Abstract
For efficient generation of high-affinity protein-based binding molecules, fast and reliable downstream characterization platforms are needed. In this work, we have explored the use of staphylococcal cell surface display together with flow cytometry for affinity characterization of candidate affibody molecules directly on the cell surface. A model system comprising three closely related affibody molecules with different affinities for immunoglobulin G and an albumin binding domain with affinity for human serum albumin was used to investigate advantages and differences compared to biosensor technology in a side-by-side manner. Equilibrium dissociation constant (KD) determinations as well as dissociation rate analysis were performed using both methods, and the results show that the on-cell determinations give both KD and dissociation rate values in a very fast and reproducible manner and that the relative affinities are very similar to the biosensor results. Interestingly, the results also show that there are differences between the absolute affinities determined with the two different technologies, and possible explanations for this are discussed. This work demonstrates the advantages of cell surface display for directed evolution of affinity proteins in terms of fast postselectional, on-cell characterization of candidate clones without the need for subcloning and subsequent protein expression and purification but also demonstrates that it is important to be aware that absolute affinities determined using different methods often vary substantially and that such comparisons therefore could be difficult.
Specific binding proteins, e.g., antibodies, are becoming increasingly important in almost all areas of life science, from large-scale proteome projects (47, 49) to in vivo imaging (26, 31, 34, 50) and biotherapy (1, 10). To meet the increasing demands, much effort has been put into developing and improving different methods for the generation of such binders. The generation is performed either in vivo through immunization of animals or in vitro using various combinatorial library display systems. As new and more efficient methods have emerged and the construction of extremely large combinatorial libraries now is possible, the need for fast and reliable downstream characterization technologies is increasing. The bottleneck in the process of discovering novel binders is today just as much in the characterization of the binding and biophysical properties of the selected protein candidates as in the actual selection process. A faster and less laborious characterization method would allow for an increased number of candidates to be analyzed, which also increases the probability of finding a candidate with the required properties. Although phage display has been available for more than two decades (23, 32, 43), it is still the in vitro selection method of choice for the majority of laboratories working in the field of combinatorial protein engineering. Nevertheless, today there are a number of more or less established competing technologies, including, among others, ribosome display (13, 21, 57), other cell-free selection systems (3, 18, 28, 29, 41), protein complementation assays (16), and various formats of cell display (4, 7-9, 14, 33, 56), all with their respective advantages and disadvantages. We have previously described a system for display of proteins and peptides on the cell surface of the gram-positive bacterium Staphylococcus carnosus (17, 36-39, 45, 51-55). The staphylococcal display system has recently been improved for protein engineering purposes (20), and optimization of the electroporation protocol has increased the transformation frequency to approximately 106 transformants per transformation (19), enabling the construction of large displayed combinatorial protein libraries. A 58-amino-acid, three-helical-bundle protein, derived from staphylococcal protein A (24), has been used as a protein engineering scaffold, and the randomized and selected affinity proteins are denoted affibody molecules (12, 26, 27, 30). The staphylococcal display system has been described to expose approximately 10,000 recombinant surface proteins per bacterium (2). Since the scaffold is of staphylococcal origin, a staphylococcus-based system should increase the probability of functional display on the cell surface.
The main advantage of cell-based display systems is that the cell is large enough to be analyzed and sorted using flow cytometry. In addition, the high polyvalency, with expression levels from a few hundred to several hundred thousand proteins displayed per cell (2), allows for sorting in a truly quantitative manner (5, 56). Furthermore, in phage display selections, elution of the binders from the target is typically required to collect bound phages, and it is not evident that the strongest binders are properly eluted. In addition to the many advantages over phage display in the selection process, staphylococcal cell display should offer the possibility to carry out a very rapid on-cell affinity determination to rank a large number of selected candidates using flow cytometry, based on which a few top candidates can be further characterized in more detail. However, since the dominating in vitro selection system still is phage display, where on-particle affinity determination is not possible, the affinity of the majority of binders today is determined using surface plasmon resonance (SPR) technology (15) and not on cells using flow cytometry. It is also known that different affinity determination techniques often result in different absolute values of affinity, making direct comparisons difficult. This is not necessarily a problem, since early in a selection process the relative ranking among the different candidates is more crucial than the absolute affinity. Furthermore, if the difference between two methods is reproducible and known, this knowledge can be taken into account when a comparison is made. It is therefore important to investigate whether a new method is reproducible and to determine any differences in comparison to the benchmark technology.
In order to do this for the novel staphylococcal cell surface display method, we have determined the equilibrium dissociation constants (KD) of three related affibody molecules (Zwt, ZN28A, and ZK35A) (20) with affinity for human immunoglobulin G (IgG) by cell display together with flow cytometry and by biosensor analysis. Furthermore, the KD and the dissociation rate constant (koff) of an albumin binding domain (ABDwt) (44) for human serum albumin (HSA) have been determined using both methods.
MATERIALS AND METHODS
Bacterial strains, vectors, and growth medium.
Staphylococcus carnosus strain TM300 (11) containing the surface display vector pSCX:Zwt, pSCX:ZN28A, or pSCX:ZK35A (20) or pSC:ABDwt is hereafter denoted Sc:Zwt, Sc:ZN28A, Sc:ZK35A, and Sc:ABDwt, respectively.
The surface display vector pSC:ABDwt was constructed from the surface display vector pSCXm (55) in two steps. First, the coding sequence for the albumin binding protein (55), which functions as a spacer and normalization tag, was cleaved out from the parental vector pSCXm using the restriction enzymes XhoI (Fermentas, Vilnius, Lithuania) and HindIII (Fermentas) and replaced with the coding sequence for a head-to-tail dimer of the Zwt domain (24, 25). The vector was thereafter digested with BamHI (Fermentas) and SalI (Fermentas) and purified. The albumin binding domain (ABDwt) was amplified by PCR, restricted with BamHI and SalI, and ligated into the vector using T4 DNA ligase (Invitrogen, Carlsbad, CA) according to the supplier's recommendations. The plasmid was transformed into competent Escherichia coli RR1ΔM15 cells (35), and the vector sequence was confirmed using DNA sequencing with BigDye Thermo Cycle sequencing reactions and an ABI Prism 3700 instrument (Applied Biosystems, Foster City, CA). Plasmid preparations were performed with the JETSTAR kit (Genomed, Bad Oeynhausen, Germany) according to the supplier's recommendations, and the plasmid was thereafter transformed using electroporation into electrocompetent S. carnosus TM300 cells as previously described (19).
