Abstract
Stenoxybacter acetivorans is a newly described, obligately microaerophilic β-proteobacterium that is abundant in the acetate-rich hindgut of Reticulitermes. Here we tested the hypotheses that cells are located in the hypoxic, peripheral region of Reticulitermes flavipes hindguts and use acetate to fuel their O2-consuming respiratory activity in situ. Physical fractionation of R. flavipes guts, followed by limited-cycle PCR with S. acetivorans-specific 16S rRNA gene primers, indicated that cells of this organism were indeed located primarily among the microbiota colonizing the hindgut wall. Likewise, reverse transcriptase PCR of hindgut RNA revealed S. acetivorans-specific transcripts for acetate-activating enzymes that were also found in cell extracts (acetate kinase and phosphotransacetylase), as well as transcripts of ccoN, which encodes the O2-reducing subunit of high-affinity cbb3-type cytochrome oxidases. However, S. acetivorans strains did not possess typical enzymes of the glyoxylate cycle (isocitrate lyase and malate synthase A), suggesting that they may use an alternate pathway to replenish tricarboxylic acid cycle intermediates or they obtain such compounds (or their precursors) in situ. Respirometric measurements indicated that much of the O2 consumption by R. flavipes worker larvae was attributable to their guts, and the potential contribution of S. acetivorans to O2 consumption by extracted guts was about 0.2%, a value similar to that obtained for other hindgut bacteria examined. Similar measurements obtained with guts of larvae prefed diets to disrupt major members of the hindgut microbiota implied that most of the O2 consumption observed with extracted guts was attributable to protozoans, a group of microbes long thought to be “strict anaerobes.”
In termite hindguts, fermentative production of acetate—a major carbon and energy source for the insect—depends on efficient removal of inwardly diffusing O2 by microbes residing on and near the hindgut wall (8, 52). However, not much is known about the nature of such organisms or the substrate(s) that fuels their O2-consuming respiratory activity, although the level of acetate in hindgut fluid (60 to 80 mM) (38) makes it a reasonable default candidate. In a companion paper, we report on the isolation and identification of bacterial isolates from guts of Reticulitermes flavipes that may be important O2 consumers in situ (55). Abundant among such isolates were strains of a novel acetate-oxidizing, obligately microaerophilic β-proteobacterium, Stenoxybacter acetivorans gen. nov., sp. nov., that appeared to be autochthonous in hindguts of R. flavipes and were present in genetically distinct populations of Reticulitermes collected from different geographical locations. Owing to their abundance, their obligately microaerophilic nature, and their restriction to a relatively narrow range of substrates, including acetate, as oxidizable energy sources, it seemed reasonable to hypothesize that S. acetivorans cells reside in the hypoxic, peripheral region of R. flavipes hindguts and use a high-affinity terminal oxidase for acetate-fueled respiratory activity in situ. Among the information needed to test such hypotheses, however, was the nature of S. acetivorans enzymes (and their encoding genes) specific to acetate utilization and oxygen consumption, as well as a demonstration that such genes (and, by inference, their gene products) were actually expressed in situ.
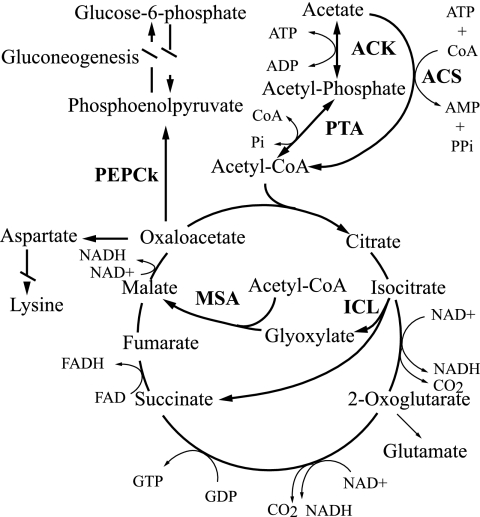
The key enzymes specific to acetate utilization by microbes are the enzymes that activate acetate to acetyl coenzyme A (acetyl-CoA) prior to its entry into dissimilatory and assimilatory pathways. Among these are acetate kinase (EC 2.7.2.1) and phosphotransacetylase (EC 2.3.1.8), which catalyze the ATP-dependent phosphorylation of acetate to acetyl-phosphate and then its transacetylation to acetyl-CoA, respectively (Fig. 1). Acetate kinase and phosphotransacetylase are the primary enzymes of acetate activation in organisms such as Corynebacterium glutamicum and Methanosarcina thermophila (1, 18). By contrast, in many other organisms, such as members of the Enterobacteriaceae, the acetate kinase-phosphotransacetylase enzyme pair functions in the reverse direction, primarily during anaerobic growth, and acetate is a major fermentation product and is excreted. This allows additional ATP to be made by substrate-level phosphorylation (11, 39, 58). When grown aerobically on acetate as a carbon and energy source, members of the Enterobacteriaceae and many other microbes express an acetate-inducible AMP-forming acetyl-CoA synthetase (EC 6.2.1.1), which catalyzes the conversion of acetate, ATP, and CoA to acetyl-CoA, AMP, and inorganic pyrophosphate (PPi) (7, 26, 27) (Fig. 1). However, the AMP-forming acetyl-CoA synthetase is a high-affinity acetate-activating enzyme (Km for acetate, ∼200 μM) (58) and consumes the equivalent of two high-energy phosphate bonds owing to production, from ATP, of both AMP and PPi, the latter of which is usually hydrolyzed by endogenous pyrophosphatase to pull the reaction. By contrast, use of the low-affinity acetate kinase (Km for acetate, 7 to 10 mM) (58) with phosphotransacetylase consumes only one high-energy phosphate bond and is a more energetically efficient way to activate acetate if the ambient concentration of acetate is relatively high. Other, less widely distributed acetate-activating enzymes include a pyrophosphate-dependent acetylkinase (EC 2.7.2.12) found in Entamoeba histolytica (59) and an ADP-forming acetyl-CoA synthetase (EC 6.2.1.13) found in Archaea and some Eukarya, which is structurally and evolutionarily unrelated to AMP-forming acetyl-CoA synthetase (23, 36, 58).
FIG. 1.
Common pathways for acetate activation and metabolism. Enzymes involved in acetate activation are acetate kinase (ACK) together with phosphotransacetylase (PTA) or AMP-forming acetyl-CoA synthetase (ACS). Once activated, acetyl-CoA proceeds through the TCA cycle. Typically, if acetate is the sole carbon and energy source, replenishment of TCA cycle intermediates drawn off for biosynthesis (such as 2-oxoglutarate or oxaloacetate) is accomplished by the combination of isocitrate lyase (ICL) and malate synthase A (MSA). For gluconeogenesis from acetate, oxaloacetate is drawn off the TCA cycle via phosphoenolpyruvate carboxykinase (PEPCk). FAD, flavin adenine dinucleotide; FADH, reduced flavin adenine dinucleotide.
When acetate is the sole or primary carbon and energy source for a bacterium such as Escherichia coli, further catabolism of acetyl-CoA occurs via the tricarboxylic acid (TCA) cycle. However, the TCA cycle alone does not allow for net assimilation of acetate carbon. Under these conditions, the anaplerotic glyoxylate cycle is derepressed and serves to replenish TCA cycle intermediates derived from acetate catabolism and drawn off for biosynthesis (11, 15) (Fig. 1). The two key enzymes of the glyoxylate cycle are isocitrate lyase (EC 4.1.3.1) and malate synthase A (EC 2.3.3.9). Each turn of the glyoxylate cycle results in the net formation of one molecule of malate from two molecules of acetyl-CoA (14).
Among the high-affinity oxidases used to mediate terminal electron transfer to oxygen, especially during aerobic respiration at low ambient oxygen concentrations, are the cbb3-type cytochrome oxidases. These enzymes have been referred to as “specialized enzymes at the heart of microaerobic metabolism” (41). Many so-called microaerophilic microorganisms or organisms that spend at least part of their life in low-O2 environments, such as the N2-fixing root nodule symbiont Bradyrhizobium japonicum or the gastric mucosa-colonizing pathogen Helicobacter pylori, utilize a cbb3-type terminal cytochrome oxidase (32, 42). In contrast to the well-studied cytochrome aa3 oxidases, which have an estimated Km for oxygen of 0.1 to 1 μM, the cbb3 oxidases have estimated Km values in the nanomolar range (e.g., 7 nM for B. japonicum) (42). The ability of organisms utilizing these high-affinity oxidases to “pull” oxygen down to very low concentrations presumably minimizes the production of reactive oxygen species (to which microaerophiles may be particularly sensitive) and also allows O2-sensitive processes such as N2 fixation to occur, as it does in B. japonicum bacteroids in root nodules (42). The typically greater energy yield of aerobic metabolism than of anaerobic metabolism may give a competitive edge to organisms with a cbb3-type terminal oxidase in low-O2 environments. However, physiological studies with cells, as well as experiments with purified cbb3 oxidase reconstituted into phospholipid vesicles, have suggested that the cbb3-type oxidase is significantly less efficient at transducing energy than cytochrome aa3 (48).
In this study, a combination of molecular and physiological approaches were used to (i) determine the primary location of S. acetivorans cells in situ; (ii) identify key genes and gene products involved in acetate utilization and oxygen consumption by S. acetivorans in vitro; and (iii) determine whether the same genes (and, by inference, their gene products) were expressed by cells of S. acetivorans in termite hindguts. Experiments were also done to estimate the potential contribution of S. acetivorans to overall O2 consumption in hindguts of R. flavipes, as well as the relative contributions of major microbial groups (i.e., bacteria and protozoans) to O2 consumption by extracted guts.
MATERIALS AND METHODS
Termites.
Worker larvae of R. flavipes (Kollar) (Rhinotermitidae) were collected in Dansville and Spring Arbor, MI, as described previously (6, 38).
Microorganisms and growth conditions.
S. acetivorans strains, including type strain TAM-DN1, and Citrobacter sp. strain RFC-10 were isolated from guts of R. flavipes (55). Routine cultivation was performed in an acetate-yeast extract-salts broth (BYA medium) under an hypoxic atmosphere (2% O2, 5% CO2, 93% N2), and growth was monitored by measuring the optical density of cultures, which in turn was related to cell density, cell protein, and dry cell mass, as previously described (55).
