and a prestained protein ladder (PageRuler Ladder Plus; Fermentas) was included on each gel. After electrophoresis, gels were fixed in ethanol-acetic acid overnight and stained with a periodic acid-silver stain (60). Specificity of the staining was ensured by monitoring silver staining of LPS from the internal control (P. fluorescens Ag1) without obtaining staining of the included protein ladder. The strains were grouped based on similar LPS profiles. The profiles were reproducible between independent replications, yet the intensities of individual bands, especially in the low-molecular-weight region, varied between runs.
Specificity of phage infection.
To determine phage host range and the bacterial susceptibility to specific phages, a cross infectivity test where all bacterial strains were exposed to all individual phage stocks was performed. Three microliters of phage stock (108 to 1011 PFU ml−1) was spotted onto a lawn of host bacteria in top agar, and plates were examined for cell lysis after 1, 3, and 5 days. These spot tests were performed three independent times. Infection was considered positive when lysis (i.e., a clearing zone) was seen in all three tests. In spot tests, spots may originate from inhibition of bacterial growth, often seen as turbid spots, rather than direct viral lysis (39). To verify the presence of infectious phages, plaque assays in dilution series were performed in some cases where turbid spots were seen. In all cases plaques were obtained, suggesting that phage lysis caused the clearing zones. An unweighted-pair group method using average linkages (UPGMA) tree was constructed using the software Quantity One 4.2.1 (Bio-Rad), where the lysis/no lysis matrix was converted to pairwise distances using the Dice similarity coefficient.
Since several bacterial strains were susceptible to the same phage, we sought to determine whether phage infection was equally efficient in different hosts. Three hosts were exposed to the same phage titer, and infectivity was quantified by plaque assay. PFU were examined after 1, 2, and 3 days. This was performed for three different phages.
Phage genome sizes.
Phage genome sizes were determined by pulsed-field gel electrophoresis (PFGE). Freshly made phage stocks (see above) were concentrated by ultracentrifugation (222,000 × g, 2.5 h, 4°C; Beckman). DNA was obtained according to the protocol for λ DNA (3) and quantified fluorometrically (PicoGreen; Molecular Probes). Some phage genomes disintegrated when extracted using this protocol. These were therefore lysed in agar plugs. Equal amounts of phage concentrate and melted 1.5% low-melting-point agarose (Sigma) in MSM buffer without gelatin (53) (450 mM NaCl, 50 mM MgSO4, 50 mM Tris, pH 8.0) were mixed, transferred to plug molds, and left to solidify at 4°C. The plugs were incubated in lysis solution (0.25 M EDTA [pH 8], 1% SDS, 1 mg ml−1 proteinase K) overnight, washed three times in TE buffer (10 mM Tris-HCl, 1 mM EDTA, pH 8), and stored in TE at 4°C.
For PFGE analysis, 20 ng DNA or 5 × 107 phage particles were loaded in each well. PFGE was performed on a CHEF-DR III system (Bio-Rad) using 1% PFGE certified agarose (Sigma). The gel was run for 11 h in 0.5× TBE buffer (1× TBE is 89 M Tris, 2 mM EDTA, and 89 mM boric acid, pH 8.3), at a 0.5- to 5.0-s switch time, 6 V/cm, and an included angel of 120°. The gel was stained with SYBR gold (Molecular Probes) for 30 min, destained in 0.5× TBE for 30 min, and photographed (Gel Doc XR; Bio-Rad). Genome sizes were determined manually using an 8- to 48-kb standard (Bio-Rad) and a λ PFGE marker (Amersham) as molecular size markers.
Restriction digests of phage genomes.
Some phages with identical genome sizes showed differences in host range. To examine whether these phages were genetically different, they were compared by RFLP analysis. HindIII was chosen after testing four different restriction enzymes (NdeII, HaeIII, EcoRI, and HindIII; Boehringer Mannheim), and phage DNA (100 ng) was cleaved according to the manufacturer's recommendation. The restriction digests were analyzed on an agarose gel (0.7%, 90 V, 2 h), stained with ethidium bromide, and photographed as described above. A GeneRuler 1-kb DNA ladder (Fermenta) was used as a molecular size marker.
TEM.
Selected phages representing a range in genome size and host range were examined by transmission electron microscopy (TEM). Freshly made lysates were obtained from fully lysed plates (see above). Grids were prepared by placing 8 μl of lysate onto 200-mesh Formvar-coated copper grids (Ted Pella) for 2 min. The phage solution was removed with filter paper, and grids were subsequently stained with 8 μl 2% sodium phosphotungstate (phages φ40:2, φ18:4, φ18:4, φ38:1, φ39:1, φ4:1, and φ3:2) or 2% uranyl acetate (phages φ13:1, φ18:1, φ12:1, φSM, and φ10:1) for 2 min. The grids were examined using a Zeiss EM 900 microscope with an accelerating voltage of 80 kV.
Nucleotide sequence accession numbers.
The partial 16S rRNA gene sequences and ITS sequences obtained in this study have been deposited in GenBank and assigned accession numbers EF667101 to EF667143 (Table 1).
TABLE 1.
Genetic and morphological characteristics of bacterial strains used in this study
| Taxon | Strain | Isolation yr | Isolation location | % DNA homologyb | % Similarityc (GenBank accession no.)
