Abstract
The occurrence of taste and odor problems in drinking water supplies is a widespread phenomenon. From a Saxonian water reservoir we isolated a cyanobacterial species which was classified as Phormidium sp. Under laboratory conditions it produced an earthy-musty smell due to the synthesis of geosmin. The only genes shown to be involved in geosmin biosynthesis are cyc2 and geoA of Streptomyces. Based on the alignment of Cyc2 with a putative sesquiterpene synthase of Nostoc punctiforme, a degenerate primer pair was designed. By PCR, we could amplify two similar genes in Phormidium sp., which we named geoA1 and geoA2. Their expression was studied by reverse transcription-PCR. This revealed that both genes are expressed at 20°C and a light-dark cycle of 12 h. Expression was not detectable at the end of a 24-h dark period. To analyze the prevalence of geoA1 and geoA2 in samples from the phytobenthos, we generated PCR fragments with the same degenerate primer pair. Fifty-five different sequences that might represent geoA variants were obtained. The GC content ranged from 42% to 67%, suggesting that taxonomically very different bacteria might contain such genes.
The occurrence of taste and odor problems (TO problems) in drinking water supplies is a widespread and worldwide issue (9, 11, 12, 18, 27, 29, 32). It is a general demand in all countries that drinking water should be free from flavor and smell. The odor is described mostly as an earthy-musty smell, which may be caused by the volatile sesquiterpene geosmin. This bicyclic molecule has a very low odor threshold concentration of 4 ng/liter (33). Many microorganisms, such as actinomycetes (6), cyanobacteria (9), myxobacteria (25), or fungi (15), can produce it (30).
For TO episodes in water supply systems, actinomycetes and cyanobacteria are most relevant. Both planktonic and benthic forms of cyanobacteria were found to be odor producers (10, 14, 22, 23, 26, 28). Benthic cyanobacteria are often present in large quantities in drinking water reservoirs of Saxony where TO episodes have been observed. We were interested in identifying benthic cyanobacteria that are likely to be involved in such odor problems and to characterize the corresponding genes. We analyzed different isolates for geosmin production. The isolate with highest production was chosen for genetic characterization.
Useful reports on the genetic background of geosmin production deal with Streptomyces. Bentley and Meganathan (1) showed in Streptomyces antibioticus that geosmin derives from the isoprenoid pathway. Later it was found that the gene cyc2 is involved in the biosynthesis of geosmin in Streptomyces coelicolor (2, 7, 13). The homologue sesquiterpene synthase in Streptomyces avermitilis was named geoA (3). Based on this, we designed primers that were used for the identification of two similar genes in the above-mentioned isolate. Their expression was studied by reverse transcription-PCR (RT-PCR).
MATERIALS AND METHODS
Cultivation and isolation of benthic cyanobacteria.
Material from the phytobenthic mats of the Saidenbach and Klingenberg (Ore Mountains) drinking water reservoirs was used to prepare single trichomes of cyanobacteria. These were cultivated on solid BG-11 medium (24) in a plant chamber (MLR-351 [Sanyo Electric Co., Japan], equipped with a Philips TLD36W/965 fluorescent tube) at 20°C and a light-dark-cycle of 12 h.
For isolation of RNA, Phormidium sp. was grown in 40 ml liquid BG-11-medium in 100-ml flasks supplemented with 15 g of glass beads (3 mm in diameter). If not otherwise stated, the temperature and light-dark cycle were as described above. The medium was exchanged after 4 to 6 weeks. RNA was isolated after additional incubation for 1 to 2 weeks.
Heterotrophic bacteria that were contaminating the Phormidium sp. culture were grown in Luria-Bertani (LB) medium (21) and on starch-casein agar (containing [per liter] 10 g soluble starch, 0.3 g casein, 2 g K2HPO4, 2 g KNO3, 2 g NaCl, 0.05 g MgSO4·7H2O, 0.02 g CaCO3, 0.01 g FeSO4·7H2O, 15 g agar).