Tryptic soy broth (Merck, Darmstadt, Germany) supplemented with yeast extract and 20 μg ml−1 chloramphenicol was used as the growth medium in all further experiments.
Labeling of human IgG and HSA.
Human IgG was biotinylated and purified using the FluoReporter biotin-XX labeling kit (Invitrogen) according to the manual. HSA was labeled with fluorophore using Alexa Fluor 647 succinimidyl ester or Alexa Fluor 488 succinimidyl ester (Invitrogen) according to the supplier's recommendations. The concentration of labeled and purified IgG was determined using the bicinchoninic acid protein assay kit (Pierce Biotechnology Inc., Rockford, IL) according to the supplier's recommendations. The concentration of labeled and purified HSA was determined using amino acid analysis.
Preparation of cells for labeling.
Cultures were inoculated with Sc:Zwt, Sc:ZN28A, Sc:ZK35A, or Sc:ABDwt to an optical density at 578 nm of approximately 0.005 and incubated at 37°C and 150 rpm over night. After 16 h, 1 ml of each culture was pelleted and resuspended in 1 ml phosphate-buffered saline (PBS) with 0.1% Pluronic F108 NF surfactant (PBSP) (BASF Corporation, Mount Olive, NJ) and diluted to an optical density at 578 of 1.0 in PSBP. Finally, 100 μl was transferred to a new 1.5-ml tube for subsequent labeling and analysis.
Rapid on-cell affinity ranking of affibody molecules using flow cytometry.
A rapid affinity ranking of Zwt, ZN28A, and ZK35A was carried out by flow cytometric analysis of fluorescently labeled cells. Following preparation as described above, pellets of Sc:Zwt, Sc:ZN28A, and Sc:ZK35A were resuspended in 1 ml PBSP containing 4.9, 2.1, and 0.7 nM biotinylated human IgG. The cells were incubated at room temperature for 1 hour with gentle mixing to reach equilibrium binding and then washed in 100 μl ice-cold PBSP. Cells were resuspended in 100 μl PBSP containing 1.25 μg ml−1 streptavidin (R-phycoerythrin conjugate) (Invitrogen) and 225 nM Alexa Fluor 647 (HSA conjugate) and incubated for 40 min on ice in the dark. After a last washing step in 100 μl ice-cold PBSP, cells were resuspended in 300 μl ice-cold PBSP. The mean fluorescence intensity (MFI) was measured using a FACS Vantage SE (BD Biosciences, San Jose, CA) flow cytometer. The experiment was carried out in triplicates on different days, using freshly prepared solutions.
Extraction and purification of surface-anchored proteins for immobilization on Biacore chips.
In order to obtain the cell wall-anchored affibody molecules and ABDwt for immobilization on Biacore chips for kinetic analysis using SPR, the proteins were produced and purified. The recombinant staphylococci, Sc:Zwt, Sc:ZN28A, Sc:ZK35A, and Sc:ABDwt, were cultivated overnight, and the cells were pelleted by centrifugation, resuspended in 10 ml PBS, and subjected to lysostaphin treatment (100 units; 2 h at 37°C) followed by sonication to release the protein. The extracted cell wall proteins were purified using HSA affinity chromatography as described previously (46), and the purified proteins were analyzed using 12.5% sodium dodecyl sulfate-polyacrylamide gel electrophoresis under reducing conditions. The different proteins were immobilized to carboxymethylated dextran chips (CM5; BIAcore AB, Uppsala, Sweden) by amine coupling using a Biacore 2000 instrument (BIAcore AB) according to the manufacturer's suggestions. On chip one, 500 Biacore resonance units (RU) of Zwt, ZN28A, and ZK35A were immobilized to flow cells 2, 3, and 4, respectively, and with IgG as a negative control on flow cell 1. On chip two, 400 RU of ABDwt was immobilized to flow cells 3 and 4. The blank surface was treated like the other surfaces on the chip but with no protein injected.
KD determination using SPR.
SPR analysis was carried out using a Biacore 2000 instrument (BIAcore AB) to determine the KD of the three affibody molecules and the ABDwt. All experiments were performed using HEPES-buffered saline (HBS) (10 mM HEPES, 0.15 M NaCl, 3.4 mM EDTA, and 0.005% Surfactant P20 [BIAcore AB], pH 7.4) as running buffer and 10 mM HCl for regeneration of the chip surface. A series of different concentrations of IgG (550, 413, 275, 138, 69, 34, 17, 8.6, 4.3, and 2.1 nM) were injected over the flow cells of chip one with the immobilized affibody molecules, and different concentrations of HSA (7,500, 2,500, 750, 500, 100, 50, 25, 1, and 0.75 nM) were injected over the flow cells of chip two with the immobilized ABDwt. The injections were performed in duplicates in a random order and at a flow rate of 20 μl min−1, and the responses at equilibrium binding were collected. The entire experiment was carried out in duplicates on different days, using freshly prepared solutions.
On-cell KD determination using flow cytometry.
To determine the KD on cells using flow cytometry, cells were prepared as described above and pellets of Sc:Zwt, Sc:ZN28A, and Sc:ZK35A were resuspended in 1 ml PBSP containing different concentrations of biotinylated human IgG (21, 14, 7, 4.9, 4.2, 2.1, 1.4, 0.7, 0.35, and 0.14 nM). The cells were incubated at room temperature for 1 hour with gentle mixing to reach equilibrium binding and then washed in 100 μl ice-cold PBSP. Cells were thereafter resuspended in 100 μl PBSP containing 1.25 μg ml−1 streptavidin (R-phycoerythrin conjugate) (Invitrogen) and incubated for 40 min on ice in the dark. After a last washing step in 100 μl ice-cold PBSP, cells were resuspended in 300 μl ice-cold PBSP and kept on ice until flow cytometric analysis. Cells displaying ABDwt were incubated with different concentrations of Alexa Fluor 488 (HSA conjugate) (1,000, 126, 63, 32, 16, 7.9, 3.9, 2.0, 0.98, and 0.42 nM) for 1 hour at room temperature in the dark with gentle mixing to reach equilibrium binding, followed by washing and resuspension in 300 μl ice-cold PBSP. The MFI was measured using a FACS Vantage SE (BD Biosciences) flow cytometer. The experiment was carried out in triplicates on different days, using freshly prepared solutions.