|
Group no. according to:
|
|||
|---|---|---|---|---|---|---|---|---|---|
| 16S rRNA gene | ITS | UP-PCR | LPS | Colony morphology | |||||
| Cellulophaga | NN015840a | 1994 | Svaneke | 100 | 100.0 (AJ005972) | 100.0 (EF667121) | 10 | 1 | 7 |
| baltica | NN016038 | 1994 | Svaneke | 100 | 100.0 (EF667101) | 98.7 (EF667122) | 9 | 1 | 7 |
| NN015839 | 1994 | Svaneke | 100 | 100.0 (EF667102) | 99.1 (EF667123) | 10 | 1 | 7 | |
| NN014845 | 1994 | Svaneke | 100 | 100.0 (EF667103) | 99.6 (EF667124) | 10 | 1 | 6 | |
| NN014847 | 1994 | Svaneke | 102 | 100.0 (EF667104) | 98.7 (EF667125) | 9 | 1 | 8 | |
| NN014850 | 1994 | Svaneke | 100 | 100.0 (EF667105) | 99.9 (EF667126) | 10 | 1 | 6 | |
| NN014873 | 1994 | Svaneke | 102 | 100.0 (EF667106) | 99.9 (EF667127) | 10 | 1 | 6 | |
| NN016046 | 1994 | Ellekilde Hage | 78 | 100.0 (EF667107) | 98.8 (EF667128) | 2 | 1 | 10 | |
| NN016048 | 1994 | Ellekilde Hage | 71 | 100.0 (EF667108) | 98.1 (EF667129) | 3 | 1 | 10 | |
| MM#3 | 2000 | Øresund | 100.0 (AF497997) | 98.4 (EF667130) | 2 | 1 | 2 | ||
| #4 | 2000 | Øresund | 100.0 (EF667109) | 98.5 (EF667131) | 4 | 1 | 2 | ||
| #10 | 2000 | Øresund | 100.0 (EF667110) | 98.7 (EF667132) | 5 | 1 | 7 | ||
| #12 | 2000 | Øresund | 100.0 (EF667111) | 98.5 (EF667133) | 6 | 1 | 3 | ||
| #13 | 2000 | Øresund | 99.5 (EF667112) | 98.8 (EF667134) | 1 | 1 | 9 | ||
| #14 | 2000 | Øresund | 99.5 (EF667113) | 98.5 (EF667135) | 2 | 1 | 4 | ||
| #17 | 2000 | Øresund | 99.5 (EF667114) | 98.1 (EF667136) | 7 | 1 | 4 | ||
| #18 | 2000 | Øresund | 100.0 (EF667115) | 99.0 (EF667137) | 8 | 1 | 5 | ||
| #19 | 2000 | Øresund | 99.5 (EF667116) | 98.2 (EF667138) | 2 | 1 | 4 | ||
| OL12a | 2005 | Off Öland | 99.5 (EF667117) | 98.9 (EF667139) | 10 | 1 | 8 | ||
| #3 r φSM | Laboratory | 100.0 (EF667118) | 98.4 (EF667140) | 2 | 1 | 1 | |||
| #3 r φST | Laboratory | 100.0 (EF667119) | 98.4 (EF667141) | 2 | 1 | 2 | |||
| Cellulophaga fucicola | NN015860 | 1994 | Hirsholm | 0 | 92.3 (AJ005973) | 68.4 (EF667142) | 11 | 2 | 11 |
| Flavobacteriaceae bacterium IE-7 | #8 | 2000 | Øresund | 88.2 (EF667120) | 75.1 (EF667143) | 12 | 1 | 12 | |
RESULTS
Bacterial characterization.
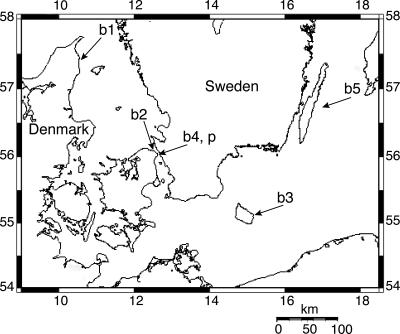
A total of 23 bacterial strains obtained from five localities (Fig. 1) in 1994, 2000, and 2005 were characterized and grouped according to the parameters shown in Table 1. The C. baltica strains formed yellow colonies on nutrient agar plates and could be divided into 10 morphological groups. Only four of the C. baltica strains were regarded as having unique colony morphology. The remaining isolates were identical to up to three other isolates. For the phage-resistant strains, strain #3 r φST had morphology identical to that of MM#3, while strain #3 r φSM differed (Table 1).
FIG. 1.
Sampling sites for bacterial (b) and phage (p) isolations. Exact locations are marked by arrows. b1, Hirsholm; b2, Ellekilde Hage; b3, Svaneke; b4, Øresund; b5, off Öland. The map was generated using the Online Map Creation software (http://www.aquarius.geomar.de/omc/).
Sequencing of the 16S rRNA gene and ITS.
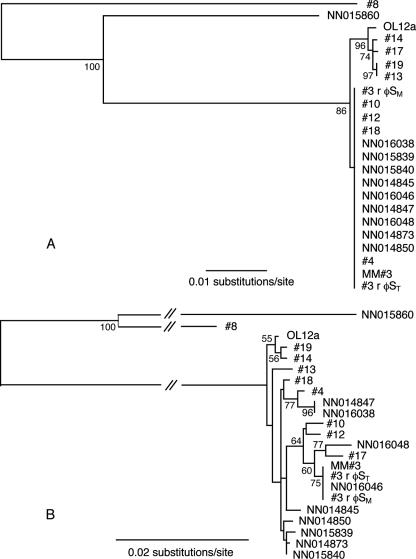
Partial sequencing of the 16S rRNA gene (∼800 bp) showed that 15 strains were identical to the C. baltica type strain NN015840, while 5 strains showed a 4-bp difference from this bacterium (Table 1; Fig. 2A). Strains NN015860 (Cellulophaga fucicola, accession no. AJ005973) and #8 (98% similar to Flavobacteriaceae bacterium IE-7, accession no. EF105392) showed only 93% and 88% 16S rRNA gene similarity to the other strains (Fig. 2A). These strains also had shorter ITS (800 and 700 bp, respectively) than the C. baltica strains (900 bp). All the C. baltica strains showed high ITS sequence similarity (98 to 100%) (Table 1) and formed a tight phylogenetic cluster (Fig. 2B).
FIG. 2.
Neighbor-joining trees showing the phylogenetic relationships between bacteria used in this study. Trees are based on 16S rRNA gene sequences (∼800 bp) (A) and ITS gene sequences (∼675 bp) (B). Phylogenetic relationships were bootstrapped 1,000 times, and values greater than 50% are shown.
UP-PCR and LPS profiling.
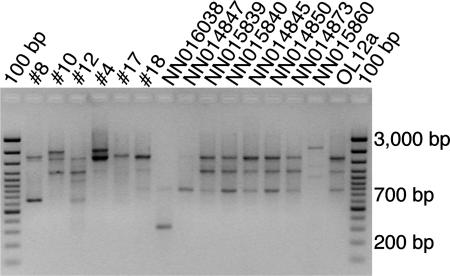
Due to the high sequence similarities of 16S rRNA genes and ITS, we sought to distinguish the C. baltica strains by means of UP-PCR fingerprinting. Accordingly, the C. baltica strains could be divided into 10 different UP-PCR groups (Table 1). Still, several of the strains had identical UP-PCR fingerprints, and three of the UP-PCR groups contained two to six identical isolates (Fig. 3).