General molecular techniques.
DNA was isolated from samples of the phytobenthic belt of the drinking water reservoir Klingenberg (from a depth of about 4 m) or from unialgal cyanobacterial cultures using the Fast DNA spin kit for soil (MP Biomedicals, Eschwege, Germany). The oligonucleotides used in this study are depicted in Table 1. Specific primers were designed using the primer3 program (19).
TABLE 1.
Oligonucleotides used in this study
| Primer | Sequence (5′→3′)a |
|---|---|
| CycFW | TGGTAYGTITGGGTITTYTTYTTYGAYGAYCAYTT |
| CycRW | CATRTGCCAYTCRTGICCICCISWYTGCCARTCYTG |
| Vect57 | GATCGAAGGAGAGGACGCTGTCTGTCGAAGGTAAGAACGGACGAGAGAAGGGAGAG |
| Vect53 | CTCTCCCTTCTCGAATCGTAACCGTTCGTACGAGAATCGCTGTCCTCTCCTTC |
| ForCycA | GGATTGCTAACCCCATTGAATA |
| RevCycA | GATAAGCCTTAGCCCCTACCA |
| Primer AF | GCAACAGTTTCAACACATCGTTAAG |
| Primer AR | CCTAAACCCAAAGTAGCGTATAACGA |
| Primer BF | TTAAGACTGAATTACCAGCCCTTTTC |
| Primer BR | TAAACCTGTAGGCCAGCCTAAAATC |
| Primer CF | AAGTACCCGCGAAAAACTAATGAAA |
| Primer CR | CTTCCTGTTGATTCATGTAGCGACTC |
| GeoA1_for | CGCTATATGAACGAAGAAGC |
| GeoA1_rev | ACGCTTGGTATCTCTTGGTA |
| GeoA2_for | ACGATGAGTTTGTCTCGACT |
| GeoA2_rev | ATATCATTTGTAAAACAGGCAA |
| rbcL_for | ATCGACCGTCAAAAGAACC |
| rbcL_rev | CTTGGGTGAAGTAGATACCG |
Nucleotides are abbreviated according to the International Union of Biochemistry code.
For amplification of specific fragments from genomic DNA, the HotMasterMix (Eppendorf, Hamburg, Germany) was used. Two hundred picomoles of degenerate primers (CycFW and CycRW) or 50 pmol of nondegenerate primers was used in a final volume of 50 μl, containing about 150 ng of total DNA. Prior to amplification, DNA was denatured for 2 min at 95°C. For the primer pair CycFW and CycRW, the cycling parameters were 95°C for 1 min, 50°C for 1 min, and 68°C for 90 s (35 cycles). After amplification, the sample was incubated for 10 min at 68°C.
To determine the 5′ end of geoA1, the vectorette approach (16) was used. Annealing of vectorette oligonucleotides Vect57 and Vect53 resulted in a single-stranded overhang compatible to BamHI- or BglII-restricted DNA. About 100 ng of total DNA of Phormidium sp. was digested with BamHI and BglII. About 30 ng of digested DNA was ligated with 6.6 pmol vectorette units. Subsequently, vectorette and geoA1 primers were used to amplify the 5′ end of geoA1.
For amplification of the 3′ end of geoA2, 100 ng of the genomic DNA of Phormidium sp. was digested with PagI (Fermentas, St. Leon-Rot, Germany). Restricted DNA was circularized and used for nested PCR with specific primer pairs (AF/AR, BF/BR, and CF/CR).
Amplified sesquiterpene synthase DNA fragments were purified using the QIAquick PCR purification kit (QIAGEN, Hilden, Germany). PCR products were cloned into vector PCR2.1-TOPO (Invitrogen, Karlsruhe, Germany).
Isolation and purification of RNA.