In situ location of S. acetivorans.
To determine the primary location of S. acetivorans cells in hindguts, four worker larvae were degutted, and the guts were pooled in a 100-μl drop of basal salts solution (BSS) (55) in a sterile petri dish. Midguts were separated from the hindguts, and the hindguts were then transferred into a fresh 100-μl drop of BSS, sliced longitudinally, and washed by gentle agitation using sterile forceps. The 100 μl of BSS (now visibly turbid with gut microbes) was transferred into a sterile, 10-ml conical centrifuge tube. The remaining sliced gut fragments were transferred to a separate 1.5-ml centrifuge tube containing 0.5 ml of BSS and were vigorously agitated for 60 s with the aid of a Vortex mixing device. The gut fragments were allowed to settle by gravity, and the supernatant liquid was removed and added to the 10-ml conical tube; this fraction was referred to as the “fluid fraction.” The remaining gut fragments (termed the “wall fraction”) were transferred with sterile forceps to a 1-ml Bead tube (Mo-Bio Ultraclean fecal DNA isolation kit; Mo-Bio Laboratories, Carlsbad, CA) containing 500 μl of Bead solution for DNA extraction. This process was repeated for a total of 50 guts. The pooled “fluid fraction” was centrifuged at 4,000 × g for 10 min to pellet cells, the supernatant was removed, and the pellet was suspended in 500 μl of Bead solution and transferred to a dry 1-ml Bead tube for DNA extraction.
DNA was obtained from the “fluid fraction” and “wall fraction” using a Mo-Bio Ultraclean fecal DNA isolation kit after bead homogenization by following the manufacturer's recommendations. Stenoxybacter- and spirochete-specific 16S rRNA genes were PCR amplified from samples of each fraction by using forward primers TAM203f (55) and 63f (5′-CAT GTC GAC GTY TTA AGC ATG CAA GT-3′; domain Spirochaetes [30]), respectively, each combined with general reverse primer 1492r. The PCR conditions were those described previously (55), except that samples were subjected to various numbers of PCR cycles (13 to 27 cycles) before the final extension. Each reaction mixture contained one gut equivalent of DNA made up to 50 ng with calf thymus DNA. After PCR, 5-μl samples of each reaction mixture were analyzed by electrophoresis on 1.0% agarose-0.5× Tris-borate-EDTA gels stained with 1× Gelstar nucleic acid stain. Fluorescent bands of the PCR products were digitally captured by using a Kodak electrophoresis documentation and analysis system 290 (Eastman Kodak), and the amount of PCR product in each band was quantified with the system software by dividing the total, background-subtracted intensity of each band by the total band area (in pixels).
Enzyme assays.
Dialyzed, crude cell extracts of S. acetivorans TAM-DN1 were prepared from cells grown with shaking (250 rpm) in 100 ml BYA medium under hypoxia, as described previously (55).
AMP-forming acetyl-CoA synthetase (EC 6.2.1.1) activity was assayed (i) by measurement of acetate- and CoA-dependent formation of AMP from ATP as described by Oberlies et al. (37) and (ii) by measurement of acetate- and CoA-dependent formation of PPi from ATP using the protocol supplied with a kit for enzymatic detection of pyrophosphate (catalog no. P7275; Sigma-Aldrich, St. Louis, MO). Pyrophosphate-acetate phosphotransferase (EC 2.7.2.12) activity was assayed by the same method, except that the rate of acetate-dependent disappearance of PPi was monitored. ADP-forming acetyl-CoA synthetase (EC 6.2.1.13) activity was measured by determining the rate of acetate- and CoA-dependent formation of ADP (from ATP) by a coupled assay (45). Acetate kinase (EC 2.7.2.1) activity was determined by the hydroxylamine method (22) as modified by Brown et al. (7). Phosphotransacetylase (EC 2.3.1.8) (56) and malate synthase A (EC 2.3.3.9) (10) activities were assayed as described previously. Isocitrate lyase (EC 4.1.3.1) activity was assayed like malate synthase activity, except that isocitrate was substituted for glyoxylate. The protein content of cell extracts was measured by the bicinchoninic acid assay (49) with bovine serum albumin as a standard.
PCR primer design. (i) Broad-specificity primers.
Deduced amino acid sequences for genes encoding known acetate kinase (ack), phosphotransacetylase (pta), AMP-forming acetyl-CoA synthetase (acs), isocitrate lyase (aceA), malate synthase A (aceB), malate synthase G (glcB), and the O2-reducing subunit of the cbb3-type cytochrome oxidase (ccoN) were obtained from GenBank (2), and complete, annotated microbial genomes were obtained from the Comprehensive Microbial Resource at The Institute for Genomic Research (TIGR) (http://cmr.tigr.org/tigr-scripts/CMR/CmrHomePage.cgi). The amino acid sequences for each enzyme were aligned using ClustalW, and the alignments were uploaded into the Blocks Multiple Alignment Processor of the CODEHOP online primer design tool (44). The blocks were then imported into the CODEHOP program. For all primers, the default search criteria were used, except that Ralstonia eutropha replaced Homo sapiens as a model for codon bias. Suggested primers from the CODEHOP output were selected with an effort to obtain the maximum distance between the forward and reverse primers in order to allow amplification of as much genetic information as possible (Table 1).
TABLE 1.
Gene-targeted broad-specificity and S. acetivorans-specific forward and reverse PCR primers used in this study
| Primera | Sequence (5′-3′)b | Amplicon size (bp)c | Product of targeted gene | Annealing temp (°C)d |
|---|---|---|---|---|
| Broad-specificity primers | ||||
| Ack312F | TCC CGC TGG CCC CNY YNC AYA AYC | 657 | Acetate kinase (EC 2.7.2.1) | 56 |
| Ack969R | GAG TTC TCG CCG ATG CCN SCN GTR AA | |||
| Pta1138F | CCG ACA AGC GCA TCG TGY TNC CNG ARG G | 848 | Phosphotransacetylase (EC 2.3.1.8) | 56 |
| Pta1986R | GCG CAT GCC CTG CAR CAT NGG NCC | |||
| Acs840F | GAC CCG CTG TTC ATC CTG TAY CAN WSN GG | 735 | Acetyl-CoA synthetase (EC 6.2.1.1) | 52 |
| Acs1575R | TGG CCG GAC ACG TTG AND ACR TCR TC | |||
| AceA328F | TGG CCG GCC ACA TGT AYC CNG AYC A | 728 | Isocitrate lyase (EC 4.1.3.1) | 56 |
| AceA1056R | TGT GGA AGC CGG CCA GNG TRA TRA AC | |||
| AceBGenF | CGG CCT GAA TTG CGG NVG NTG GGA | 315 | Malate synthase A (EC 2.3.3.9) | 56 |
| AceBGenR | CGG AAC CAG ACC CGG ATG NGC NRY CCA | |||
| GlcBGenF | CGC GTT GGG GCT CCY TNT AYG AYG C | 1,155 | Malate synthase Ge | 56 |
| GlcBGenR | CCA TCA GAT CCG GCA TCG SCC ACA YNC C | |||
| CcoNGenF | CTC CAA GCG CAC GGG NCC NGA YYT | 2,000 | N subunit of cbb3-type cytochrome oxidase | 53 |
| CcoNGenR | CGT GCA CGT GGC CGA YNR TCC ART C | |||
| S. acetivorans-specific primers | ||||
| AckTAMF | CAA CAC SCT TTT CCT GAA C | 600 | Acetate kinase (EC 2.7.2.1) | 56 |
| AckTAMR | CCA CAY GCC ACA GTG ATG GC | |||
| PtaTAMF | GAC ACC ATT CGC ASC AAT T | 450 | Phosphotransacetylase (EC 2.3.1.8) | 58 |
| PtaTAMR | CAA TTC CWC TSC TTT GGC TAC | |||
| GlcBTAMF | GCC CGA CGG CAA AAC CAC ATT CAA ATT GC | 500e | Malate synthase Gf | 56 |
| CcoNTAMF | GTG CCT TCT ACC TTC ACC | 700 | N subunit of cbb3-type cytochrome oxidase | 58 |
| CcoNTAMR | GCG ATA GAA AAT CAT ACT TTG |
The suffix F indicates a forward primer, and the suffix R indicates a reverse primer.
The designations for bases follow IUPAC/IUBMB recommendations.
Estimated amplicon size based on the enzyme sequence for E. coli.
Annealing temperatures were optimized with E. coli genomic DNA for broad-specificity primers and with strain TAM-DN1 genomic DNA for S. acetivorans-specific primers.
Amplicon size when GlcBTAMF is paired with GlcBGenR.
An EC number has not been established yet for malate synthase G.
(ii) S. acetivorans-specific primers.
The ClustalW protein alignment tool in the ARB software package (http://www.arb-home.de/) (31) was used to align the deduced amino acid sequences for homologous functional genes from known organisms with the sequences for the genes amplified by PCR from S. acetivorans strains using the broad-specificity primers. Primer pairs were then designed to target conserved areas specific to the S. acetivorans clade (Table 1). The sequence specificity of candidate primers was checked by searching the ARB database and performing a BLAST search.
PCR.
The optimal annealing temperature for each primer pair was determined by temperature gradient PCR with DNA from S. acetivorans TAM-DN1 or E. coli DNA. Each 25-μl reaction mixture contained 25 to 100 ng DNA template, 1× reaction buffer (Invitrogen, Carlsbad, CA), 1.5 mM MgCl2, 0.25 mM of each deoxynucleoside triphosphate, 0.2 μM of each primer, and 0.625 U Taq polymerase (Invitrogen). PCR mixtures were incubated in a model PTC-200 DNA Engine gradient thermal cycler (MJ Research, Watertown, MA) as follows: (i) 3 min at 95°C, (ii) 30 cycles of 45 s at 95°C, 45 s at 50 to 62°C, and 45 s at 72°C, and (iii) 5 min at 72°C. PCR products were then visualized on a 0.5× Tris-borate-EDTA-1% agarose gel with ethidium bromide stain. Once the optimal annealing temperature was determined, the amplicon was cloned by using a TA cloning kit (Invitrogen) and transformed into E. coli TOP10. Several clones were selected for sequencing.