FIG. 3.
Genomic profiling of bacterial isolates. An agarose gel showing UP-PCR banding patterns for 15 of the 23 bacterial isolates analyzed is shown. Identical strains (e.g., strains NN015839 and NN015840) and strains with differences (e.g., strains 10 and 12) are displayed.
Acquisition of phage resistance may yield detectable changes in LPS profiles (see, e.g., references 18, 37, and 56); however, we observed no differences in LPS profiles among the C. baltica isolates despite their pronounced variation in phage susceptibility (see below). Only isolate NN015860 (C. fucicola) had a different LPS profile (Table 1; see Fig. S1 in the supplemental material).
Phage characterization.
Phages were obtained from Øresund in 2000 and 2005 using plaque assays (Fig. 1). Plaques were isolated for all bacterial host strains except for strains #3 r φSM, #8, and NN014845, and a total of 45 phages infecting the C. baltica strains and 1 phage infecting C. fucicola were obtained. From a single water sample, up to four different plaque morphologies could be distinguished for a single bacterial host (e.g., φ48:1 to φ48:4 [Table 2]).
TABLE 2.
Phages isolated and analyzed in this study
| Phage | Original host strain | Isolation yr | Genome size (kb)a | Phage familyb |
|---|---|---|---|---|
| φ40:1 | NN015840 | 2005 | 68 | ND |
| φ40:2 | NN015840 | 2005 | >242 | Myoviridae |
| φ38:1 | NN016038 | 2005 | 70 | Podoviridae |
| φ38:2 | NN016038 | 2005 | 54 | ND |
| φ39:1 | NN015839 | 2000 | 29 | Siphoviridae |
| φ39:2 | NN015839 | 2005 | >242 | ND |
| φ47:1 | NN014847 | 2005 | 54 | ND |
| φ50:1 | NN014850 | 2005 | >242 | ND |
| φ73:1 | NN014873 | 2005 | >242 | ND |
| φ46:1 | NN016046 | 2000 | 38 | ND |
| φ46:2 | NN016046 | 2005 | 21/85 | ND |
| φ46:3 | NN016046 | 2005 | 78 | ND |
| φ46:4 | NN016046 | 2005 | 145 | ND |
| φ48:1 | NN016048 | 2005 | 9/19 | ND |
| φ48:2 | NN016048 | 2005 | 18/72 | ND |
| φ48:3 | NN016048 | 2005 | 18/72 | ND |
| φ48:4 | NN016048 | 2005 | 145 | ND |
| φSM | MM#3 | 2000 | 54 | Myoviridae |
| φST | MM#3 | 2000 | 97 | ND |
| φ3:1 | MM#3 | 2000 | 54 | ND |
| φ3:2 | MM#3 | 2005 | 18/76 | Myoviridae |
| φ4:1 | #4 | 2005 | 145 | Myoviridae? |
| φ10:1 | #10 | 2000 | 54 | Myoviridae |
| φ12:1 | #12 | 2000 | 40 | Siphoviridae |
| φ12:2 | #12 | 2005 | 8/17 | ND |
| φ12:3 | #12 | 2000 | 40 | ND |
| φ12:4 | #12 | 2000 | 40 | ND |
| φ12:5 | #12 | 2000 | 40 | ND |
| φ13:1 | #13 | 2000 | 97 | Siphoviridae |
| φ13:2 | #13 | 2005 | 78 | ND |
| φ14:1 | #14 | 2005 | 9/19 | ND |
| φ14:2 | #14 | 2005 | 110 | ND |
| φ17:1 | #17 | 2000 | 42 | ND |
| φ17:2 | #17 | 2005 | 145 | ND |
| φ18:1 | #18 | 2000 | 40 | Podoviridae? |
| φ18:2 | #18 | 2000 | 40 | ND |
| φ18:3 | #18 | 2005 | 78 | ND |
| φ18:4 | #18 | 2005 | 9/19 | Siphoviridae/Siphoviridae |
| φ19:1 | #19 | 2000 | 60 | ND |
| φ19:2 | #19 | 2000 | 97 | ND |
| φ19:3 | #19 | 2005 | 70/80 | ND |
| φ19:4 | #19 | 2005 | >242 | ND |
| φ12a:1 | OL12a | 2005 | 9/19 | ND |
| φ3ST:1 | #3 r φST | 2005 | 18/76 | ND |
| φ3ST:2 | #3 r φST | 2005 | 54 | ND |
| φ60:1 | NN015860 | 2000 | 40 | ND |
The presence of two values indicates two bands in the PFGE.
ND, no data.
Phage genome size and morphology.
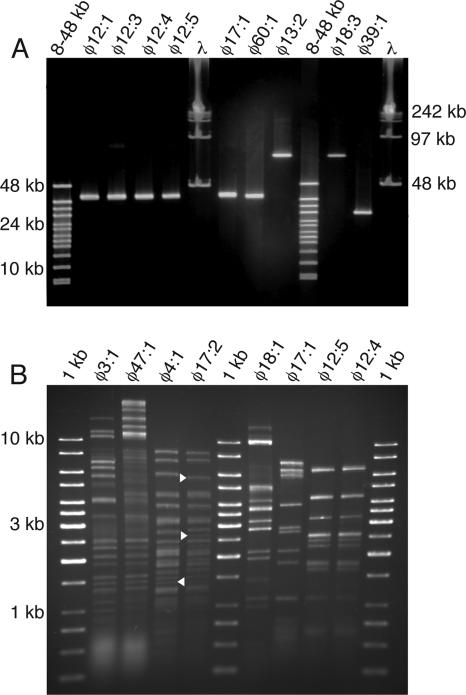
To genetically distinguish the phages, genome sizes were determined by PFGE analysis. Phage genome sizes ranged from 8 to >242 kb and represented 18 different sizes (Table 2; Fig. 4A). Several phages had identical genome sizes, and only 10 phages were unique in size (Table 2). Some phages showed double bands on the PFGE with up to a >4-fold difference in molecular size. For example φ3:2 had bands of 18 kb and 76 kb. Some phages with very different genome sizes were infectious to the same bacterial strain. For instance, strain NN015839 could be infected by both φ40:2 (> 242 kb) and φ39:1 (29 kb).
FIG. 4.