Determination of koff using SPR.
SPR analysis of the dissociation rate for the ABDwt and HSA pair was performed by injecting HSA (250, 125, 63, and 31 nM) in duplicates at a flow rate of 50 μl min−1 over the flow cells of chip two, containing immobilized ABDwt. After association, the chip was washed for 15 min with HBS buffer and the dissociation response data were collected. The entire experiment was carried out in duplicates on different days, using freshly prepared solutions.
On-cell determination of koff using flow cytometry.
To determine the dissociation rate for the ABDwt and HSA pair on cells using flow cytometry, cells were prepared as described above and then resuspended and incubated in the dark at room temperature in 1 ml 42 μM Alexa Fluor 488 (HSA conjugate) for 1 hour at room temperature to saturate all binding sites. After washing with PBSP at room temperature, cells were resuspended and incubated in 1 ml PBSP containing 420 μM of unlabeled HSA to avoid reassociation of the labeled HSA molecules to the ABDwt on the cell surface. To determine the dissociation rate of the complex, samples were taken every 5 minutes for 45 min, and the MFI was measured using a FACS Vantage SE (BD Biosciences) flow cytometer. The experiment was carried out in triplicates on different days, using freshly prepared solutions.
RESULTS
Model system.
Three previously described recombinant staphylococcal strains, denoted Sc:Zwt, Sc:ZK35A, and Sc:ZN28A (20) (Fig. 1A) and expressing single-amino-acid mutants of the protein A-derived Z domain with different affinities for IgG, were used together with a newly constructed strain, Sc:ABDwt. In the new strain, a 46-amino-acid-residue albumin binding domain (ABDwt) (42, 44, 46), derived from streptococcal protein G, was introduced into a modified variant of the general surface display vector pSCXm. The newly constructed strain, Sc:ABDwt, contains the cell wall-anchoring region from staphylococcal protein A for functional display of the bacterial domains (22, 40, 48, 55) but lacks the albumin binding protein (42, 55), which is replaced by a head-to-tail dimer of Zwt functioning as a spacer to minimize sterical hindrance from the cell surface and as a surface expression normalization tag for parallel monitoring of the surface expression level (Fig. 1B). Functional cell surface expression of both the ABDwt and the Zwt domain was verified using flow cytometry (data not shown).
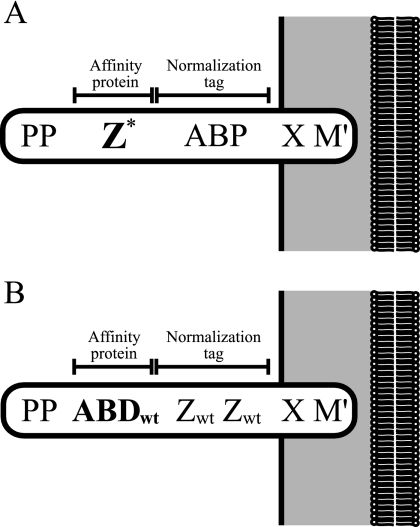
FIG. 1.
(A) Schematic picture of the processed gene fusion products anchored to the cell wall in strains Sc:Zwt, Sc:ZN28A, and Sc:ZK35A (20). Z* represents the three different single-amino-acid mutants, Zwt, ZN28A, and ZK35A. PP, propeptide from Staphylococcus hyicus lipase; Z, engineered IgG binding domain derived from staphylococcal protein A; ABP, albumin binding protein derived from streptococcal protein G and used as a normalization tag (20); X, charged repetitive region derived from staphylococcal protein A, postulated to interact with the peptidoglycan cell wall; M′, the processed and covalently cell wall-anchored form of the M sequence of staphylococcal protein A. (B) Schematic picture of the processed gene fusion products anchored to the cell wall in the new strain Sc:ABDwt. ABDwt, albumin binding domain derived from streptococcal protein G (44); Zwt, engineered IgG binding domain derived from staphylococcal protein A and used as a dimer for normalization, according to the previously described principle (20).
Rapid on-cell affinity ranking of affibody molecules using flow cytometry.
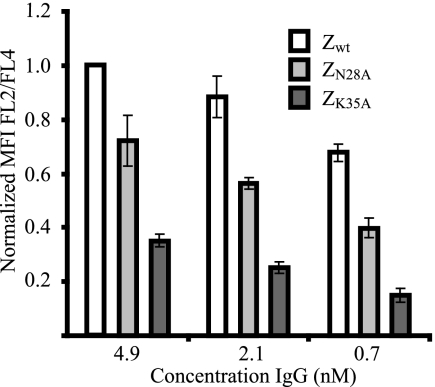
Briefly, for a rapid affinity ranking of the three surface-displayed affibody variants, Zwt, ZN28A, and ZK35A, cells were incubated with three concentrations of biotinylated human IgG, followed by incubation with streptavidin (R-phycoerythrin conjugate) and Alexa Fluor 647 (HSA conjugate). The MFI was measured using flow cytometry, and the signal corresponding to the IgG binding (R-phycoerythrin, FL-2) was normalized with the signal corresponding to the expression level (Alexa Fluor 647, FL-4). The results show a clear ranking of the three affibody molecules, with Zwt as the strongest binder, followed by ZN28A and ZK35A as the weakest (Fig. 2), which correlates with previously published data (6).
FIG. 2.
Histogram representation of the results from the rapid on-cell affinity ranking of the three different affibody molecules, Zwt, ZN28A, and ZK35A, using flow cytometry. y axis, MFI corresponding to the IgG binding (FL-2) normalized to the MFI corresponding to the surface expression level (FL-4). x axis, concentration of biotinylated human IgG. Note that due to differences in signal intensities between days, all MFIs within one experiment (performed on one day) are normalized to the intensity corresponding to Zwt incubated with 4.1 nM human IgG. Error bars indicate standard deviations.