Sequencing and phylogenetic analysis.
Prior to sequencing, unreacted deoxynucleoside triphosphates and primers from PCRs were removed using ExoSAP-IT (USB) according to the supplier's protocol.
Sequences were determined with Applied Biosystems cycle sequencing technology (Applied Biosystems), using the broad-specificity or S. acetivorans-specific forward primer corresponding to the gene of interest (Table 1). Sequences were quality checked, and the initial identification of each sequence was performed by using the BLASTx search tool in the GenBank protein database. Each nucleotide sequence was converted to the deduced amino acid sequence by using Transeq (www.ebi.ac.uk/emboss/transeq).
For phylogenetic analyses, sequences were aligned against a database populated with protein sequences from TIGR and GenBank by using the ClustalW protein alignment algorithm in the ARB software package (31). Ambiguities in sequence alignments were corrected manually. Phylogenetic trees were constructed in ARB using the maximum likelihood routine for protein sequences, and a bootstrap analysis with 1,000 samplings was done using MEGA3 (25).
RNA extraction and purification.
All RNA work was done with RNase-free reagents and materials. Guts were removed from ca. 100 worker larvae and immediately placed in a 1.5-ml centrifuge tube on dry ice. One milliliter of RNA Protect reagent was then added, and the contents were immediately transferred into a sterile glass tissue homogenizer and homogenized for 3 min. RNA was purified from the homogenate using the protocol for bacteria with an RNeasy RNA purification kit (QIAGEN, Valencia, CA). The RNA was quantified by determining the optical density at 260 nm. After quantification, 1 μg RNA was treated with DNase I (amplification grade; Invitrogen) according to the manufacturer's instructions. The DNA-free RNA was stored at −80°C until it was used.
RT-PCR.
First-strand cDNA synthesis was done by following the protocol described for Superscript III reverse transcriptase (RT) (Invitrogen).
PCR amplification of the first-strand cDNA was done using the PCR methods described above with 2 to 4 μl of the first-strand cDNA synthesis reaction mixture and the annealing temperature of the specific primer set (Table 1) and extending the total number of PCR cycles to 35.
Oxygen uptake measurements.
S. acetivorans TAM-DN1 and Citrobacter sp. strain RFC-10 were grown in BYA medium or BYA medium modified to contain 0.75 mM NH4Cl instead of 4.7 mM NH4Cl (the former concentration is known to limit the cell yield to approximately one-half the cell yield otherwise attained) and 20 mM sodium acetate instead of 10 mM sodium acetate. These modifications were made in an effort to mimic the relatively low-nitrogen, acetate-rich conditions in termite guts, especially when growth of cells was obviously becoming limited by available N. Cells were harvested just after entry into N limitation-induced stationary phase by centrifugation (10,000 × g, 10 min, 4°C), washed twice in insect Ringer's solution (containing [per liter] 7.5 g NaCl, 0.35 g KCl, and 0.21 g CaCl2; pH 7.0), and then resuspended to obtain 10 times the initial cell density in the same buffer. Oxygen uptake rates were measured with rapid stirring in a 2-ml glass high-performance liquid chromatography vial fitted with a screw cap having a central hole through which a narrow Clark-type oxygen electrode (Diamond General, Ann Arbor, MI) was inserted and sealed with dental wax. Prior to use, the oxygen electrode was calibrated by immersion in insect Ringer's solution that had been vigorously bubbled with air for >15 min (100% air saturation) or degassed under a vacuum and bubbled with 100% N2 (0% air saturation). Cell suspensions were vigorously aerated by shaking prior to transfer into the O2 uptake chamber, and oxygen consumption was measured before and after the addition of a substrate (sodium acetate, sodium succinate, sodium lactate, or d-glucose; final concentration, 10 mM).
For measurement of oxygen uptake by termite guts, guts were removed from worker larvae that had been maintained in the laboratory for ca. 8 months. The termites either were not treated (controls) or were prefed diets to eliminate major components of the gut microbiota (i.e., bacteria and/or cellulolytic protozoans [see below]) before gut removal. For each experiment, 10 to 20 extracted guts were placed in 2 ml of insect Ringer's solution in the O2 uptake chamber, as described above. The guts were first allowed to settle, and then the overlying buffer was aspirated and immediately replaced with fresh, fully aerated buffer and O2 uptake was measured as described above with and without addition of 10 mM (final concentration) sodium acetate.
Elimination of gut microbes.
To determine the relative contributions of cellulolytic protozoans and bacteria to oxygen consumption in termite guts, either or both groups of microbes were largely eliminated from guts by prefeeding the termites for 11 days using 1% agarose food blocks (2 by 2 by 0.5 in.) containing either microgranular CC41 cellulose powder (Whatman, Brentford, United Kingdom) or corn starch (which is known to eliminate cellulolytic protozoans [54]), with or without a mixture of three antibiotics targeted at disruption of bacterial cell walls (ampicillin, cefoperazone, and vancomycin; final concentration, 800 μg/ml each). Food blocks were individually distributed into sterile petri dishes (100 by 15 mm). For each treatment, a total of six replicate dishes were prepared, and 20 to 40 worker larvae of R. flavipes were added to each dish. The petri dishes were then placed in a humid chamber and incubated at room temperature for 11 days, during which time the food blocks were replaced every 3 days.
To determine the efficacy of such treatments for removing gut microbes, viable cell counts of bacteria were obtained every second day of treatment, as were direct microscopic counts of protozoans. For viable cell counts, the guts of 12 termites from a single dish in each treatment were placed into 2 ml of 1× basal salts solution (50) buffered with 10 mM morpholinopropanesulfonic acid (MOPS) (pH 7.0). The guts were thoroughly homogenized with a sterile glass tissue homogenizer, and the homogenate was serially diluted in 10-fold increments in the same buffer. Samples from each dilution were plated in triplicate onto a complex medium containing (per liter) 0.2 g KH2PO4, 0.25 g NH4Cl, 0.5 g KCl, 0.15 g CaCl2 · 2H2O, 1.0 g NaCl, 0.62 g MgCl2 · 6H2O, 2.84 g Na2SO4, 3.7 g brain heart infusion medium (BD Franklin Lakes, NJ), and 1.0 g Casamino Acids, as well as 1 mM cellobiose, 1 mM d-glucose, 1 mM d-xylose, 1 mM maltose, 1 mM sodium pyruvate, 1 mM sodium lactate, and 1 mM sodium acetate. The medium was buffered by inclusion of MOPS at a final concentration of 10 mM, and the pH was adjusted to 7.0 prior to autoclaving. Plates were incubated at 23°C under hypoxia (93% N2, 5% CO2, 2% O2) for 15 days before colonies were counted.
For quantification of protozoans, the guts of four termites from a single dish in each treatment were placed in a 100-μl pool of insect Ringer's solution on a sterile petri dish under a dissecting microscope. Each gut was sliced longitudinally with a razor blade, and while the gut was held with forceps, the gut contents were washed out of the gut and into the Ringer's solution by agitation. The 100-μl suspension containing gut fluid and microorganisms, but not sliced guts, was added to a sterile, 1.5-ml centrifuge tube and briefly centrifuged to pellet the cells. The pellet was resuspended in 10% neutral buffered formalin (containing [per liter] 100 ml of 37% formalin, 6.5 g of Na2HPO4, and 4.0 g of NaH2PO4; pH 7.0) and incubated at 4°C overnight. The fixed cells were again collected by centrifugation and then resuspended in a mixture of equal volumes of insect Ringer's solution and absolute ethanol and kept at −20°C until they were counted. For counting, 10 μl of the cell suspension was transferred into three separate wells of an eight-well Teflon-coated slide (area of each well, 28.26 mm2), allowed to dry, and washed briefly in water. The cells were stained with freshly prepared 5-(4,6-dichlorotriazin-2-yl)aminofluorescein (DTAF) (0.2 mg/ml; prepared in a buffer containing 0.05 M Na2HPO4 and 0.15 M NaCl [pH 9.0]) for 30 min, and this was followed by three washes in the same buffer for 30 min each. The slides were air dried, coverslips were mounted with Entellan (Merck) preservative, and cells were visualized at a magnification of ×100 by UV epifluorescence with a Zeiss Axioskop equipped with a DTAF-specific filter set. Protozoans in at least 20 fields of view per well were counted.
Nucleotide sequence accession numbers.
Partial nucleotide and deduced amino acid sequences for the ack, pta, ccoN, and glcB genes obtained in this study have been deposited in the EMBL, GenBank, and DDBJ databases under accession numbers EF364565 to EF364726. Specific accession numbers are given below.
RESULTS
In situ location of S. acetivorans.
Repeated attempts to identify cells of S. acetivorans in guts of R. flavipes by fluorescent in situ hybridization (3, 51) with an S. acetivorans-specific 16S rRNA gene probe (validated with Neisseria sicca as a negative control) were unsuccessful, owing mainly to a high level of nonspecific background fluorescence from hindgut tissue and from cells of protozoans, coupled with the relatively small diameter (ca. 0.5 μm) of the target S. acetivorans cells (55). Attempts to decrease the autofluorescence with various fixatives, blocking reagents, HCl, and H2O2 washes (4, 51) were unsuccessful, as were attempts to amplify the Stenoxybacter-specific signal by tyramide signal amplification (40) or hybridization with dual fluorescently labeled probes (4). Accordingly, a limited-cycle PCR-based approach was used as an alternative with template DNA extracted from hindguts that were physically fractionated to obtain a hindgut “fluid fraction” rich in spirochetes, which are among the most abundant bacteria in R. flavipes hindguts and are known to occur primarily in termite hindgut fluid either as free-swimming cells or as ectosymbionts attached to the surface of protozoans (5), and a hindgut “wall fraction” containing an adherent heterogeneous microbiota that was resistant to dislodgement by vigorous vortex mixing. Results of this alternative approach revealed that compared to spirochetes, cells of S. acetivorans were more closely associated with the hindgut wall. DNA extracted from the “wall fraction” and amplified with S. acetivorans-specific primers yielded a quantifiable product after 24 cycles, whose abundance increased steadily through 27 cycles (Fig. 2A). Clone libraries prepared from such PCR products confirmed that they were S. acetivorans specific (data not shown). However, 25 cycles were required before an S. acetivorans-specific product could be detected with DNA from the nonadherent and/or loosely wall-associated microbes comprising the “fluid fraction,” and the abundance of this product increased only moderately through 27 cycles.