Genomic characterization of phages. (A) PFGE analysis of genomic DNAs from 9 of the 46 phage isolates used in this study. The gel displays phages with identical (e.g., φ12:4 and φ12:5) and different (e.g., φ18:3 and φ39:1) genomes sizes. (B) HindIII restriction digests of eight phage genomes. Cleavage patterns for phages with identical (φ12:4 and φ12:5) and different (e.g., φ17:1 and φ18:1) infectivity patterns are shown along with patterns for phages with almost identical infectivity patterns (φ4:1 and φ17:2; see Fig. 6). Arrowheads indicate differences in banding patterns for φ4:1 and φ17:2. See Table 2 for phage genome sizes.
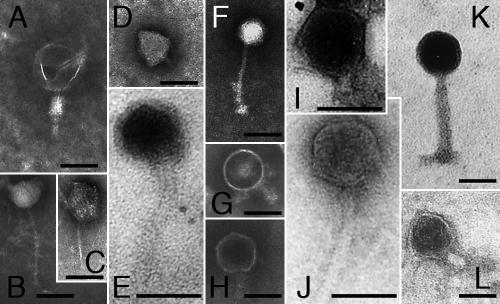
Selected phages showing a large range in genome size (8 to >242 kb) and host range (1 to 20 hosts) represented a wide variety of morphotypes (Fig. 5) and belonged to the Myoviridae, Siphoviridae, and Podoviridae families (1) (Table 2). Broad- and narrow-host-range phages were found in all three families. Two phages showing two bands each in the PFGE analysis were examined by TEM. For phages φ3:2 and φ18:4, one and two morphotypes could be observed, respectively (Fig. 5). Interestingly, some phages with identical genome sizes, e.g., φ12:1 and φ18:1, belonged to different families (Table 2; Fig. 5).
FIG. 5.
TEM of selected C. baltica phages. Bars, 50 nm. (A) φ40:2; (B) φ18:4; (C) φ18:4; (D) φ38:1; (E) φ13:1; (F) φ39:1; (G) φ4:1; (H) φ3:2; (I) φ18:1; (J) φ12:1; (K) φSM; (L) φ10:1. The phage φ18:4 lysate included two morphotypes (B and C). Examples of phages with identical genome sizes but with different morphotypes are shown (e.g., I versus J and K versus L). Genome sizes and host ranges for the phages shown are given in Table 2 and Fig. 6.
Host range and diversity of obtained phages.
To examine whether the phages were specific for C. baltica, infectivity was tested on five Bacteroidetes isolates, one alphaproteobacterium, and one gammaproteobacterium. No infections were observed (data not shown). Also, no cross-infectivity was observed for C. baltica and C. fucicola phages, and no phages infected strain #8 (Fig. 6). Within C. baltica, the phages showed a large variation in infectivity (Fig. 6). Six phages showed narrow host specificity, infecting only the bacterial strain with which they were originally isolated, but a broader host range was more common. A few phages were able to infect all bacterial strains except strains #3 r φSM and #3 r φST. No relationship between host range and time of isolation was found.
FIG. 6.
Host ranges of the 46 phages. Gray squares indicate lysis and white squares indicate no lysis.
In some cases there were no or only small differences in host range between phages with identical genome size. To determine whether these phages were genetically different, they were examined by RFLP analysis. The RFLP patterns showed that phages with even minor differences in host range were genetically different and therefore should be regarded as unique (Fig. 4B). For instance, phages φ4:1 and φ17:2 had a genome size of 145 kb (Table 2), differed with respect to infection of a single bacterial strain (Fig. 6), and showed slightly different RFLP profiles (Fig. 4B). Phages with identical genome sizes, host ranges, and RFLP patterns were considered identical (e.g., phages φ12:1 and φ12:3 to φ12:5 [Table 2; Fig. 4 and 6]). Of the 45 isolated phages, 40 were concluded to be unique. In all cases, identical phages originated from the same water sample.
Susceptibility and resistance to phage infection.
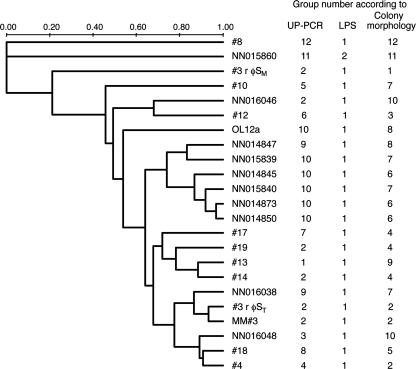
The phage susceptibility of the C. baltica strains was highly variable. For instance, the most (strain #13) and the least (strain NN014845) susceptible strains could be infected by 27 and 12 different phages, respectively (Fig. 6). All strains showed unique susceptibility patterns. To facilitate a comparison with other bacterial characteristics, the phage susceptibility data were analyzed by UPGMA and converted to a dendrogram (Fig. 7). In some cases the clustering was consistent with genetic or morphological characteristics. For instance, for strains NN015840, NN014845, NN014850, and NN014873, the phage susceptibility was consistent with high similarity of the 16S rRNA gene, UP-PCR, and colony morphology as well as with a 100% match in DNA homology (23). Strains #13, #14, #17, and #19 grouped according to 16S rRNA gene sequences and phage susceptibility but showed small differences in UP-PCR pattern and colony morphology (Fig. 2A and 7). For most strains, no consistency was observed between phage susceptibility and any of the analyzed strain characteristics.
FIG. 7.
UPGMA tree based on susceptibility to phages infection. The data shown in Fig. 6 were scored and converted to pairwise distances using the Dice similarity coefficient. Groupings of bacterial strains according to UP-PCR and LPS profiling and colony morphology are inserted to facilitate comparison.
The effects of resistance acquisition were examined for strain MM#3. When resistance against φSM was acquired, MM#3 became resistant towards 21 phages, while resistance against φST conferred resistance against only 3 phages (Fig. 6).
To determine whether phage infection was equally efficient in different hosts, infectivity was examined for three phages by exposing three susceptible hosts to the same phage titer (Table 3). The number of PFU varied up to 6 orders of magnitude between host strains.
TABLE 3.
Efficiency of phage infection in three bacterial strains
| Phage | Infection efficiencya in strain:
|
||
|---|---|---|---|
| MM#3 | #4 | #10 | |
| φST | 100 | 14 | 0.001 |
| φ38:1 | 100 | 0.2 | 0.003 |
| φ40:1 | 100 | 1 | 0.0008 |
Strains were exposed to the same phage titer (∼108 PFU). Efficiency is expressed in relative PFU, where the highest PFU was set to 100%.