Cells were harvested at the end of the light phase. The flasks with the Phormidium sp. cultures were shaken to detach biomass. The biomass was transferred into centrifuge tubes containing 4 ml stop solution (10% buffered phenol in ethanol). After centrifugation (2,400 × g, 4°C, 4 min), the supernatant was decanted and the cell pellet was flash frozen with liquid nitrogen and stored at −70°C for at least 1 hour. Subsequently, the cell pellet was resuspended in 1.5 ml cold buffer A (330 μl 3 M sodium acetate [pH 5.5], 100 μl 0.5 M EDTA [pH 8.0], 49.57 ml diethyl pyrocarbonate-treated double-distilled water). The suspension was then quickly mixed with a preheated phenol solution (3.5 ml phenol equilibrated with buffer A, 160 μl of 10% sodium dodecyl sulfate, and 2 ml of buffer A, 65°C). The mixture was vortexed for 1 min, incubated for 2 min at 65°C, vortexed again for 1 min, and incubated for 5 min at 65°C. After centrifugation (8,400 × g, 4°C, 5 min), the upper phase was taken and the purification procedure with preheated phenol solution was repeated. The upper phase was transferred into 3 ml of a cold solution of phenol-chloroform-isoamyl alcohol (25:24:1, vol/vol/vol). After mixing vigorously, the suspension was centrifuged (8,400 × g, 4°C, 5 min), and the upper phase was extracted with 3 ml chloroform. After centrifugation (8,400 × g, 4°C, 5 min), the upper phase was transferred to a 15-ml centrifuge tube and RNA was precipitated by addition of sodium acetate (0.3 M final concentration) and 2 volumes of cold ethanol and incubation at −70°C for at least 1 hour. RNA was harvested by centrifugation (15.500 × g, 4°C, 1 h) and washed with 1 ml of 80% ethanol. After drying, RNA was resuspended in 50 μl H2O (GIBCO ultraPURE; Invitrogen).
For removal of DNA, the RNA sample was treated with DNase I. Up to 60 μg of RNA was mixed with 100 U SUPERaseIn (Ambion, Huntingdon, United Kingdom) and 20 U of DNase RQ1 (Promega, Mannheim, Germany) in a total volume of 200 μl. After incubation for 25 min at 37°C, the RNA was transferred to a new microcentrifuge tube. RQ1 DNase (20 U) was added again, and the mixture was incubated at 37°C for 25 min. The RNA was purified with the RNeasy minikit (QIAGEN) according to the manufacturer's protocol with the exception that 700 μl of RLT buffer and 500 μl of 96% ethanol were used. For elution of RNA, 32 μl of RNase-free water was used. To increase the amount of recovered RNA, the elution was repeated with the first eluate. To check for the presence of DNA in the RNA sample, a PCR with 1 μl of the RNA sample as the template was done.
Expression analyses by RT-PCR.
For cDNA synthesis, 750 ng random hexamer primers (Invitrogen) was added to 5 to 10 μg of RNA in a total volume of 30 μl. For primer annealing, the mixture was incubated for 10 min at 70°C, 10 min at 25°C, and 5 min at 4°C. For reverse transcription, 12 μl of 5× Moloney murine leukemia virus first-strand buffer, 3 μl of 10 mM deoxynucleoside triphosphates, 1.5 μl of SUPERaseIn (20 U/μl), 7.5 μl of Moloney murine leukemia virus reverse transcriptase (200 U/μl; Promega), and 6 μl distilled water (Invitrogen) were added and incubated for 10 min at 25°C, 60 min at 37°C, 60 min at 42°C, and 10 min at 70°C. The remaining RNA was degraded by addition of 20 μl 1 M NaOH and incubation at 65°C for 30 min. For neutralization, 20 μl of 1 M HCl was added. cDNA was purified with the MinElute PCR purification kit (QIAGEN) with the exception that 10 μl of 3 M sodium acetate (pH 5.5) was added as a first step. cDNA was eluted with 12 μl of elution buffer. Finally, 24 μl of H20 were added. For PCR, 1 μl of cDNA solution was used.
Analyses of odorous substances by solid-phase microextraction (SPME).