FIG. 2.
Primary in situ location of S. acetivorans as inferred from limited-cycle PCRs done with S. acetivorans-specific (A) or spirochete-specific (B) 16S rRNA gene primers and template DNA extracted from the hindgut wall fraction (black bars) or hindgut fluid fraction (gray bars). The ordinate indicates the fluorescence intensity of the amplification products, quantified after electrophoretic separation on Gelstar dye-containing agarose gels. The error bars indicate standard deviations (n = 3).
By contrast, analogous PCRs performed with spirochete-specific primers yielded a quantifiable PCR product after only 13 cycles from the “fluid fraction,” and its abundance increased at a higher rate with additional cycles than the abundance of the spirochete-specific product from the “wall fraction,” which did not appear until cycle 14 (Fig. 2B). The smaller, but still significant, amount of spirochete-specific PCR product in the “wall fraction” was presumably derived from spirochete cells that, through their motility, managed to insinuate themselves among members of the wall-associated nonspirochetal microbiota, as commonly seen in electron micrographs (6).
Enzyme activities and genes relevant to acetate oxidation in vitro.
In order to identify enzymes (and the encoding genes) that are specific to acetate oxidation by S. acetivorans and whose transcripts could be sought in situ, crude extracts of acetate-grown cells were examined for enzyme activities associated with the activation of acetate to acetyl-CoA. Results revealed the presence of acetate kinase (3.7 ± 0.7 U/mg protein [mean ± standard deviation; n = 3]) and phosphotransacetylase (1.1 ± 0.5 U/mg protein) but not AMP-forming acetyl-CoA synthetase or the less widely distributed ADP-forming synthetase or PPi-acetate phosphotransferase. However, AMP-forming acetyl-CoA synthetase activity could be readily detected in reaction mixtures to which authentic acetyl-CoA synthetase (purified from Saccharomyces cerevisiae) was added. Acetate kinase activity was dependent on the presence of both acetate and ATP in the reaction mixture, and phosphotransacetylase activity was dependent on the presence of both acetyl-phosphate and CoA. Robust acetate kinase activity was also observed in crude extracts of succinate-grown cells (9.4 ± 1.9 U · mg protein−1), and it is conceivable that this enzyme is constitutive in S. acetivorans.
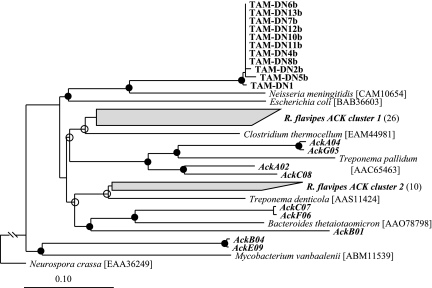
PCR amplification of S. acetivorans genomic DNA with broad-specificity primers readily revealed the presence of genes encoding acetate kinase (ack) and phosphotransacetylase (pta) but not a gene encoding AMP-forming acetyl-CoA synthetase. However, the acs gene was readily amplified from E. coli genomic DNA (control) by using the same primers. On average, the deduced amino acid sequences for the ack PCR products from 11 different strains of S. acetivorans were 99.8% identical to each other (based on 202 amino acid positions shared among all members used in the phylogeny), and they contained the “Connect 1” and “Phosphate 2” domains characteristic of acetate kinase (34). However, these sequences were only 69% identical to their closest known relative, an acetate kinase from Neisseria meningitidis (Fig. 3).
FIG. 3.
Maximum likelihood-based phylogenetic analysis of the deduced amino acid sequences (202 positions) of acetate kinases from selected S. acetivorans isolates (designations beginning with “TAM”) and from R. flavipes gut homogenates (boldface italics). The numbers in parentheses after R. flavipes clusters are the numbers of sequences in the clusters. The GenBank accession numbers are EF364701 to EF364711 for sequences obtained from S. acetivorans isolates and EF364565 to EF364609 for clones obtained from R. flavipes guts. The GenBank numbers for reference species are shown in brackets. Branch points with >75% conservation are indicated by filled circles; branch points with 50 to 74% conservation are indicated by open circles. The acetate kinase from Neurospora crassa was used as an outgroup. Scale bar = 0.1 change per amino acid.
The same broad-specificity acetate kinase primers were also used to PCR amplify R. flavipes gut homogenate DNA, and a clone library was constructed from the amplimers. Phylogenetic analysis of the deduced amino acid sequences from 45 clones revealed that a majority of these sequences (26 clones) formed a distinct cluster (R. flavipes acetate kinase cluster 1) whose closest relative was an acetate kinase from Clostridium thermocellum (Fig. 3). Ten other clones grouped together with an acetate kinase from Treponema denticola (R. flavipes acetate kinase cluster 2). Less abundant were clones related to Treponema pallidum, Bacteroides thetaiotaomicron, and Mycobacterium vanbaalenii. However, no clones corresponding to S. acetivorans ack were found among these 45 clones.
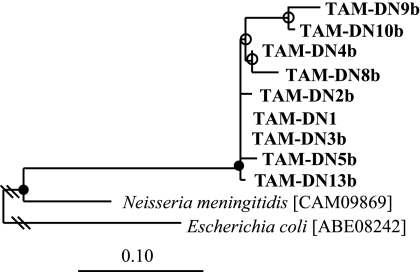
The product of an ∼800-nucleotide amplicon (of which 678 bp was sequenced) from S. acetivorans TAM-DN1 DNA obtained with broad-specificity phosphotransacetylase primers exhibited 75% deduced amino acid identity to its closest known relative, a phosphotransacetylase from N. meningitidis (Fig. 4), and contained characteristic essential arginine residues at positions 87, 133, and 287 (43). On average, the deduced amino acid sequences of the phosphotransacetylases from different S. acetivorans strains were 93% identical (based on the 201 amino acid positions shared among all members used in the phylogeny).
FIG. 4.
Maximum likelihood-based phylogenetic analysis of the deduced amino acid sequences (201 positions) of phosphotransacetylases from selected S. acetivorans isolates (boldface). These sequences have been deposited in the GenBank database under accession numbers EF364712 to EF364720. GenBank accession numbers for reference species are shown in brackets. Branch points with >75% conservation are indicated by filled circles; branch points with 50 to 74% conservation are indicated by open circles. E. coli phosphotransacetylase was used as an outgroup. Scale bar = 0.1 change per amino acid.
As additional potential targets for exploring in situ acetate-oxidizing activity of S. acetivorans, two key enzymes (and their encoding genes) of the glyoxylate cycle, isocitrate lyase and malate synthase A, were sought (Fig. 1). Although glyoxylate- and acetyl-CoA-dependent putative malate synthase A activity was detected (0.5 ± 0.3 U/mg protein; n = 3), isocitrate lyase activity was not detected. Moreover, despite multiple attempts to amplify the genes encoding these two enzymes, including a redesign of primers and use of less stringent amplification conditions, no amplification products were generated with primers targeted to isocitrate lyase or malate synthase A, although amplification of these genes always occurred with E. coli control DNA. However, S. acetivorans TAM-DN1 DNA amplified with malate synthase G-targeting primers resulted in a ∼1,100-nucleotide product, 906 bp of which was sequenced (GenBank accession number EF364721). The sequenced product exhibited 64% deduced amino acid sequence identity to malate synthase G from Pseudomonas syringae pv. tomato strain DC3000 (accession number AAO54024) (data not shown). Phylogenetic analysis revealed that the deduced amino acid sequences for malate synthase G PCR products from eight different S. acetivorans strains were, on average, 99.5% identical and grouped specifically with malate synthase G enzymes and not with the phylogenetically related malate synthase A and malyl-CoA lyase enzymes (not shown) (33, 35).
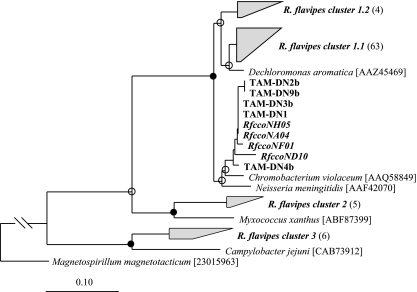
Inasmuch as S. acetivorans is an obligate microaerophile (55), it was hypothesized that cells of this organism possess a high-affinity cbb3-type oxidase as their terminal respiratory enzyme. To explore this possibility, broad-specificity primers were designed that targeted ccoN, the gene encoding the O2-reducing subunit of cbb3 cytochrome oxidases (Table 1). PCR amplification of S. acetivorans TAM-DN1 genomic DNA yielded a 2-kb product, 1,110 to 1,131 bp of which was sequenced. The deduced amino acid sequence was 96% identical (based on 169 amino acid positions shared among all members used in the phylogeny) to a cbb3-type cytochrome oxidase from Chromobacterium violaceum (Fig. 5). Sequencing and phylogenetic analysis also revealed that the ccoN subunit was highly conserved among the other S. acetivorans strains in the tree (99.8% sequence identity).
FIG. 5.
Maximum likelihood-based phylogenetic analysis of the deduced amino acid sequences (169 positions) of the O2-reducing (ccoN-encoded) subunits of cbb3-type cytochrome oxidases from selected S. acetivorans isolates (designations beginning with “TAM”) and from R. flavipes guts (boldface italics). The GenBank accession numbers are EF364722 to EF364726 for sequences obtained from S. acetivorans isolates and EF364610 to EF364691 for clones obtained from R. flavipes guts. The GenBank accession numbers for reference species are shown in brackets. Branch points with >75% conservation are indicated by filled circles; branch points with 50 to 74% conservation are indicated by open circles. The ccoN subunit from Magnetospirillum magnetotacticum was used as an outgroup. Scale bar = 0.1 change per amino acid.