DISCUSSION
Similarity of the bacterial strains.
C. baltica strains are members of the Flavobacteria class within the Bacteroidetes, which is one of the dominant phyla in marine waters (16, 25). Flavobacteria in particular are thought to be important during phytoplankton blooms (42, 46). Therefore, the ecology of the C. baltica-phage system could be an important part of carbon cycling associated with phytoplankton blooms. According to 16S rRNA gene sequences, the C. baltica strains were closely related. Therefore, sequencing of the ITS was performed to differentiate the strains. The ITS is known to differ in length and/or nucleotide composition within species (47, 49), and recently it was shown that bacterial groups showing >99% 16S rRNA gene similarity may have ITS sequences with <93% identity (8). Still, the C. baltica strains showed >98% ITS sequence similarity, confirming that there is no overall relationship between 16S rRNA gene and ITS sequence similarity (8). Since the different strains were not widely distinguishable by 16S rRNA gene and ITS sequencing, a whole-genome PCR typing analysis (UP-PCR) was performed. Indeed, using this method the strains could be divided into 10 groups based on banding patterns, but three groups still contained two to six isolates. Combined with an earlier analysis showing 100% hybridization-based DNA-homology for a number of the strains (Table 1) (23), our molecular characterizations show that the C. baltica strains are highly homologous at the genomic level. Therefore, these bacterial strains were considered to be well suited for this study of phage-host interactions.
Phage diversity and host range.
The C. baltica phages included representatives of Myoviridae, Siphoviridae, and Podoviridae and ranged in genome size from 8 to >242 kb. This is a significantly wider range than for other isolated aquatic phages (∼30 to 125 kb) (12, 51, 67) or for marine phage communities (∼25 to 70 kb), where genomes of >100 kb often are assumed to originate from algal viruses (54, 68). Nine of the 45 C. baltica phages isolated in the present study were >100 kb, and recently a phage genome of >200 kb from polluted seawater was characterized (36). Hence, obviously large genome viruses in natural assemblages cannot be assumed to be phytoplankton viruses.
In some cases the PFGE analysis yielded double bands for a phage stock. This phenomenon has previously been recognized as multiple-phage infection according to electron microscopy (12). Accordingly, two phage morphotypes were observed for φ18:4. However, for φ3:2 only one morphotype was detected. It remains unclear whether a second phage was present at low abundance in the φ3:2 lysate and therefore was not detected or whether the two bands originate from a single phage with a fragmented genome (see, e.g., reference 33). Of the 45 C. baltica phages, 40 were unique according to genome size and host range. Selected phages with identical genome sizes were found to belong to different families according to TEM analysis. Further, RFLP digests demonstrated that even phages that differed only with respect to infection of a single host strain could be distinguished. Hence, along with other recent studies (2, 12), our data highlight that whole-genome fingerprinting analyses of viruses with similar genome sizes may reveal an otherwise concealed genetic and morphological variation. This indicates that analyses of natural viral assemblages based on genome size (i.e., PFGE) severely underestimate genetic diversity and viral community dynamics.
Infection by the C. baltica phages appeared to be species specific, with a wide intraspecies host range (Fig. 6) and a tremendous variation in infectivity of specific hosts (Table 3). This illustrates how phage community composition will have a large impact on the way that phages influence mortality as well as the species and strain compositions of natural bacterioplankton communities. Further, since phages with up to 10-fold differences in genome size were infectious to the same host, it is possible that the extent and/or efficiency of transduction events (41) and phage conversion (9) in the environment will be highly dependent on infection by specific phages.
Strain-specific susceptibility and resistance to phage infection.
While the C. baltica strains appeared to be genetically similar, they all showed a unique combination of susceptibility to the tested phages. This was seen not only as the number of phages infective to a given bacterial strain but also in relative efficiency of infection when different hosts were exposed to the same phage and titer (Table 3). Similarly, variable infectivity has been observed for phages infecting different bacterial species (22). Therefore, the impact of phage infection and lysis on a bacterial population is dependent not only on whether the phage can infect the host but, just as importantly, on how susceptible the host is to infection. In E. coli, acquisition of resistance, often associated with modification or loss of receptor molecules to which the phage initially binds (5, 28, 43), may occur in one or two steps leading to the presence of partially resistant mutants (29). Hence, the specific mutation and receptor in question determine the extent of resistance. This complexity is also illustrated by acquisition of resistance in the C. baltica strain MM#3. While resistance against φSM conferred resistance against a large number of phages, resistance against φST gave resistance against only three phages with similar genome size (Table 2; Fig. 6). This suggests that different receptors are used by φSM and φST.
In E. coli the acquisition of resistance is often coupled to changes in the LPS structure, which are sometimes detectable by SDS-polyacrylamide gel electrophoresis (37, 56). Using this method, we found no differences between C. baltica strains, despite the fact that all showed unique susceptibility patterns, or between strain 8 and the C. baltica strains. This calls the sensitivity of the method into question; however, it cannot be excluded that susceptibility was determined by cellular features other than LPS (e.g., cell surface proteins) (29).
Temporal and spatial co-occurrence of phage-host systems.
In the present study, hosts were isolated from five different locations during a 10-year period, while phages were obtained from a single location on two occasions with 5 years in between (Fig. 1; Tables 1 and 2). The isolated assemblages consisted of bacterial strains with variable susceptibility and phages with large host range diversity. Comparable results have been obtained for phage-host systems in other coastal regions. For instance, in several successive years, Wichels et al. (64) isolated Pseudoalteromonas phage-host systems from locations in the German Bight and observed complex interactions between phages and hosts at the strain level. Also, Comeau et al. (12) showed that the vibriophage community of the Strait of Georgia (Canada) consisted of a highly diverse mixture of phenotypes and genotypes.
We obtained phages infectious to bacterial strains that were isolated from localities hundreds of kilometers apart (e.g., see locations p, b2, and b5 in Fig. 1) encompassing salinities from ∼7 to ∼18 practical salinity units. Interestingly, despite the fact that one C. baltica strain was isolated from the Baltic Sea (location b5 in Fig. 1), we were not able to isolate phages infective to any of the C. baltica strains from this brackish environment (data not shown). Hence, as also indicated by the study by Wichels and coauthors (64), salinity may function as a natural barrier for the distribution of some phage-host systems. In oceanic waters, genetically related phages may be distributed over thousands of kilometers (24).