Samples of the epipelic cyanobacteria layer from the sediment (about 4 m depth) were taken from the Klingenberg drinking water reservoir by using a Van Veen grab sampler. Transport was in a box under dark and cool conditions. Samples were immediately analyzed or stored at −18°C.
The following chemicals were used to verify the results of gas chromatography/mass spectrometry (GC/MS) screening analysis: geosmin, 2-methylisoborneol, 2-isobutyl-3-methoxypyrazine, and 2-isopropyl-3-methoxypyrazine (all from Supelco, Munich, Germany); β-ionone (ABCR, Karlsruhe, Germany); and borneol, 1,4-cineole, octanal, and geranyl acetone (all from Fluka, Munich, Germany). Each chemical had a purity of >95%. Biphenyl D10 (Ehrenstorfer, Augsburg, Germany) and 2,4,6-trichloroanisole (Supelco) were used as internal standards.
A HP5890 series 2 GC and the mass-selective detector 5971 series 2 (both from Hewlett-Packard) were utilized for the determination of odor compounds. The column used was an SPB-1 column (Supelco; 60 m by 0.25 mm [inner diameter]; film thickness, 1 μm). The injection and detector temperature were 250°C and 275°C, respectively. The SPME fibers consisted of 2-cm DVB/Carboxen/PDMS fibers (Supelco; film thickness, 50/30 μm) and were used in a manual sorption and desorption mode.
An aliquot of 1 ml biomass/water suspension was filled up to 20 ml with pure water. After addition of 3 g sodium chloride, the SPME vial was sealed with a Teflon-lined silicon rubber septum cap. The fiber was positioned in the headspace of the vial. The sample thermostated at 60°C was stirred at highest rotation (1280 rpm) for 60 min. For desorption, the fiber was inserted into the GC injector at 250°C for 15 min.
The injection and detector temperatures were 250°C and 275°C, respectively. The column temperature was set at 100°C for 2 min and then increased to 160°C at a rate of 12°C/min, then to 230°C at 7°C/min, then to 275°C at 25°C/min, and then 9 min isothermal. The carrier gas was helium with a flow rate of 0.8 ml/min. The mass range in scan mode was selected from 50 to 200; the scan cycle time was approximately 5 scans per second.
The identification of volatile components was based both on the comparison of their retention times with retention times of standard compounds and on the matching of mass spectra of the compounds with the reference mass spectra of the library (NIST98).
Nucleotide sequence accession numbers.
Sequences have been deposited in the NCBI nucleotide sequence database under accession numbers EF626645 to EF626656.
RESULTS
Isolation of a benthic cyanobacterium involved in geosmin synthesis.
A small sample of phythobenthic material from the Saidenbach drinking water reservoir was analyzed under a binocular microscope. Single trichomes were picked and grown on BG-11 agar plates. In this way, unialgal cultures could be obtained. One culture produced geosmin in a large quantity (>5,000 ng/mg ash-free dry weight). Microscopic analysis as described by Komárek and Anagnostidis (17) indicated that the isolated cyanobacterium is a Phormidium sp. strain (data not shown).
Identification of putative sesquiterpene synthase genes of Phormidium sp.