Extraction and purification of surface-anchored proteins for biosensor analysis.
The cell wall-anchored affibody molecules, Zwt, ZN28A, and ZK35A, and ABDwt were extracted from the cell wall by lysostaphin treatment and sonication of overnight cell cultures followed by HSA affinity chromatography and low-pH elution. Eluted fractions were evaluated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis under reducing conditions, which showed that the extracted and affinity-purified cell wall proteins were pure and that no degradation products were present (data not shown). The purified cell wall proteins were then immobilized as follows on CM5 Biacore chips. On chip one, Zwt, ZN28A, and ZK35A were immobilized, on one surface each, to a final response of around 500 RU. To account for unspecific binding, IgG was immobilized on one flow cell surface, serving as a blank reference surface. On chip two. ABDwt was immobilized on two of the flow cell surfaces to a final response of around 400 RU. The blank surface was treated like the other surfaces on the chip but with no protein injected.
Determination of KD of affibody molecules using SPR.
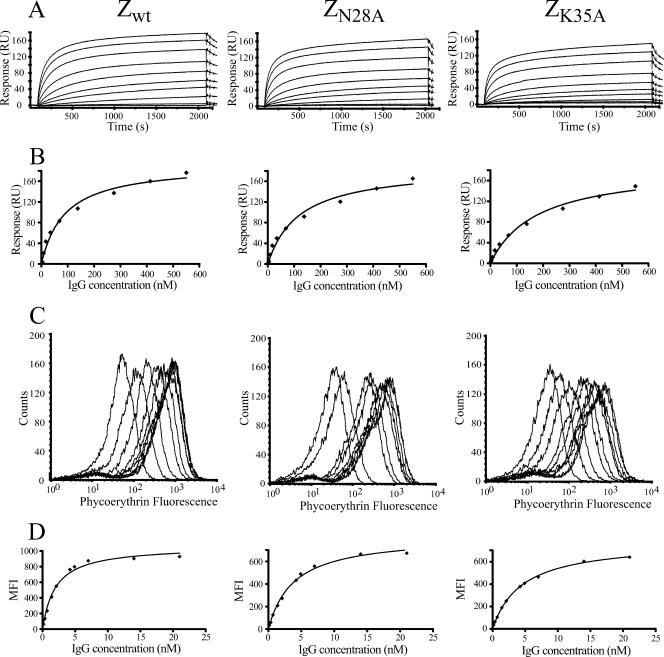
In order to determine the apparent KD of the affibody molecules using SPR, different concentrations of IgG were injected over the previously described chip one. The response at equilibrium binding was collected for each concentration, and the data were fitted to a monovalent binding model to determine the apparent KD (Table 1; Fig. 3A and B). The results were in accordance with the rapid on-cell relative ranking and also with previously published data (Fig. 2) (6).
TABLE 1.
KD determined both on cells using flow cytometry and by SPR
| Binding pair |
KD (nM [mean ± SD])
|
KD ratio (SPR/on cells) | |
|---|---|---|---|
| SPRa | On cellsb | ||
| Zwt/IgG | 92 ± 6 | 2.1 ± 0.3 | 43 |
| ZN28A/IgG | 104 ± 2 | 2.8 ± 0.8 | 37 |
| ZK35A/IgG | 169 ± 4 | 3.7 ± 0.8 | 46 |
Performed in duplicate on different days with duplicates in each run.
Performed in triplicate on different days.
FIG. 3.
Determination of the KD for three different affibody molecules (Zwt, ZN28A, and ZK35A). (A) Determination of KD values using SPR analysis. Sensorgrams show the response after injection of 10 different concentrations of IgG. (B) Binding isotherms, with the response at equilibrium from the SPR analysis on the y axis and the IgG concentration on the x axis. The solid line is the equation obtained from fitting the data to a monovalent binding model. (C) On-cell determination of KD values for the three different affibody molecules using flow cytometry. Histograms showing the IgG binding signal after incubation with 10 different IgG concentrations and flow cytometric analysis. (D) Binding isotherms, with the MFI from the flow cytometric analysis on the y axis and the IgG concentration on the x axis. The solid line is the equation obtained from fitting the data to a monovalent binding model.
On-cell determination of KD of affibody molecules using flow cytometry.
The apparent KD of the affibody molecules were determined on cells by immunofluorescent labeling of cells by incubation with various concentrations of biotinylated IgG for 1 hour to reach equilibrium, followed by a washing step and incubation with streptavidin (R-phycoerythrin conjugate). The MFI was measured by flow cytometry, and the fluorescence data were fitted, as for the SPR data, to a monovalent binding model to determine the apparent KD (Table 1; Fig. 3C and D). The results were reproducible and show that the relative values of the KD obtained for Zwt, ZN28A, and ZK35A agree with the rapid on-cell ranking (Fig. 2) and that the relative affinities are very similar to the results obtained using SPR analysis (Table 1; Fig. 3A and B). However, the absolute affinities differ around 40-fold between the two methods (Table 1).
Determinations of KD and koff of ABDwt using SPR.
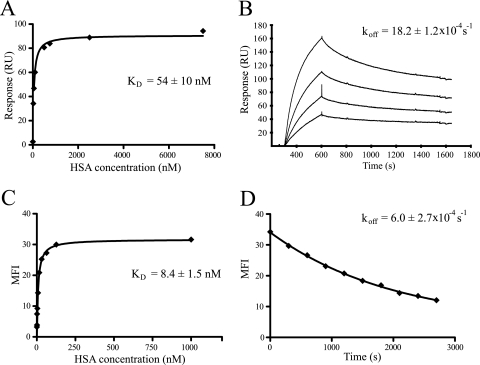
In order to determine the KD of ABDwt using SPR, different concentrations of HSA were injected over the previously described chip two. The response at equilibrium binding was collected for each concentration, and the data were fitted to a monovalent binding model to determine the KD (Fig. 4A).
FIG. 4.