Major findings and implications.
This study shows that there is a large variation in phage susceptibility within a well-defined phylogenetic group of marine bacteria, the C. baltica strains, as well as large host range diversity among C. baltica phages. Despite the fact that only some of the 21 C. baltica strains were distinguishable through genomic/membrane profiling and DNA sequencing, they all showed unique phage susceptibility patterns, thus indicating an extremely complex web of phage-host interactions even within this restricted group of marine bacteria.
If our data on C. baltica-phage interactions are representative of marine bacteria in general, the results add completely new perspectives to the role of phages in bacterial diversity and function. One potential implication is that phage-host interactions may have gone undetected in studies concerning natural communities of phages and bacteria (see, e.g., references 44 and 50) if they were based on the phylogenetic resolution provided by the 16S rRNA gene or ITS. However, a few studies based on 16S rRNA gene methodology have been able to demonstrate viral control of single operational taxonomic units (terminal RFLP) (66) or bacterial classes (fluorescent in situ hybridization) (6). The observed variations in susceptibility among our strains and in infectivity of individual phages predict that the specific strain-level composition of co-occurring bacterial and phage assemblages determines to what extent infection and lysis affect bacterial community composition as analyzed by 16S rRNA gene- or ITS-based methods. Conceivably, the large diversity at the strain level implies that strains exposed to phages are replaced by strains that are less susceptible to the co-occurring phages and, consequently, that phage activity is a driving force in the strain-specific diversity and continuous succession in bacterial populations. Indeed, we found that susceptible bacterial strains may differ by up to 6 orders of magnitude in sensitivity to the same titer of phage. Hence, we speculate that marine bacterioplankton species are susceptible to multiple co-occurring phages but that the sensitivity to phage infection exists as a continuum between two extremes, highly sensitive and resistant. The data presented contribute to our emerging view of phage-host interactions in marine waters but also illustrate that the link between phage action and bacterial diversification is complex and poorly understood. Future studies of basic phage-host interactions and dynamics in their natural environment are, therefore, fundamental for improving existing conceptual models on the role of phages in bacterial diversity and population dynamics.
Supplementary Material
Acknowledgments
We thank J. E. Johansen and P. Nielsen for providing strains NN014847 to NN016048, B. Olsen for use of PFGE equipment, and S. Iversen for excellent technical assistance with LPS and UP-PCR analyses. We also thank two anonymous reviewers for thoughtful comments on the manuscript.
This study was supported by Kalmar University (L.R.), the Danish Natural Sciences Research Council and the Directorate for Food Fisheries and Agribusiness (M.M.), and the Faculty of Life Sciences, University of Copenhagen (O.N.).
Footnotes
Published ahead of print on 31 August 2007.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Ackermann, H. W. 2007. 5500 phages examined in the electron microscope. Arch. Virol. 152:227-243. [DOI] [PubMed] [Google Scholar]
- 2.Allen, M. J., J. Martinez-Martinez, D. C. Schroeder, P. J. Somerfield, and W. H. Wilson. 2007. Use of microarrays to assess viral diversity: from genotype to phenotype. Environ. Microbiol. 9:971-982. [DOI] [PubMed] [Google Scholar]
- 3.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.). 1995. Short protocols in molecular biology, 3rd ed. John Wiley & Sons, Inc., New York, NY.
- 4.Bergh, O., K. Y. Børsheim, G. Bratbak, and M. Heldal. 1989. High abundance of viruses found in aquatic environments. Nature 340:467-468. [DOI] [PubMed] [Google Scholar]
- 5.Bohannan, B. J. M., and R. E. Lenski. 2000. Linking genetic change to community evolution: insights from studies of bacteria and bacteriophage. Ecol. Lett. 3:362-377. [Google Scholar]
- 6.Bouvier, T., and P. A. del Giorgio. 2007. Key role of selective viral-induced mortality in determining marine bacterial community composition. Environ. Microbiol. 9:287-297. [DOI] [PubMed] [Google Scholar]
- 7.Brandt, K. K., A. Petersen, P. E. Holm, and O. Nybroe. 2006. Decreased abundance and diversity of culturable Pseudomonas spp. populations with increasing copper exposure in the sugar beet rhizosphere. FEMS Microbiol. Ecol. 56:281-291. [DOI] [PubMed] [Google Scholar]
- 8.Brown, M. V., and J. A. Fuhrman. 2005. Marine bacterial microdiversity as revealed by internal transcribed spacer analysis. Aquat. Microb. Ecol. 41:15-23. [Google Scholar]
- 9.Brüssow, H., C. Canchaya, and W. D. Hardt. 2004. Phages and the evolution of bacterial pathogens: from genomic rearrangements to lysogenic conversion. Microbiol. Mol. Biol. Rev. 68:560-602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chiura, H. X. 1997. Generalized gene transfer by virus-like particles from marine bacteria. Aquat. Microb. Ecol. 13:75-83. [Google Scholar]
- 11.Comeau, A. M., E. Buenaventura, and C. A. Suttle. 2005. A persistent, productive, and seasonally dynamic vibriophage population within Pacific oysters (Crassostrea gigas). Appl. Environ. Microbiol. 71:5324-5331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Comeau, A. M., A. M. Chan, and C. A. Suttle. 2006. Genetic richness of vibriophages isolated in a coastal environment. Environ. Microbiol. 8:1164-1176. [DOI] [PubMed] [Google Scholar]
- 13.Fischer, C. R., M. Yoichi, H. Unno, and Y. Tanji. 2004. The coexistence of Escherichia coli serotype O157:H7 and its specific bacteriophage in continuous culture. FEMS Microbiol. Lett. 241:171-177. [DOI] [PubMed] [Google Scholar]
- 14.Fuhrman, J. A. 1999. Marine viruses and their biogeochemical and ecological effects. Nature 399:541-548. [DOI] [PubMed] [Google Scholar]
- 15.Giovannoni, S. J. 1991. The polymerase chain reaction, p. 177-201. In E. Stackebrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. John Wiley & Sons, Inc., New York, NY.