The same broad-specificity ccoN primers were also used to construct a clone library from R. flavipes gut homogenate DNA. Phylogenetic analysis of 82 clones from this library revealed that a majority fell within two distinct groups (R. flavipes clusters 1.1 and 1.2 containing 63 and 4 clones, respectively), whose closest relative was the CcoN from Dechloromonas aromatica (Fig. 5). Five other clones formed a cluster most closely related to the CcoN from Myxococcus xanthus (R. flavipes cluster 2); six formed a cluster related to Campylobacter jejuni (R. flavipes cluster 3); and four (prefix RfccoN) grouped closely with the CcoN genes from various S. acetivorans isolates.
Genes expressed by S. acetivorans in situ.
Owing to the high degree of within-species similarity of S. acetivorans genes relevant to acetate-supported aerobic respiration (i.e., ack, pta, and ccoN [see above]) coupled with their substantial sequence divergence from homologues present in other known organisms or PCR amplified from termite gut homogenates, S. acetivorans-specific primers were designed to examine (by RT-PCR) whether these genes were expressed in situ as well as in vitro. To do this, presumed S. acetivorans-specific primers for these genes (Table 1) were first tested by conventional PCR amplification with purified R. flavipes gut homogenate DNA as the template. As determined by cloning and sequencing of the PCR products, all the genes were identical or nearly identical to corresponding ack, pta, and ccoN genes from S. acetivorans isolates (data not shown).
When the same primers were used in individual RT-PCRs with R. flavipes gut homogenate RNA as the template, each reaction usually yielded a single PCR product that was the same size as the analogous product obtained by using DNA from acetate-grown S. acetivorans TAM-DN1 as the template (Fig. 6). Importantly, no products were obtained if RT was omitted from the RT-PCR mixtures (Fig. 6, lane 3), confirming that products formed in the complete reaction mixtures were derived from RNA (presumably mRNA) and not from DNA contamination of the template. After cloning and sequencing of the RT-PCR products that were the correct size, phylogenetic analysis revealed that they grouped specifically with the ack, pta, and ccoN gene products from S. acetivorans isolates (Fig. 7). These results indicated that expression of ack, pta, and ccoN is relevant not only to acetate-dependent growth of S. acetivorans in vitro but presumably also to growth and/or survival in situ.
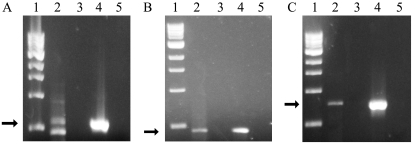
FIG. 6.
Gelstar-stained agarose gel electropherograms of RT-PCR products for (A) acetate kinase (ack), (B) phosphotransacetylase (pta), and (C) the O2-binding subunit of cbb3 terminal oxidase (ccoN). The template for RT-PCR was RNA purified from R. flavipes gut homogenates, and amplification was done with S. acetivorans-specific, gene-targeted primers. Lane 1, 1-kb DNA ladder (the bands represent, from the bottom to the top, 0.5, 1.0, 1.5, 2.0, 3.0, 4.0, 5.0, 6.0, 8.0, and 10.0 kb); lane 2, RT-PCR product obtained using R. flavipes gut homogenate RNA as the template; lane 3, same as lane 2, but without addition of RT; lane 4, RT-PCR with S. acetivorans TAM-DN1 genomic DNA as the template; lane 5, RT-PCR without a template. The arrows indicate the correct size of the anticipated RT-PCR product.
FIG. 7.
Maximum likelihood-based phylogenetic analysis of the deduced amino acid sequences of cloned RT-PCR products shown in Fig. 6. All RT-PCR products (boldface italics) clustered specifically with the corresponding genes from S. acetivorans isolates (designations beginning with “TAM”) encoding (A) acetate kinase (170 positions), (B) phosphotransacetylase (182 positions), and (C) the ccoN subunit of cbb3 oxidase (124 positions). N. meningitidis was used as an outgroup for all phylogenetic analyses (not shown in panel C). (A and C) Scale bar = 0.025 change per amino acid; (B) scale bar = 0.1 change per amino acid.
It was noticed that in RT-PCRs with the ack-targeting primer pair, an additional product was made that was slightly smaller (450 bp) than the authentic ack fragment (600 bp) (Fig. 6A, lane 2). However, cloning and sequencing of material from this band revealed that it shared 45% identity (53/116 amino acids) to a gene encoding a hypothetical protein in Drosophila melanogaster (GenBank accession no. AY061833) and probably arose from nonspecific amplification of mRNA derived from R. flavipes gut tissue.
Oxygen consumption by R. flavipes guts attributable to S. acetivorans and other components of the gut microbiota.
To estimate the potential contribution of S. acetivorans to O2 consumption in guts of R. flavipes, per-cell rates of O2 consumption were determined for S. acetivorans TAM-DN1 grown in vitro without and with NH4+ limitation in a medium that contained excess (20 mM) acetate. The latter condition was imagined to possibly mimic what the cells might experience in situ, as the concentration of N in the food source of R. flavipes (wood) is relatively low (ca. 0.05% [wt/wt] for sound wood). When cells were harvested from batch cultures at a stage when growth had just ceased owing to NH4+ limitation, the O2 consumption rate of S. acetivorans TAM-DN1 was 1.5 × 10−5 ± 0.4 × 10−5 pmol · min−1 · cell−1 in buffer supplemented with acetate and 1.0 × 10−5 ± 0.2 × 10−5 pmol · min−1 · cell−1 in succinate-supplemented buffer (Table 2). The O2 consumption rates exhibited by cells grown in medium with no NH4+ limitation were nearly identical, 1.0 × 10−5 ± 0.6 × 10−5 and 9.5 × 10−6 ± 0.4 × 10−6 pmol · min−1 · cell−1 in acetate-supplemented buffer and succinate-supplemented buffer, respectively. Not surprisingly, little or no oxygen consumption was observed when glucose or lactate was added to the buffer, as S. acetivorans does not use these compounds as energy sources (55), or in buffer without added substrate. The per-cell rates of O2 consumption by Citrobacter sp. strain RFC-10, another organism isolated along with S. acetivorans (55), were about one-half those of S. acetivorans on the same substrates. However, Citrobacter sp. strain RFC-10 did exhibit lactate- and glucose-supported O2 consumption (Table 2).
TABLE 2.
Substrate-specific oxygen consumption rates of S. acetivorans TAM-DN1, Citrobacter sp. strain RFC-10, and extracted guts of R. flavipes
| Substratea | Oxygen consumption rateb
|
||
|---|---|---|---|
| S. acetivorans TAM-DN1c | Citrobacter sp. strain RFC-10c | R. flavipes guts | |
| None (endogenous) | 0.0 | 0.0 | 969 ± 256 |
| Acetate | 1.5 ± 0.4 | 0.6 ± 0.2 | 1,013 ± 141 |
| Succinate | 1.0 ± 0.2 | 0.5 ± 0.2 | ND |
| Lactate | 0.0 | 1.0 ± 0.3 | ND |
| Glucose | 0.0 | 0.8 ± 0.2 | ND |
The initial substrate concentration was 10 mM in fully aerated insect Ringer's solution.
The values are means ± standard deviations (n = 3) and are expressed in 10−5 pmol · min−1 · cell−1 for bacteria and in pmol · min−1 · gut−1 for R. flavipes guts. ND, not determined.
The strain was harvested from batch culture after NH4+-limited growth in a medium that contained excess acetate.
The O2 consumption rate for intact extracted guts of R. flavipes in acetate-supplemented buffer was 1,013 ± 141 pmol · min−1 · gut−1. However, the oxygen consumption rate for guts incubated in buffer without acetate (969 ± 256 pmol · min−1 · gut−1) was not significantly different (P = 0.80). These data suggested that respiration by intact guts was probably largely, if not entirely, supported by endogenous substrates such as acetate, which is present at relatively high concentrations in R. flavipes hindgut fluid (up to 80 mM) (38).
To estimate the relative contribution of bulk bacteria and bulk protozoans to O2 uptake by guts, termites were fed diets intended to eliminate all or most members of these microbial communities from the gut, after which whole-gut O2 consumption was measured (Table 3). Compared to the normal diet of wood, even a seemingly innocuous shift to a short-term diet of pure cellulose in agarose resulted in moderate, but statistically significant decreases in both bacterial viable cell counts (P = 0.04) and numbers of protozoans (P = 0.008), as well as a decrease in whole-gut O2 consumption (P = 0.04). Incorporation of a three-antibiotic cocktail (800 μg/ml each of ampicillin, cefoperazone, and vancomycin) into such a diet drastically reduced the bacterial CFU by 3 orders of magnitude but decreased the O2 consumption rate of extracted guts by slightly less than 50%, roughly the same magnitude as the decrease in total numbers of protozoans.
TABLE 3.
Effect of diet on gut microbiota and O2 consumption by extracted guts of R. flavipes
| Dieta | Concn of cultivable bacteria (105 CFU/gut)b | Concn of protozoans (103 cells/gut)c | Whole-gut O2 consumption (pmol · min−1 · gut−1)d |
|---|---|---|---|
| Wood (control) | 4.5 ± 0.5 | 9.6 ± 1.5 | 1,013 ± 141 |
| Cellulose | 3.2 ± 0.7 | 6.6 ± 0.3 | 740 ± 86 |
| Cellulose + Ab | 0.004 ± 0.002 | 3.3 ± 1.5 | 348 ± 65 |
| Corn starch | 4.0 ± 0.5 | 2.3 ± 0.3 | 436 ± 108 |
| Starch + Ab | 0.003 ± 0.001 | 0.7 ± 0.1 | 286 ± 92 |
Termites were maintained on diets for 11 days prior to assays. Ab, antibiotic mixture (800 μg/ml each of ampicillin, cefoperazone, and vancomycin).
Colonies were counted 14 days after plating. The values are means ± standard deviations (n = 4).
Determined by direct counting of DTAF-stained cells. The values are means ± standard deviations (n = 3).
Consumption in buffer supplemented with 10 mM acetate (pH 7.0). The values are means ± standard deviations (n = 3).