Determination of both KD and koff for ABDwt. (A) Determination of KD using SPR analysis. Binding isotherms show the response at equilibrium from the SPR analysis on the y axis and the HSA concentration on the x axis. The solid line is the equation obtained from fitting the data to a monovalent binding model. (B) Determination of koff values using SPR analysis. Dissociation data from the four different concentrations were fitted to a first-order kinetic model. (C) On-cell determination of KD using flow cytometry. Binding isotherms show the MFI from the flow cytometric analysis on the y axis and the HSA concentration on the x axis. The solid line is the equation obtained from fitting the data to a monovalent binding model. (D) On-cell determination of koff values using flow cytometry. The MFI from the flow cytometric analysis on the y axis is plotted against time on the x axis. The solid line is the equation obtained from fitting the data to a first-order kinetic model.
The koff of the ABDwt and HSA pair was determined using SPR by applying different concentrations of HSA to the flow cells of chip two. After association, HBS buffer was injected for 15 min for the collection of dissociation data. The data were fitted to a first-order kinetic model to determine the koff for the complex (Fig. 4B).
On-cell determinations of KD and koff of ABDwt using flow cytometry.
The KD of ABDwt were determined on cells by immunofluorescent labeling of cells by incubation in various concentrations of labeled HSA for 1 hour to reach equilibrium, followed by a washing step. The MFI was measured by flow cytometry, and the fluorescence data were fitted, as for the SPR data, to a monovalent binding model to determine the KD (Fig. 4C). The results were reproducible and show that the values of the KD obtained for ABDwt are similar to the results obtained using SPR analysis (Fig. 4A), but still with a sixfold difference between the two methods.
To investigate the possibility of on-cell determination the koff of the ABDwt and HSA pair using flow cytometry, Sc:ABDwt cells were incubated in Alexa Fluor 488 (HSA conjugate) to saturate all surface-displayed binding sites. After washing and incubation with unlabeled HSA to minimize reassociation, samples were taken at different time points and the MFI was measured for each sample in the flow cytometer. The data were thereafter fitted to a first-order kinetic model to determine the koff of the ABDwt/HSA complex (Fig. 4D). The results showed a high reproducibility and were in agreement with the results obtained using SPR analysis (Fig. 4B), but they did differ by around threefold.
DISCUSSION
In this study we have investigated the use of staphylococcal cell surface display together with flow cytometry as an alternative to more traditional SPR technology (i.e., Biacore) for rapid on-cell affinity determination of binding molecules previously selected from a staphylococcus-displayed combinatorial protein engineering library. A model system consisting of three affibody variants (Zwt, ZN28A, and ZK35A) with different affinities for IgG and an albumin binding domain (ABDwt) with affinity for HSA was used to compare the on-cell method with the established SPR technology. The three affibody molecules were expressed on the surface of S. carnosus cells using our general surface display vector pSCXm, incubated with three different concentrations of IgG, and analyzed in order to perform a very rapid ranking of the relative affinities. To avoid biases due to cell-to-cell variations in surface expression levels, a dual-labeling technique (described previously [20]) was employed for parallel monitoring of the expression levels of individual cells and the subsequent normalization of the binding signal. The normalized fluorescence intensities showed an affinity ranking that was in accordance with previously published data. This result is of significant importance, since the ability to perform a rapid affinity screen, with few target concentrations, of a large set of candidate binders obtained from a cell surface-displayed protein library is essential to obtain a high throughput in the binding analysis. After the initial ranking, the affinities were determined directly on the cells by incubating the cells with different concentrations of the target followed by flow cytometric analysis. The equilibrium mean fluorescence intensities were then fitted using nonlinear regression to a monovalent binding model, and the apparent KD were calculated. All experiments were performed in triplicates and showed a very high reproducibility. In order to compare the results to those of the well-established SPR technology, the different affibody variants were extracted from the staphylococcal cell wall using enzymatic digestion, purified, and immobilized on a Biacore chip surface. Producing and purifying the affibody molecules in this manner minimized variability between the methods due to differences in production host and fusion environment. Equilibrium data were collected and fitted to the binding model using the same software as for the flow cytometry data. Interestingly, when comparing the affinities obtained using the two methods, a 40-fold difference was observed. However, the difference was highly reproducible for all three affibody variants, making the relative order and ratios almost identical for the two methods. The rather large difference between the absolute affinities observed here may have several possible explanations. First, the proteins on the Biacore chip surface have been extracted, purified, and immobilized, and there is a risk that at least a portion of the proteins has lost some binding functionality. Furthermore, since the proteins are immobilized using amine coupling chemistry, resulting in a random orientation on the chip surface, a number of proteins will obtain a conformation not optimal for target binding. In contrast to the Biacore system, when analyzing the staphylococcus-derived affibody molecules on cells using the anchoring system from staphylococcal protein A, it is more likely that they are displayed in a functional manner. In addition, the affibody variants bind to an epitope on the Fc part and thereby have two possible binding sites per IgG molecule. This will lead to avidity effects, which are more or less pronounced depending on the conformation and density of the proteins on the surface. In order to determine how large a part of the overall difference between the methods was due to avidity, the ABDwt and HSA pair, a binding pair with true monovalent binding, was analyzed using both techniques. As expected, the difference was now much smaller, approximately sixfold, confirming that the avidity effect accounted for a large part of the difference when analyzing the affibody/IgG pairs. The observed difference in absolute affinity is not surprising, since differences between alternative techniques are known to be common due to the possibilities discussed above. However, it is easier to underestimate than to overestimate an affinity due to loss in functionality, and a higher affinity would therefore more often be closer to the affinity of the fully functional pair. In either case, most important are that the technique is robust and results in a reproducible output that can accurately rank a large set of candidates and that the difference from another technique is known when a comparison is made. Finally, to show the versatile use of the cell display platform for binding analysis of selected binders, an experiment was performed with the ABDwt and HSA pair to determine the koff of the binding reaction. The results were again compared to results obtained with the SPR technology, showing a threefold lower dissociation rate using the on-cell technique, in accordance with the previous KD determinations. This also indicates that the differences between the two techniques affect the association rate as much as the dissociation rate of the reaction. The possibility of performing on-cell determinations of the koff and thereby also be able to calculate the association rate constant is very useful for thorough characterization of interesting binders.