- 16.Glöckner, F. O., B. M. Fuchs, and R. Amann. 1999. Bacterioplankton compositions of lakes and oceans: a first comparison based on fluorescence in situ hybridization. Appl. Environ. Microbiol. 65:3721-3726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hahn, M. W., and M. Pockl. 2005. Ecotypes of planktonic actinobacteria with identical 16S rRNA genes adapted to thermal niches in temperate, subtropical, and tropical freshwater habitats. Appl. Environ. Microbiol. 71:766-773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hancock, R. E., and P. Reeves. 1976. Lipopolysaccharide-deficient, bacteriophage-resistant mutants of Escherichia coli K-12. J. Bacteriol. 127:98-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hennes, K. P., C. A. Suttle, and A. M. Chan. 1995. Fluorescently labeled virus probes show that natural virus populations can control the structure of marine microbial communities. Appl. Environ. Microbiol. 61:3623-3627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Horne, M. T. 1970. Coevolution of Escherichia coli and bacteriophages in chemostat culture. Science 168:992-993. [DOI] [PubMed] [Google Scholar]
- 21.Jaspers, E., and J. Overmann. 2004. Ecological significance of microdiversity: identical 16S rRNA gene sequences can be found in bacteria with highly divergent genomes and ecophysiologies. Appl. Environ. Microbiol. 70:4831-4839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jensen, E. C., H. S. Schrader, B. Rieland, T. L. Thompson, K. W. Lee, K. W. Nickerson, and T. A. Kokjohn. 1998. Prevalence of broad-host-range lytic bacteriophages of Sphaerotilus natans, Escherichia coli, and Pseudomonas aeruginosa. Appl. Environ. Microbiol. 64:575-580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johansen, J. E., P. Nielsen, and C. Sjøholm. 1999. Description of Cellulophaga baltica gen. nov., sp. nov. and Cellulophaga fucicola gen. nov., sp. nov. and reclassification of [Cytophaga] lytica to Cellulophaga lytica gen. nov., comb. nov. Int. J. Syst. Bacteriol. 49:1231-1240. [DOI] [PubMed] [Google Scholar]
- 24.Kellogg, C. A., J. B. Rose, S. C. Jiang, J. M. Thurmond, and J. H. Paul. 1995. Genetic diversity of related vibrophages isolated from marine environments around Florida and Hawaii, USA. Mar. Ecol. Prog. Ser. 120:89-98. [Google Scholar]
- 25.Kirchman, D. L. 2002. The ecology of Cytophaga-Flavobacteria in aquatic environments. FEMS Microbiol. Ecol. 39:91-100. [DOI] [PubMed] [Google Scholar]
- 26.Lane, D. J., B. Pace, G. J. Olsen, D. A. Stahl, M. L. Sogin, and N. R. Pace. 1985. Rapid determination of 16S ribosomal RNA sequences for phylogenetic analyses. Proc. Natl. Acad. Sci. USA 82:6955-6959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lenski, R. E. 1984. Coevolution of bacteria and phage: are there endless cycles of bacterial defenses and phage counterdefenses? J. Theor. Biol. 108:319-325. [DOI] [PubMed] [Google Scholar]
- 28.Lenski, R. E. 1988. Experimental studies of pleiotropy and epistasis in Escherichia coli. 1. Variation in competitive fitness among mutants resistant to vitus T4. Evolution 42:425-432. [DOI] [PubMed] [Google Scholar]
- 29.Lenski, R. E. 1984. Two-step resistance by Escherichia coli B to bacteriophage T2. Genetics 107:1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lenski, R. E., and B. R. Levin. 1985. Constraints on the coevolution of bacteria and virulent phage: a model, some experiments, and predictions for natural communities. Am. Nat. 125:585-602. [Google Scholar]
- 31.Lübeck, M., and P. S. Lübeck. 2005. Universally primed PCR (UP-PCR) and its application in mycology, p. 409-438. In S. K. Deshmukh and M. A. Rai (ed.), The biodiversity of fungi. Their role in human life. Science Publishers Inc., Enfield, NH.
- 32.Lübeck, P. S., I. A. Alekhina, M. Lübeck, and S. A. Bulat. 1998. UP-PCR genotyping and rDNA analysis of Ascophyta pisi. J. Phytopathol. 146:51-55. [Google Scholar]
- 33.Martin, W. J. 1996. Genetic instability and fragmentation of a stealth viral genome. Pathobiology 64:9-17. [DOI] [PubMed] [Google Scholar]
- 34.Middelboe, M. 2000. Bacterial growth rate and marine virus-host dynamics. Microb. Ecol. 40:114-124. [DOI] [PubMed] [Google Scholar]
- 35.Middelboe, M., Å. Hagström, N. Blackburn, B. Sinn, U. Fischer, N. H. Borch, J. Pinhassi, K. Simu, and M. G. Lorenz. 2001. Effects of bacteriophages on the population dynamics of four strains of pelagic marine bacteria. Microb. Ecol. 42:395-406. [DOI] [PubMed] [Google Scholar]
- 36.Miller, E. S., J. F. Heidelberg, J. A. Eisen, W. C. Nelson, A. S. Durkin, A. Ciecko, T. V. Feldblyum, O. White, I. T. Paulsen, W. C. Nierman, J. Lee, B. Szczypinski, and C. M. Fraser. 2003. Complete genome sequence of the broad-host-range vibriophage KVP40: comparative genomics of a T4-related bacteriophage. J. Bacteriol. 185:5220-5233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mizoguchi, K., M. Morita, C. R. Fischer, M. Yoichi, Y. Tanji, and H. Unno. 2003. Coevolution of bacteriophage PP01 and Escherichia coli O157:H7 in continuous culture. Appl. Environ. Microbiol. 69:170-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moebus, K. 1992. Further investigations on the concentration of marine bacteriophages in the water around Helgoland, with reference to the phage-host systems encountered. Helgoländer. Meeresunters. 46:275-292. [Google Scholar]
- 39.Moebus, K. 1983. Lytic and inhibition responses to bacteriophages among marine bacteria, with special reference to the origin of phage-host systems. Helgoländer. Meeresunters. 36:375-391. [Google Scholar]
- 40.Moebus, K., and H. Nattkemper. 1981. Bacteriophage sensitivity patterns among bacteria isolated from marine waters. Helgoländer. Meeresunters. 34:375-385. [Google Scholar]
- 41.Paul, J. H. 1999. Microbial gene transfer: an ecological perspective. J. Mol. Microbiol. Biotechnol. 1:45-50. [PubMed] [Google Scholar]