Termites fed a diet of corn starch for 11 days exhibited a 25% increase in the number of bacterial CFU per gut but lost about two-thirds of their protozoans compared to cellulose-fed controls. This was expected; starch is known to eliminate cellulolytic protozoans from termite guts (12, 54), probably because it is so readily hydrolyzed, even by salivary amylases present in the termite itself, that protozoans no longer have a selective advantage for retention in the gut. Moreover, it is a substrate on which wood-feeding termites can survive indefinitely. However, the O2 consumption rate of whole guts from starch-fed termites (436 ± 108 pmol · min−1 · gut−1) was only about 60% that of the cellulose-fed controls (P = 0.003). Incorporation of antibiotics into the starch diet caused a large decrease in the number of bacterial CFU, and it also further reduced the numbers of protozoans and the whole-gut O2 consumption rate to the lowest value seen in the entire experiment (286 ± 92 pmol · min−1 · gut−1) (Table 3).
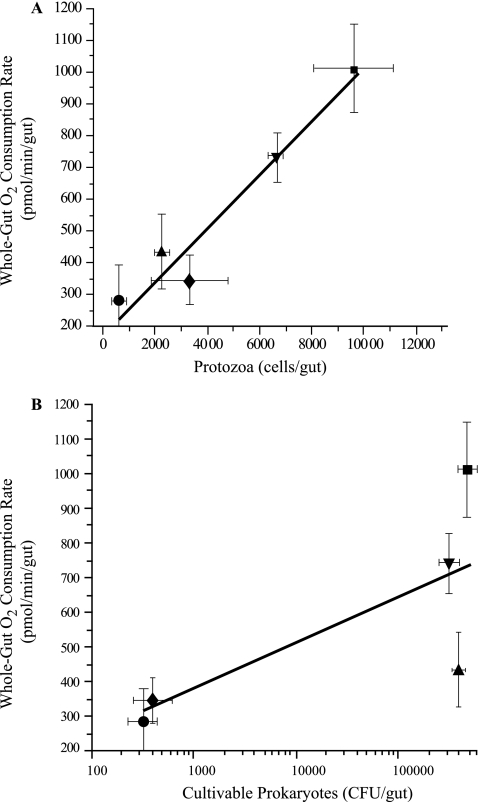
When these data were plotted, there was a strong correlation between the number of protozoans per gut and the whole-gut O2 consumption rate (r2 = 0.95) (Fig. 8A), but there was a weak correlation between the number of bacterial CFU and the whole-gut O2 consumption rate (r2 = 0.56) (Fig. 8B).
FIG. 8.
Correlation between the R. flavipes whole-gut oxygen consumption rate and the number of gut protozoans (A) or cultivable prokaryotes (B). The r2 values for the best-fit regression lines are 0.95 (A) and 0.56 (B). Termites were maintained for 11 days on diets consisting of cellulose (▾), cellulose with antibiotics (⧫), starch (▴), and starch with antibiotics (•). ▪, wood control.
DISCUSSION
Results reported here support the hypotheses that (i) cells of S. acetivorans occur primarily in the peripheral, hypoxic region of hindguts of R. flavipes, either attached directly to the hindgut wall or occurring among the heterogeneous microbiota situated on or near the hindgut wall; (ii) acetate is a major, if not the sole, substrate that fuels the O2-consuming respiratory activity of S. acetivorans in situ; and (iii) cells of S. acetivorans use a high-affinity, cbb3-type terminal oxidase for O2 consumption in vitro and in situ.
Detection of S. acetivorans-specific transcripts for acetate kinase and phosphotransacetylase, especially within an imagined “sea” of homologous transcripts from a microbial community using the same enzymes for fermentative acetate production instead of activation, was expected to be a major challenge. Indeed, a majority of 45 randomly selected ack clones obtained by conventional PCR from gut homogenates with broad-specificity primers clustered with ack genes of known acetate-producing anaerobes, such as Clostridium (57) and Treponema species (19, 20); not a single one corresponded to an S. acetivorans gene (Fig. 3). However, our ultimate success was facilitated by the ability to readily design S. acetivorans-specific RT-PCR primers for ack and pta genes which, like the 16S rRNA-encoding genes of S. acetivorans (55), were phylogenetically distant from homologues present in other microbes, especially those comprising the hindgut microbiota. Our previous demonstration that S. acetivorans can oxidize other substrates, such as succinate, and can do this simultaneously with acetate oxidation during in vitro growth (55) suggests that cells in situ may also utilize other oxidizable substrates, if and when such substrates become available. The ability to do this may be important during colonization of guts of newly hatched larvae and recently molted colony mates, whose guts are devoid or largely depleted of microbes, and before a robust acetate-producing fermentative community has had time to become established.
It was not altogether surprising that S. acetivorans appears to lack AMP-forming acetyl-CoA synthetase and instead uses the low-affinity (but more energetically efficient) acetate kinase to activate acetate, as the steady-state concentration of acetate in termite guts is already about eightfold greater than the Km of acetate kinase. Assuming that the phosphotransacetylase step is the rate-limiting step in acetate activation by S. acetivorans TAM-DN1, the phosphotransacetylase specific activity (1.1 ± 0.5 μmol · min−1 · mg protein−1 [see above]) is still sufficient to account for the experimentally determined rate of acetate consumption by cells during growth (ca. 0.12 μmol · min−1 · mg protein−1) (55), further supporting the notion that the acetate kinase-phosphotransacetylase pathway is the most likely route for acetate activation in S. acetivorans. The absence (or loss) of AMP-forming acetyl-CoA synthetase, together with possible constitutive expression of acetate kinase and phosphotransacetylase, may reflect an adaptation by S. acetivorans to autochthony (55) in the acetate-rich hindgut of R. flavipes.
When the protein content is converted to cell number equivalents, the potential in vitro per-cell rate of acetate consumption by S. acetivorans (see above) corresponds to 1.1 × 10−6 nmol · h−1. For an in situ density of ca. 1 × 105 S. acetivorans cells · gut−1 (55) and to the extent that in vitro rates are a rough approximation of in situ activity, the rate of acetate consumption by the S. acetivorans population in R. flavipes hindguts is estimated to be about 0.1 nmol · gut−1 · h−1. This value is 4.2% of the 2.4-nmol · gut−1 · h−1 oxidation rate measured by microinjection of radiolabeled acetate into R. flavipes hindguts (52) but only 0.5% of the 20.2-nmol · termite−1 · h−1 acetate production rate measured with laboratory-maintained R. flavipes (38). Therefore, in light of the diversity of bacteria in R. flavipes hindguts potentially capable of acetate oxidation in situ (55), the S. acetivorans population probably has a significant impact on acetate consumption within the hindgut but only a minimal impact on the acetate available to meet the daily energy requirements of its host.
In spite of evidence that acetate fuels the respiratory activity of S. acetivorans in situ, we obtained no evidence for the presence of a classical glyoxylate cycle in this bacterium. Neither isocitrate lyase nor its encoding gene was detected, and the malate synthase activity that was detected in cell extracts appeared to be attributable to malate synthase G, the product of the glcB gene, rather than to the malate synthase A isoform typically associated with the glyoxylate cycle (35) or to the related malyl-CoA lyase associated with the citramalate cycle (33). Whether S. acetivorans has an alternative anaplerotic cycle remains a topic for future research. However, it is equally possible that S. acetivorans may not need such a cycle, as amino acids and volatile fatty acids that can be used for biosynthesis or regeneration of TCA cycle intermediates are present in termite gut fluid (17, 38, 47).
The demonstration of S. acetivorans-specific in situ transcripts for CcoN, the O2-binding subunit of high-affinity (Km for oxygen, ∼7 nM) cbb3-type heme-copper terminal oxidases, the same subunit (and by inference, holoenzyme) present in cells growing in vitro under hypoxia, implies that S. acetivorans does, as expected, carry out aerobic respiration in situ, as it does not appear to be capable of anaerobic respiration or fermentation (55). The presence of a cbb3-type terminal oxidase in S. acetivorans was not surprising given its phylogenetic relationship to Eikenella corrodens and Neisseria spp., which also contain cbb3-type terminal oxidases (21), and it is entirely consistent with the obligately microaerophilic nature of S. acetivorans and its occurrence in the hypoxic periphery of hindguts. PCR amplification with broad-range primers revealed a diversity of ccoN sequences in R. flavipes guts, including several ccoN sequences whose deduced amino acid sequences corresponded to those present in S. acetivorans strains (Fig. 5). The majority of the amino acid sequences, however, were most closely related to the CcoN sequence from D. aromatica, an organism known to degrade aromatic compounds (13). Bacteria closely related to D. aromatica have not been isolated or described from termite guts; however, O2-dependent ring cleavage of aromatic lignin model compounds is known to occur in R. flavipes and other termite species (9). The possession of high-affinity terminal oxidases by various bacteria in termite hindguts may enable these bacteria to reduce the O2 concentration to a level low enough that O2-sensitive activities (methanogenesis, CO2-reductive acetogenesis, N2 fixation) can proceed in nearby cells in the hypoxic zone and not be restricted to the anoxic luminal region. Indeed, methane emission by R. flavipes originates from cells of methanoarchaea that are located primarily on or near the hindgut wall (28, 29).
The per-cell rate of acetate-supported O2 consumption by cell suspensions of S. acetivorans in vitro (1.5 × 10−5 pmol · min−1) (Table 2), together with the in situ density of S. acetivorans (ca. 1 × 105 cells · gut−1) (55), indicates that the potential rate of oxygen consumption by the S. acetivorans population in situ is on the order of 1.5 pmol · min−1 · gut−1. This value is close to the potential rate of glucose-supported O2 consumption by lactic acid bacteria in guts of R. flavipes (3.0 pmol · min−1 · gut−1), based on per-cell rates of O2 consumption by the representative isolate Enterococcus sp. strain RfL6 (53), but about fivefold greater than the O2 consumption by Enterobacteriaceae (0.3 pmol · min−1 · gut−1), based on the representative isolate Citrobacter sp. strain RFC-10 (mean, 0.7 × 10−5 pmol · min−1 · cell−1) (Table 2), together with estimates of the total concentration of Enterobacteriaceae cells in guts of R. flavipes (ca. 4.4 × 104 cells · gut−1, calculated from data presented previously [46, 53]). However, lactic acid bacteria and members of the Enterobacteriaceae are facultative anaerobes and not absolutely O2 dependent, whereas S. acetivorans is an obligate microaerophile. Based on rates of O2 consumption occurring in the bulbous “paunch” region of hindguts, as estimated from microelectrode profiles (178 pmol · min−1 · gut−1) (8), and on rates of O2 consumption exhibited by entire extracted guts (which also include a portion of attached midgut) incubated in a respirometry chamber (1,013 pmol · min−1 · gut−1) (Table 3), the S. acetivorans population could potentially account for 0.8 and 0.2%, respectively, of the O2 consumption. However, the observation that the rates of O2 consumption by extracted guts (see above) were close to the rates exhibited by individual intact larvae (860 to 1,060 pmol · min−1) (38) was surprising. Notwithstanding unknown variables introduced by extraction of guts and incubation in a buffered salts solution in a respirometry chamber, it appears that much of the O2 consumption observed with larvae actually occurs in the gut and its contents.