In conclusion, we have shown that our staphylococcal surface display system is a very rapid and useful platform for characterizing selected affinity proteins directly on the cells without any need for subcloning, protein production, and purification. In addition, to increasing the throughput even more, the system would easily be amenable to automation using liquid handling robots together with multiwell plates and analysis using a plate-reading spectrofluorometer. The results presented here show that staphylococcal surface display has several advantages over phage display, not only in the actual selection process but also in the time-consuming downstream characterization part. Ultimately, it is hoped that the increased number of selected clones that it will be possible to analyze with our cell display method will lead to a higher success rate in the discovery of novel affinity proteins.
Acknowledgments
We thank Lena Jendeberg for providing clones of the Z variants.
This work was financed by a grant (no. 621-2003-2876) from the Swedish Research Council (VR).
Footnotes
Published ahead of print on 14 September 2007.
REFERENCES
- 1.Adams, G. P., and L. M. Weiner. 2005. Monoclonal antibody therapy of cancer. Nat. Biotechnol. 23:1147-1157. [DOI] [PubMed] [Google Scholar]
- 2.Andreoni, C., L. Goetsch, C. Libon, P. Samuelson, T. N. Nguyen, A. Robert, M. Uhlén, H. Binz, and S. Ståhl. 1997. Flow cytometric quantification of surface-displayed recombinant receptors on staphylococci. BioTechniques 23:696-702:704. [PubMed] [Google Scholar]
- 3.Bertschinger, J., D. Grabulovski, and D. Neri. 2007. Selection of single domain binding proteins by covalent DNA display. Protein Eng. Des. Sel. 20:57-68. [DOI] [PubMed] [Google Scholar]
- 4.Bessette, P. H., J. J. Rice, and P. S. Daugherty. 2004. Rapid isolation of high-affinity protein binding peptides using bacterial display. Protein Eng. Des. Sel. 17:731-739. [DOI] [PubMed] [Google Scholar]
- 5.Boder, E. T., and K. D. Wittrup. 1998. Optimal screening of surface-displayed polypeptide libraries. Biotechnol. Prog. 14:55-62. [DOI] [PubMed] [Google Scholar]
- 6.Cedergren, L., R. Andersson, B. Jansson, M. Uhlén, and B. Nilsson. 1993. Mutational analysis of the interaction between staphylococcal protein A and human IgG1. Protein Eng. 6:441-448. [DOI] [PubMed] [Google Scholar]
- 7.Chen, G., A. Hayhurst, J. G. Thomas, B. R. Harvey, B. L. Iverson, and G. Georgiou. 2001. Isolation of high-affinity ligand-binding proteins by periplasmic expression with cytometric screening (PECS). Nat. Biotechnol. 19:537-542. [DOI] [PubMed] [Google Scholar]
- 8.Daugherty, P. S., G. Chen, M. J. Olsen, B. L. Iverson, and G. Georgiou. 1998. Antibody affinity maturation using bacterial surface display. Protein Eng. 11:825-832. [DOI] [PubMed] [Google Scholar]
- 9.Feldhaus, M. J., R. W. Siegel, L. K. Opresko, J. R. Coleman, J. M. Feldhaus, Y. A. Yeung, J. R. Cochran, P. Heinzelman, D. Colby, J. Swers, C. Graff, H. S. Wiley, and K. D. Wittrup. 2003. Flow-cytometric isolation of human antibodies from a nonimmune Saccharomyces cerevisiae surface display library. Nat. Biotechnol. 21:163-170. [DOI] [PubMed] [Google Scholar]
- 10.Gill, D. S., and N. K. Damle. 2006. Biopharmaceutical drug discovery using novel protein scaffolds. Curr. Opin. Biotechnol. 17:653-658. [DOI] [PubMed] [Google Scholar]
- 11.Götz, F. 1990. Staphylococcus carnosus: a new host organism for gene cloning and protein production. Soc. Appl. Bacteriol. Symp. Ser. 19:49S-53S. [DOI] [PubMed] [Google Scholar]
- 12.Grönwall, C., A. Jonsson, S. Lindström, E. Gunneriusson, S. Ståhl, and N. Herne. 2007. Selection and characterization of Affibody ligands binding to Alzheimer amyloid beta peptides. J. Biotechnol. 128:162-183. [DOI] [PubMed] [Google Scholar]
- 13.Hanes, J., and A. Plückthun. 1997. In vitro selection and evolution of functional proteins by using ribosome display. Proc. Natl. Acad. Sci. USA 94:4937-4942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harvey, B. R., G. Georgiou, A. Hayhurst, K. J. Jeong, B. L. Iverson, and G. K. Rogers. 2004. Anchored periplasmic expression, a versatile technology for the isolation of high-affinity antibodies from Escherichia coli-expressed libraries. Proc. Natl. Acad. Sci. USA 101:9193-9198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Karlsson, R. 2004. SPR for molecular interaction analysis: a review of emerging application areas. J. Mol. Recognit. 17:151-161. [DOI] [PubMed] [Google Scholar]
- 16.Koch, H., N. Grafe, R. Schiess, and A. Plückthun. 2006. Direct selection of antibodies from complex libraries with the protein fragment complementation assay. J. Mol. Biol. 357:427-441. [DOI] [PubMed] [Google Scholar]
- 17.Lehtiö, J., H. Wernérus, P. Samuelson, T. T. Teeri, and S. Ståhl. 2001. Directed immobilization of recombinant staphylococci on cotton fibers by functional display of a fungal cellulose-binding domain. FEMS Microbiol. Lett. 195:197-204. [DOI] [PubMed] [Google Scholar]
- 18.Lipovsek, D., and A. Plückthun. 2004. In-vitro protein evolution by ribosome display and mRNA display. J. Immunol. Methods 290:51-67. [DOI] [PubMed] [Google Scholar]