- 42.Pinhassi, J., M. M. Sala, H. Havskum, F. Peters, O. Guadayol, A. Malits, and C. Marrase. 2004. Changes in bacterioplankton composition under different phytoplankton regimens. Appl. Environ. Microbiol. 70:6753-6766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Prehm, P., B. Jann, K. Jann, G. Schmidt, and S. Stirm. 1976. On a bacteriophage T3 and T4 receptor region within the cell wall lipopolysaccharide of Escherichia coli B. J. Mol. Biol. 101:277-281. [DOI] [PubMed] [Google Scholar]
- 44.Riemann, L., and M. Middelboe. 2002. Stability of bacterial and viral community composition in Danish coastal waters as depicted by DNA fingerprinting techniques. Aquat. Microb. Ecol. 27:219-232. [Google Scholar]
- 45.Riemann, L., and M. Middelboe. 2002. Viral lysis of marine bacterioplankton: implications for organic matter cycling and bacterial clonal composition. Ophelia 56:57-68. [Google Scholar]
- 46.Riemann, L., G. F. Steward, and F. Azam. 2000. Dynamics of bacterial community composition and activity during a mesocosm diatom bloom. Appl. Environ. Microbiol. 66:578-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rocap, G., D. L. Distel, J. B. Waterbury, and S. W. Chisholm. 2002. Resolution of Prochlorococcus and Synechococcus ecotypes by using 16S-23S ribosomal DNA internal transcribed spacer sequences. Appl. Environ. Microbiol. 68:1180-1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed., vol. 1. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 49.Scheinert, P., R. Krausse, U. Ullmann, R. Söller, and G. Krupp. 1996. Molecular differentation of bacteria by PCR amplification of the 16S-23S rRNA spacer. J. Microbiol. Methods 26:103-117. [Google Scholar]
- 50.Schwalbach, M. S., I. Hewson, and J. A. Fuhrman. 2004. Viral effects on bacterial community composition in marine plankton microcosms. Aquat. Microb. Ecol. 34:117-127. [Google Scholar]
- 51.Shivu, M. M., B. C. Rajeeva, S. K. Girisha, I. Karunasagar, G. Krohne, and I. Karunasagar. 2006. Molecular characterisation of Vibrio harveyi bacteriophages isolated from aquaculture environments along the coast of India. Environ. Microbiol. 9:322-331. [DOI] [PubMed] [Google Scholar]
- 52.Sørensen, J., J. Skouv, A. Jørgensen, and O. Nybroe. 1992. Rapid identification of environmental isolates of Pseudomonas aeruginosa, P. fluorescens and P. putida by SDS-PAGE analysis of whole-cell protein patterns. FEMS Microbiol. Ecol. 101:41-50. [Google Scholar]
- 53.Steward, G. F. 2001. Fingerprinting viral assemblages by pulsed field gel electrophoresis (PFGE). Methods Microbiol. 30:85-104. [Google Scholar]
- 54.Steward, G. F., J. L. Montiel, and F. Azam. 2000. Genome size distributions indicate variability and similarities among marine viral assemblages from diverse environments. Limnol. Oceanogr. 45:1697-1706. [Google Scholar]
- 55.Suttle, C. A. 1994. The significance of viruses to mortality in aquatic microbial communities. Microb. Ecol. 28:237-243. [DOI] [PubMed] [Google Scholar]
- 56.Tanji, Y., T. Shimada, M. Yoichi, K. Miyanaga, K. Hori, and H. Unno. 2004. Toward rational control of Escherichia coli O157:H7 by a phage cocktail. Appl. Microbiol. Biotechnol. 64:270-274. [DOI] [PubMed] [Google Scholar]
- 57.Thingstad, T. F. 2000. Elements of a theory for the mechanisms controlling abundance, diversity, and biogeochemical role of lytic bacterial viruses in aquatic systems. Limnol. Oceanogr. 45:1320-1328. [Google Scholar]
- 58.Thingstad, T. F., and R. Lignell. 1997. Theoretical models for the control of bacterial growth rate, abundance, diversity and carbon demand. Aquat. Microb. Ecol. 13:19-27. [Google Scholar]
- 59.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tsai, C. M., and C. E. Frasch. 1982. A sensitive silver stain for detecting lipopolysaccharides in polyacrylamide gels. Anal. Biochem. 119:115-119. [DOI] [PubMed] [Google Scholar]
- 61.Waterbury, J. B., and F. W. Valois. 1993. Resistance to co-occurring phages enables marine Synechococcus communities to coexist with cyanophages abundant in seawater. Appl. Environ. Microbiol. 59:3393-3399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Weinbauer, M. G., and F. Rassoulzadegan. 2004. Are viruses driving microbial diversification and diversity? Environ. Microbiol. 6:1-11. [DOI] [PubMed] [Google Scholar]
- 63.Wichels, A., S. S. Biel, H. R. Gelderblom, T. Brinkhoff, G. Muyzer, and C. Schütt. 1998. Bacteriophage diversity in the North Sea. Appl. Environ. Microbiol. 64:4128-4133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wichels, A., G. Gerdts, and C. Schütt. 2002. Pseudoalteromonas spp. phages, a significant group of marine bacteriophages in the North Sea. Aquat. Microb. Ecol. 27:233-239. [Google Scholar]
- 65.Wilhelm, S. W., and C. A. Suttle. 1999. Viruses and nutrient cycles in the sea. Bioscience 49:781-788. [Google Scholar]
- 66.Winter, C., A. Smit, G. J. Herndl, and M. G. Weinbauer. 2004. Impact of virioplankton on archaeal and bacterial community richness as assessed in seawater batch cultures. Appl. Environ. Microbiol. 70:804-813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wolf, A., J. Wiese, G. Jost, and K.-P. Witzel. 2003. Wide geographic distribution of bacteriophages that lyse the same indigenous freshwater isolate (Sphingomonas sp. strain B18). Appl. Environ. Microbiol. 69:2395-2398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wommack, E. K., J. Ravel, R. T. Hill, J. Chun, and R. R. Colwell. 1999. Population dynamics of Chesapeake Bay virioplankton: total-community analysis by pulse-field gel electrophoresis. Appl. Environ. Microbiol. 65:231-240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wommack, E. K., J. Ravel, R. T. Hill, and R. R. Colwell. 1999. Hybridization analysis of Chesapeake Bay virioplankton. Appl. Environ. Microbiol. 65:241-250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Worm, J., and O. Nybroe. 2001. Input of protein to lake water microcosms affects expression of proteolytic enzymes and the dynamics of Pseudomonas spp. Appl. Environ. Microbiol. 67:4955-4962. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.