Given that the potential rates of O2 consumption by the S. acetivorans population and by two other major groups of bacteria in R. flavipes (lactic acid bacteria and Enterobacteriaceae) each amounted to less than 1% of the O2 consumption rate observed with extracted guts (see above), an effort was made to identify the component(s) of the hindgut that might account for the remaining 97%. The results of experiments involving placement of R. flavipes on short-term diets intended to eliminate or greatly reduce the total bacterial population and/or the total protozoan population implied that hindgut protozoans, long considered “strict anaerobes,” have the greatest impact on whole-gut O2 consumption rates (Table 3 and Fig. 8A and B). A reduction in the number of protozoans per gut was highly correlated (r2 = 0.95) with whole-gut O2 consumption rates, whereas a disturbance in the number of bacteria was much less strongly correlated (r2 = 0.56). Particularly striking was the effect of antibiotics added to the cellulose diet (Table 3). A 1,000-fold decrease in the number of bacterial CFU (the number of CFU was used as a surrogate for the total number of viable bacteria) was associated with only a 53% decrease in whole-gut O2 consumption; the latter value was essentially identical to the accompanying decrease in the number of protozoans. Conversely, a diet of starch (compared to wood) supported high numbers of viable bacteria but resulted in a 76% decrease in the number of protozoans and a 57% decrease in whole-gut O2 consumption. The lowest O2 consumption rates were observed with guts from termites fed a combination of starch and antibiotics, which after 11 days eliminated nearly all protozoans and bacteria; hence, this activity presumably approximated the respiratory activity of the gut and associated tissues alone. The observed activity (286 pmol · min−1 · gut−1) was a substantial fraction of the theoretical O2 consumption rate (666 pmol · min−1 · gut−1) required for complete oxidation of acetate produced at a rate of 20 nmol · h−1 · termite−1 (see above) and hence is consistent with the notion that much of the respiratory activity of worker larvae is attributable to the gut and its contents.
The basis for the strong correlation between the whole-gut O2 consumption rate and the number of protozoans per gut is unknown. The simplest explanation is that the protozoans themselves, with or without the assistance of prokaryotic endo- and ectosymbionts, are major O2-consuming entities in hindguts and are analogous to certain rumen ciliates that can use O2 as an electron acceptor, if O2 is present at relatively low (nontoxic) concentrations (16). In this regard, it is worth noting that hindgut protozoans occupy the bulk of the hindgut volume (6) and can account for as much as one-third the body weight of termites (24). Moreover, complete anoxia does not occur in hindguts (as measured with microelectrodes) until a distance that is about 150 to 250 μm inward from the hindgut wall (8), yet the wall-associated assemblage of prokaryotic microbiota extends only about 10 to 20 μm inward (6, 8) and so cannot be solely responsible for creation of anoxia. Furthermore, some of the protozoans (e.g., Pyrsonympha species) are often seen attached end-on to the hindgut wall (60) and appear to have attachment organelles that enable them to do this (6). Hence, it is not difficult to imagine that a significant amount of terminal electron flow in these eukaryotes involves not only proton reduction to H2 but also O2 reduction, thereby contributing to anoxia in the lumen and resulting in continuous partial oxidation of plant polysaccharides to the level of acetate, the main carbon and energy source for the insect. Certainly, the present results must be considered preliminary, as we have almost no understanding of how disruption of one or more members of the hindgut microbiota affects the nature and activity of the members that remain or the effect on physiological processes associated with the gut tissue. However, exploration and resolution of these issues seem to be worthwhile avenues for future research.
Acknowledgments
We thank T. M. Schmidt for help with primer design and phylogenetic analyses, Kwi Kim for assistance with RNA work, C. G. Arvidson for providing a culture of N. sicca, and B. Stevenson for helpful discussions and analysis of cbb3 terminal oxidases.
This research was supported by grants from the National Science Foundation (grant IBN-0114505) and Michigan State University Agricultural Experiment Station to J.A.B. and by an MSU College of Natural Science Recruiting Fellowship to J.T.W.
Footnotes
Published ahead of print on 7 September 2007.
REFERENCES
- 1.Aceti, D. J., and J. G. Ferry. 1988. Purification and characterization of acetate kinase from acetate-grown Methanosarcina thermophila. J. Biol. Chem. 263:15444-15448. [PubMed] [Google Scholar]
- 2.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 3.Berchtold, M., A. Chatzinotas, W. Schonhuber, A. Brune, R. Amann, D. Hahn, and H. König. 1999. Differential enumeration and in situ localization of microorganisms in the hindgut of the lower termite Mastotermes darwiniensis by hybridization with rRNA-targeted probes. Arch. Microbiol. 172:407-416. [DOI] [PubMed] [Google Scholar]
- 4.Bouvier, T., and P. A. Del Giorgio. 2003. Factors influencing the detection of bacterial cells using fluorescence in situ hybridization (FISH): a quantitative review of published reports. FEMS Microbiol. Ecol. 44:3-15. [DOI] [PubMed] [Google Scholar]
- 5.Breznak, J. A. 2000. Ecology of prokaryotic microbes in guts of wood- and litter-feeding termites, p. 209-231. In T. Abe, D. E. Bignell, and M. Higashi (ed.), Termites: evolution, sociality, symbiosis, ecology. Kluwer Academic, Norwell, MA.
- 6.Breznak, J. A., and S. Prankratz. 1977. In situ morphology of the gut microbiota of wood-eating termites [Reticulitermes flavipes (Kollar) and Coptotermes formosanus (Shiraki)]. Appl. Environ. Microbiol. 33:406-426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brown, T., M. Jones-Mortimer, and H. Kornberg. 1977. The enzymatic interconversion of acetate and acetyl-coenzyme A in Escherichia coli. J. Gen. Microbiol. 102:327-336. [DOI] [PubMed] [Google Scholar]
- 8.Brune, A., D. Emerson, and J. A. Breznak. 1995. The termite gut microflora as an oxygen sink: microelectrode determination of oxygen and pH gradients in guts of lower and higher termites. Appl. Environ. Microbiol. 61:2681-2687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brune, A., E. Miambi, and J. A. Breznak. 1995. Roles of oxygen and the intestinal microflora in the metabolism of lignin-derived phenylpropanoids and other monoaromatic compounds by termites. Appl. Environ. Microbiol. 61:2688-2695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carpenter, S. E., and D. J. Merkler. 2003. An enzyme-coupled assay for glyoxylic acid. Anal. Biochem. 323:242-246. [DOI] [PubMed] [Google Scholar]
- 11.Clark, D. P., and J. E. Cronan. 1996. Two-carbon compounds and fatty acids as carbon sources, p. 343-357. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, DC.
- 12.Cleveland, L. R. 1926. Symbiosis among animals with special reference to termites and their intestinal flagellates. Q. Rev. Biol. 1:51-64. [Google Scholar]
- 13.Coates, J. D., R. A. Bruce, and J. D. Haddock. 1998. Anoxic bioremediation of hydrocarbons. Nature 396:730. [DOI] [PubMed] [Google Scholar]
- 14.Cozzone, A. J. 1998. Regulation of acetate metabolism by protein phosphorylation in enteric bacteria. Annu. Rev. Microbiol. 52:127-164. [DOI] [PubMed] [Google Scholar]
- 15.Cronan, J. E., and D. Laporte. 1996. Tricarboxylic acid cycle and glyoxylate bypass, p. 206-216. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, DC.