- 19.Löfblom, J., N. Kronqvist, M. Uhlén, S. Ståhl, and H. Wernérus. 2007. Optimization of electroporation-mediated transformation: Staphylococcus carnosus as model organism. J. Appl. Microbiol. 102:736-747. [DOI] [PubMed] [Google Scholar]
- 20.Löfblom, J., H. Wernérus, and S. Ståhl. 2005. Fine affinity discrimination by normalized fluorescence activated cell sorting in staphylococcal surface display. FEMS Microbiol. Lett. 248:189-198. [DOI] [PubMed] [Google Scholar]
- 21.Mattheakis, L. C., R. R. Bhatt, and W. J. Dower. 1994. An in vitro polysome display system for identifying ligands from very large peptide libraries. Proc. Natl. Acad. Sci. USA 91:9022-9026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mazmanian, S. K., G. Liu, H. Ton-That, and O. Schneewind. 1999. Staphylococcus aureus sortase, an enzyme that anchors surface proteins to the cell wall. Science 285:760-763. [DOI] [PubMed] [Google Scholar]
- 23.McCafferty, J., A. D. Griffiths, G. Winter, and D. J. Chiswell. 1990. Phage antibodies: filamentous phage displaying antibody variable domains. Nature 348:552-554. [DOI] [PubMed] [Google Scholar]
- 24.Moks, T., L. Abrahmsén, B. Nilsson, U. Hellman, J. Sjöquist, and M. Uhlén. 1986. Staphylococcal protein A consists of five IgG-binding domains. Eur. J. Biochem. 156:637-643. [DOI] [PubMed] [Google Scholar]
- 25.Nilsson, B., T. Moks, B. Jansson, L. Abrahmsén, A. Elmblad, E. Holmgren, C. Henrichson, T. A. Jones, and M. Uhlén. 1987. A synthetic IgG-binding domain based on staphylococcal protein A. Protein Eng. 1:107-113. [DOI] [PubMed] [Google Scholar]
- 26.Nilsson, F. Y., and V. Tolmachev. 2007. Affibody molecules: new protein domains for molecular imaging and targeted tumor therapy. Curr. Opin. Drug Discov. Dev. 10:167-175. [PubMed] [Google Scholar]
- 27.Nord, K., E. Gunneriusson, J. Ringdahl, S. Ståhl, M. Uhlén, and P. Å. Nygren. 1997. Binding proteins selected from combinatorial libraries of an alpha-helical bacterial receptor domain. Nat. Biotechnol. 15:772-777. [DOI] [PubMed] [Google Scholar]
- 28.Nord, O., M. Uhlén, and P. Å. Nygren. 2003. Microbead display of proteins by cell-free expression of anchored DNA. J. Biotechnol. 106:1-13. [DOI] [PubMed] [Google Scholar]
- 29.Odegrip, R., D. Coomber, B. Eldridge, R. Hederer, P. A. Kuhlman, C. Ullman, K. FitzGerald, and D. McGregor. 2004. CIS display: in vitro selection of peptides from libraries of protein-DNA complexes. Proc. Natl. Acad. Sci. USA 101:2806-2810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Orlova, A., M. Magnusson, T. L. Eriksson, M. Nilsson, B. Larsson, I. Hoiden-Guthenberg, C. Widström, J. Carlsson, V. Tolmachev, S. Ståhl, and F. Y. Nilsson. 2006. Tumor imaging using a picomolar affinity HER2 binding affibody molecule. Cancer Res. 66:4339-4348. [DOI] [PubMed] [Google Scholar]
- 31.Orlova, A., V. Tolmachev, R. Pehrson, M. Lindborg, T. Tran, M. Sandström, F. Y. Nilsson, A. Wennborg, L. Abrahmsén, and J. Feldwisch. 2007. Synthetic affibody molecules: a novel class of affinity ligands for molecular imaging of HER2-expressing malignant tumors. Cancer Res. 67:2178-2186. [DOI] [PubMed] [Google Scholar]
- 32.Rader, C., and C. F. Barbas III. 1997. Phage display of combinatorial antibody libraries. Curr. Opin. Biotechnol. 8:503-508. [DOI] [PubMed] [Google Scholar]
- 33.Rice, J. J., A. Schohn, P. H. Bessette, K. T. Boulware, and P. S. Daugherty. 2006. Bacterial display using circularly permuted outer membrane protein OmpX yields high affinity peptide ligands. Protein Sci. 15:825-836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Robinson, M. K., M. Doss, C. Shaller, D. Narayanan, J. D. Marks, L. P. Adler, D. E. Gonzalez Trotter, and G. P. Adams. 2005. Quantitative immuno-positron emission tomography imaging of HER2-positive tumor xenografts with an iodine-124 labeled anti-HER2 diabody. Cancer Res. 65:1471-1478. [DOI] [PubMed] [Google Scholar]
- 35.Rüther, U. 1982. pUR 250 allows rapid chemical sequencing of both DNA strands of its inserts. Nucleic Acids Res. 10:5765-5772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Samuelson, P., F. Cano, A. Robert, and S. Ståhl. 1999. Engineering of a Staphylococcus carnosus surface display system by substitution or deletion of a Staphylococcus hyicus lipase propeptide. FEMS Microbiol. Lett. 179:131-139. [DOI] [PubMed] [Google Scholar]
- 37.Samuelson, P., E. Gunneriusson, P. Å. Nygren, and S. Ståhl. 2002. Display of proteins on bacteria. J. Biotechnol. 96:129-154. [DOI] [PubMed] [Google Scholar]
- 38.Samuelson, P., M. Hansson, N. Ahlborg, C. Andreoni, F. Götz, T. Bachi, T. N. Nguyen, H. Binz, M. Uhlén, and S. Ståhl. 1995. Cell surface display of recombinant proteins on Staphylococcus carnosus. J. Bacteriol. 177:1470-1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Samuelson, P., H. Wernérus, M. Svedberg, and S. Ståhl. 2000. Staphylococcal surface display of metal-binding polyhistidyl peptides. Appl. Environ. Microbiol. 66:1243-1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schneewind, O., A. Fowler, and K. F. Faull. 1995. Structure of the cell wall anchor of surface proteins in Staphylococcus aureus. Science 268:103-106. [DOI] [PubMed] [Google Scholar]
- 41.Sepp, A., D. S. Tawfik, and A. D. Griffiths. 2002. Microbead display by in vitro compartmentalisation: selection for binding using flow cytometry. FEBS Lett. 532:455-458. [DOI] [PubMed] [Google Scholar]
- 42.Sjölander, A., P. Å. Nygren, S. Ståhl, K. Berzins, M. Uhlén, P. Perlmann, and R. Andersson. 1997. The serum albumin-binding region of streptococcal protein G: a bacterial fusion partner with carrier-related properties. J. Immunol. Methods 201:115-123. [DOI] [PubMed] [Google Scholar]
- 43.Smith, G. P. 1985. Filamentous fusion phage: novel expression vectors that display cloned antigens on the virion surface. Science 228:1315-1317. [DOI] [PubMed] [Google Scholar]
- 44.Ståhl, S., J. Nilsson, S. Hober, M. Uhlén, and P. Å. Nygren. 1999. Affinity fusions, gene expression, p. 49-63. In M. C. Flickinger and S. W. Drew (ed.), Encyclopedia of bioprocess technology: fermentation, biocatalysis, and bioseparation. Wiley Biotechnology Encyclopedias, vol. 5. John Wiley, New York, NY. [Google Scholar]
- 45.Ståhl, S., A. Robert, E. Gunneriusson, H. Wernérus, F. Cano, S. Liljeqvist, M. Hansson, T. N. Nguyen, and P. Samuelson. 2000. Staphylococcal surface display and its applications. Int. J. Med. Microbiol. 290:571-577. [DOI] [PubMed] [Google Scholar]
- 46.Ståhl, S., A. Sjölander, P. A. Nygren, K. Berzins, P. Perlmann, and M. Uhlén. 1989. A dual expression system for the generation, analysis and purification of antibodies to a repeated sequence of the Plasmodium falciparum antigen Pf155/RESA. J. Immunol. Methods 124:43-52. [DOI] [PubMed] [Google Scholar]
- 47.Taussig, M. J., O. Stoevesandt, C. A. Borrebaeck, A. R. Bradbury, D. Cahill, C. Cambillau, A. de Daruvar, S. Dubel, J. Eichler, R. Frank, T. J. Gibson, D. Gloriam, L. Gold, F. W. Herberg, H. Hermjakob, J. D. Hoheisel, T. O. Joos, O. Kallioniemi, M. Koegl, Z. Konthur, B. Korn, E. Kremmer, S. Krobitsch, U. Landegren, S. van der Maarel, J. McCafferty, S. Muyldermans, P. Å. Nygren, S. Palcy, A. Plückthun, B. Polic, M. Przybylski, P. Saviranta, A. Sawyer, D. J. Sherman, A. Skerra, M. Templin, M. Ueffing, and M. Uhlén. 2007. ProteomeBinders: planning a European resource of affinity reagents for analysis of the human proteome. Nat. Methods 4:13-17. [DOI] [PubMed] [Google Scholar]
- 48.Ton-That, H., S. K. Mazmanian, K. F. Faull, and O. Schneewind. 2000. Anchoring of surface proteins to the cell wall of Staphylococcus aureus. Sortase catalyzed in vitro transpeptidation reaction using LPXTG peptide and NH(2)-Gly(3) substrates. J. Biol. Chem. 275:9876-9881. [DOI] [PubMed] [Google Scholar]
- 49.Uhlén, M., E. Björling, C. Agaton, C. A. Szigyarto, B. Amini, E. Andersen, A. C. Andersson, P. Angelidou, A. Asplund, C. Asplund, L. Berglund, K. Bergström, H. Brumer, D. Cerjan, M. Ekström, A. Elobeid, C. Eriksson, L. Fagerberg, R. Falk, J. Fall, M. Forsberg, M. G. Björklund, K. Gumbel, A. Halimi, I. Hallin, C. Hamsten, M. Hansson, M. Hedhammar, G. Hercules, C. Kampf, K. Larsson, M. Lindskog, W. Lodewyckx, J. Lund, J. Lundeberg, K. Magnusson, E. Malm, P. Nilsson, J. Ödling, P. Oksvold, I. Olsson, E. Öster, J. Ottosson, L. Paavilainen, A. Persson, R. Rimini, J. Rockberg, M. Runeson, A. Sivertsson, A. Sköllermo, J. Steen, M. Stenvall, F. Sterky, S. Strömberg, M. Sundberg, H. Tegel, S. Tourle, E. Wahlund, A. Walden, J. Wan, H. Wernérus, J. Westberg, K. Wester, U. Wrethagen, L. L. Xu, S. Hober, and F. Pontén. 2005. A human protein atlas for normal and cancer tissues based on antibody proteomics. Mol. Cell. Proteomics 4:1920-1932. [DOI] [PubMed] [Google Scholar]
- 50.Weissleder, R. 2006. Molecular imaging in cancer. Science 312:1168-1171. [DOI] [PubMed] [Google Scholar]
- 51.Wernérus, H., J. Lehtiö, P. Samuelson, and S. Ståhl. 2002. Engineering of staphylococcal surfaces for biotechnological applications. J. Biotechnol. 96:67-78. [DOI] [PubMed] [Google Scholar]
- 52.Wernérus, H., J. Lehtiö, T. Teeri, P. Å. Nygren, and S. Ståhl. 2001. Generation of metal-binding staphylococci through surface display of combinatorially engineered cellulose-binding domains. Appl. Environ. Microbiol. 67:4678-4684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wernérus, H., P. Samuelson, and S. Ståhl. 2003. Fluorescence-activated cell sorting of specific affibody-displaying staphylococci. Appl. Environ. Microbiol. 69:5328-5335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wernérus, H., and S. Ståhl. 2004. Biotechnological applications for surface-engineered bacteria. Biotechnol. Appl. Biochem. 40:209-228. [DOI] [PubMed] [Google Scholar]
- 55.Wernérus, H., and S. Ståhl. 2002. Vector engineering to improve a staphylococcal surface display system. FEMS Microbiol. Lett. 212:47-54. [DOI] [PubMed] [Google Scholar]
- 56.Wittrup, K. D. 2001. Protein engineering by cell-surface display. Curr. Opin. Biotechnol. 12:395-399. [DOI] [PubMed] [Google Scholar]
- 57.Zahnd, C., P. Amstutz, and A. Plückthun. 2007. Ribosome display: selecting and evolving proteins in vitro that specifically bind to a target. Nat. Methods 4:269-279. [DOI] [PubMed] [Google Scholar]