- 16.Ellis, J. E., P. S. Mcintyre, M. Saleh, A. G. Williams, and D. Lloyd. 1991. Influence of CO2 and low concentrations of O2 on fermentative metabolism of the ruminal ciliate Polyplastron multivesiculatum. Appl. Environ. Microbiol. 57:1400-1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fujita, A., I. Shimizu, and T. Abe. 2001. Distribution of lysozyme and protease, and amino acid concentration in the guts of a wood-feeding termite, Reticulitermes speratus (Kolbe): possible digestion of symbiont bacteria transferred by trophallaxis. Physiol. Entomol. 26:116-123. [Google Scholar]
- 18.Gerstmeir, R., V. Wendisch, S. Schnicke, H. Ruan, M. Farwick, D. Reinscheid, and B. J. Eikmanns. 2003. Acetate metabolism and its regulation in Corynebacterium glutamicum. J. Biotechnol. 104:99-122. [DOI] [PubMed] [Google Scholar]
- 19.Graber, J. R., and J. A. Breznak. 2004. Physiology and nutrition of Treponema primitia, an H2/CO2-acetogenic spirochete from termite hindguts. Appl. Environ. Microbiol. 70:1307-1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Graber, J. R., J. R. Leadbetter, and J. A. Breznak. 2004. Description of Treponema azotonutricium sp. nov. and Treponema primitia sp. nov., the first spirochetes isolated from termite guts. Appl. Environ. Microbiol. 70:1315-1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jaramillo, R. D., B. C. Barraza, A. Polo, M. Sara, M. Contreras, and J. E. Escamilla. 2002. The aerobic electron transport system of Eikenella corrodens. Can. J. Microbiol. 48:895-902. [DOI] [PubMed] [Google Scholar]
- 22.Jones, M. E., and F. Lipmann. 1955. Aceto-CoA-kinase. Methods Enzymol. 1:585-595. [Google Scholar]
- 23.Karan, D., J. R. David, and P. Capy. 2001. Molecular evolution of the AMP-forming acetyl-CoA synthetase. Gene 265:95-101. [DOI] [PubMed] [Google Scholar]
- 24.Katzin, L. I., and H. Kirby. 1939. The relative weight of termites and their protozoa. J. Parasitol. 25:444-445. [Google Scholar]
- 25.Kumar, S., K. Tamura, and M. Nei. 2004. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief. Bioinform. 5:150-163. [DOI] [PubMed] [Google Scholar]
- 26.Kumari, S., C. M. Beatty, D. F. Browning, S. J. W. Busby, E. J. Simel, G. Hovel-Miner, and A. J. Wolfe. 2000. Regulation of acetyl coenzyme A synthetase in Escherichia coli. J. Bacteriol. 182:4173-4179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kumari, S., R. Tishel, M. Eisenbach, and A. Wolfe. 1995. Cloning, characterization, and functional expression of acs, the gene which encodes acetyl coenzyme A synthetase in Escherichia coli. J. Bacteriol. 177:2878-2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leadbetter, J. R., and J. A. Breznak. 1996. Physiological ecology of Methanobrevibacter cuticularis sp. nov. and Methanobrevibacter curvatus sp. nov., isolated from the hindgut of the termite Reticulitermes flavipes. Appl. Environ. Microbiol. 62:3620-3631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Leadbetter, J. R., L. D. Crosby, and J. A. Breznak. 1998. Methanobrevibacter filiformis sp. nov., a filamentous methanogen from termite hindguts. Arch. Microbiol. 169:287-292. [DOI] [PubMed] [Google Scholar]
- 30.Lilburn, T. G., T. M. Schmidt, and J. A. Breznak. 1999. Phylogenetic diversity of termite gut spirochaetes. Environ. Microbiol. 1:331-345. [DOI] [PubMed] [Google Scholar]
- 31.Ludwig, W., O. Strunk, R. Westram, L. Richter, H. Meier, Yadhukumar, A. Buchner, T. Lai, S. Steppi, G. Jobb, W. Förster, I. Brettske, S. Gerber, A. W. Ginhart, O. Gross, S. Grumann, S. Hermann, R. Jost, A. König, T. Liss, R. Lüßmann, M. May, B. Nonhoff, B. Reichel, R. Strehlow, A. Stamatakis, N. Stuckmann, A. Vilbig, M. Lenke, T. Ludwig, A. Bode, and K.-H. Schleifer. 2004. ARB: a software environment for sequence data. Nucleic Acids Res. 32:1363-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marcelli, S. W., H.-T. Chang, T. Chapman, P. A. Chalk, R. J. Miles, and R. K. Poole. 1996. The respiratory chain of Helicobacter pylori: identification of cytochromes and the effects of oxygen on cytochrome and menaquinone levels. FEMS Microbiol. Lett 138:59-64. [DOI] [PubMed] [Google Scholar]
- 33.Meister, M., S. Saum, B. E. Alber, and G. Fuchs. 2005. l-Malyl-coenzyme A/β-methylmalyl-coenzyme A lyase is involved in acetate assimilation of the isocitrate lyase-negative bacterium Rhodobacter capsulatus. J. Bacteriol. 187:1415-1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miles, R. D., P. P. Iyer, and J. G. Ferry. 2001. Site-directed mutational analysis of active site residues in the acetate kinase from Methanosarcina thermophila. J. Biol. Chem. 276:45059-45064. [DOI] [PubMed] [Google Scholar]
- 35.Molina, I., M. Pellicer, J. Badia, J. Aguilar, and L. Baldoma. 1994. Molecular characterization of Escherichia coli malate synthase G. Differentiation with the malate synthase A isoenzyme. Eur. J. Biochem. 224:541-548. [DOI] [PubMed] [Google Scholar]
- 36.Musfeldt, M., M. Selig, and P. Schönheit. 1999. Acetyl coenzyme A synthetase (ADP forming) from the hyperthermophilic archaeon Pyrococcus furiosus: identification, cloning, separate expression of the encoding genes, acdAI and acdBI, in Escherichia coli, and in vitro reconstitution of the active heterotetrameric enzyme from its recombinant subunits. J. Bacteriol. 181:5885-5888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Oberlies, G., G. Fuchs, and R. K. Thauer. 1980. Acetate thiokinase and the assimilation of acetate in Methanobacterium thermoautotrophicum. Arch. Microbiol. 128:248-252. [DOI] [PubMed] [Google Scholar]
- 38.Odelson, D. A., and J. A. Breznak. 1983. Volatile fatty acid production by the hindgut microbiota of xylophagous termites. Appl. Environ. Microbiol. 45:1602-1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Oh, M.-K., L. Rohlin, K. C. Kao, and J. C. Liao. 2002. Global expression profiling of acetate-grown Escherichia coli. J. Biol. Chem. 277:13175-13183. [DOI] [PubMed] [Google Scholar]
- 40.Pernthaler, A., J. Pernthaler, and R. Amann. 2002. Fluorescence in situ hybridization and catalyzed reporter deposition for the identification of marine bacteria. Appl. Environ. Microbiol. 68:3094-3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pitcher, R. S., T. Brittain, and N. J. Watmough. 2002. Cytochrome cbb3 oxidase and bacterial microaerobic metabolism. Biochem. Soc. Trans. 30:653-658. [DOI] [PubMed] [Google Scholar]
- 42.Preisig, O., R. Zufferey, L. Thony-Meyer, C. Appleby, and H. Hennecke. 1996. A high-affinity cbb3-type cytochrome oxidase terminates the symbiosis-specific respiratory chain of Bradyrhizobium japonicum. J. Bacteriol. 178:1532-1538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rasche, M. E., K. S. Smith, and J. G. Ferry. 1997. Identification of cysteine and arginine residues essential for the phosphotransacetylase from Methanosarcina thermophila. J. Bacteriol. 179:7712-7717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rose, T., J. Henikoff, and S. Henikoff. 2003. CODEHOP (COnsensus-DEgenerate Hybrid Oligonucleotide Primer) PCR primer design. Nucleic Acids Res. 31:3763-3766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schafer, T., and P. Schönheit. 1991. Pyruvate metabolism of the hyperthermophilic archaebacterium Pyrococcus furiosus. Arch. Microbiol. 155:366-377. [Google Scholar]
- 46.Schultz, J. E., and J. A. Breznak. 1978. Heterotrophic bacteria present in hindguts of wood-eating termites [Reticulitermes flavipes (Kollar)]. Appl. Environ. Microbiol. 35:930-936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Slaytor, M. 2000. Energy metabolism in the termite and its gut microbiota, p. 307-332. In T. Abe, D. E. Bignell, and M. Higashi (ed.), Termites: evolution, sociality, symbiosis, ecology. Kluwer Academic, Norwell, MA.
- 48.Smith, M. A., M. Finel, V. Korolik, and G. L. Mendz. 2000. Characteristics of the aerobic respiratory chains of the microaerophiles Campylobacter jejuni and Helicobacter pylori. Arch. Microbiol. 174:1-10. [DOI] [PubMed] [Google Scholar]
- 49.Smith, P. K., R. I. Krohn, G. T. Hermanson, A. K. Mallia, F. H. Gartner, M. D. Provenzano, E. K. Fujimoto, N. M. Goeke, B. J. Olson, and D. C. Klenk. 1985. Measurement of protein using bicinchoninic acid. Anal. Biochem. 150:76-85. [DOI] [PubMed] [Google Scholar]
- 50.Stevenson, B. S., S. A. Eichorst, J. T. Wertz, T. M. Schmidt, and J. A. Breznak. 2004. New strategies for cultivation and detection of previously uncultured microbes. Appl. Environ. Microbiol. 70:4748-4755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Thimm, T., and C. C. Tebbe. 2003. Protocol for rapid fluorescence in situ hybridization of bacteria in cryosections of microarthropods. Appl. Environ. Microbiol. 69:2875-2878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tholen, A., and A. Brune. 2000. Impact of oxygen on metabolic fluxes and in situ rates of reductive acetogenesis in the hindgut of the wood-feeding termite Reticulitermes flavipes. Environ. Microbiol. 2:436-449. [DOI] [PubMed] [Google Scholar]
- 53.Tholen, A., B. Schink, and A. Brune. 1997. The gut microflora of Reticulitermes flavipes, its relation to oxygen, and evidence for oxygen-dependent acetogenesis by the most abundant Enterococcus sp. FEMS Microbiol. Ecol. 24:137-149. [Google Scholar]
- 54.Veivers, P. C., R. W. O'Brien, and M. Slaytor. 1983. Selective defaunation of Mastotermes darwiniensis and its effect on cellulose and starch metabolism. Insect Biochem. 13:95-101. [Google Scholar]
- 55.Wertz, J. T., and J. A. Breznak. 2007. Stenoxybacter acetivorans gen. nov., sp. nov., an acetate-oxidizing obligate microaerophile among diverse O2-consuming bacteria from termite guts. Appl. Environ. Microbiol. 73:6819-6828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Whiteley, H. R., and R. A. Pelroy. 1972. Purification and properties of phosphotransacetylase from Veillonella alcalescens. J. Biol. Chem. 247:1911-1917. [PubMed] [Google Scholar]
- 57.Winzer, K., K. Lorenz, and P. Dürre. 1997. Acetate kinase from Clostridium acetobutylicum: a highly specific enzyme that is actively transcribed during acidogenesis and solventogenesis. Microbiology 143:3279-3286. [DOI] [PubMed] [Google Scholar]
- 58.Wolfe, A. J. 2005. The acetate switch. Microbiol. Mol. Biol. Rev. 69:12-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wood, H. G. 1977. Some reactions in which inorganic pyrophosphate replaces ATP and serves as a source of energy. Fed. Proc. 36:2197-2205. [PubMed] [Google Scholar]
- 60.Yang, H., D. Schmitt-Wagner, U. Stingl, and A. Brune. 2005. Niche heterogeneity determines bacterial community structure in the termite gut (Reticulitermes santonensis). Environ. Microbiol. 7:916-932. [DOI] [PubMed] [Google Scholar]