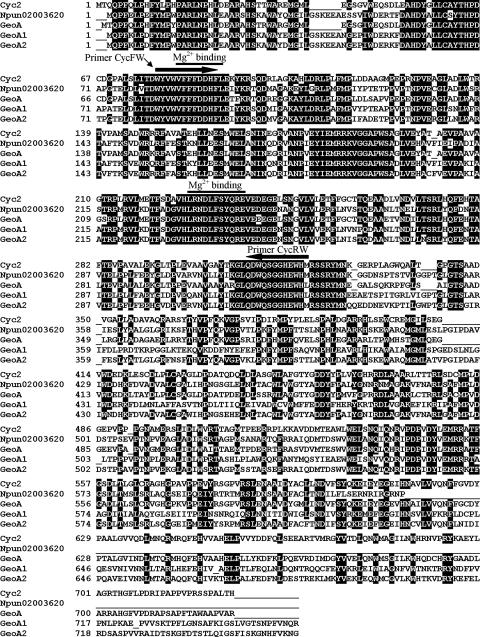
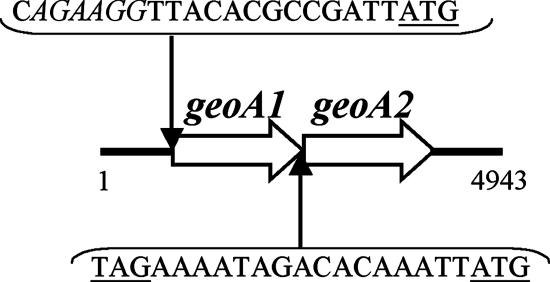
A database search with Cyc2 of Streptomyces coelicolor indicated the presence of similar proteins in Nostoc punctiforme PCC73102 (ZP_00109187) and S. avermitilis (NP_823339; this sequence became available after we had finished the cloning experiments). Alignment of the proteins revealed highly conserved regions (Fig. 1). To search for a similar gene in Phormidium sp., we designed degenerate primers (CycFW and CycRW) based on two highly conserved regions. By PCR from total DNA, we amplified a fragment that exhibited similarity to cyc2. Using gene-specific primers (ForCycA and RevCycA), we obtained a 2-kb PCR fragment. Its nucleotide sequence showed that there are two tandemly oriented open reading frames (ORFs) with high similarity (Fig. 2), which we named geoA1 and geoA2. The short distance between the two ORFs suggests that they form an operon. To identify the 5′ end, the vectorette approach was successfully used (see Materials and Methods). For sequencing the 3′ end, total DNA was cut with PagI and ligated for circularization. Primers specific for the ends of the known sequence were used to amplify the intervening DNA. This led to a total sequence of 4.9 kb encompassing two ORFs with upstream and downstream flanking regions (EF619621) (Fig. 2). Downstream, no rho-independent transcriptional terminator was identified. A sequence similar to a Shine Dalgarno sequence was found 13 nucleotides upstream of the putative start codon of geoA1.
FIG. 1.
Amino acid sequence alignment of sesquiterpene synthases. Cyc2, protein from S. coelicolor (Q9X839); Npun02003620, hypothetical protein from N. punctiforme (ZP_00109187); GeoA, protein from S. avermitilis (NP_823339); GeoA1 and GeoA2, hypothetical proteins identified in this study from Phormidium sp. Possible Mg2+ binding sites (7) are marked by lines.
FIG. 2.
Map of geoA1 and geoA2 of Phormidium sp. The geoA1 gene encodes a 753-amino-acid protein, and geoA2 encodes a 755-amino-acid protein. A sequence similar to a Shine Dalgarno sequence upstream of geoA1 is indicated in italic. Putative start and stop codons are underlined.
The deduced GeoA1 and GeoA2 amino acid sequences are highly similar to each other (62% identity) (Fig. 1). The N-terminal domains of both proteins are more conserved than the C-terminal domains. Furthermore, GeoA1 and GeoA2 show the typical repeated domain structure of Cyc2 (7) (Fig. 3).
FIG. 3.
Amino acid sequence alignment of the N- and C-terminal domains of GeoA1 and GeoA2, revealing repeated motifs.
Although the culture was derived from single trichome separations, we could not eliminate heterotrophic bacteria, a common problem with cyanobacterial cultures from environmental samples (5). To make sure that geoA1 and geoA2 originate from Phormidium sp., several experiments were done. Heterotrophic bacteria that were grown in LB medium were analyzed by PCR with geoA1-specific primers. No PCR product could be obtained. Furthermore, bacteria with mycelial growth could not be found on starch-casein agar. This suggests that actinomycetes that might contain genes similar to geoA were not present. Moreover, no odor production was observed. In contrast, geoA1 could be amplified from samples containing 10 to 20 trichomes (data not shown).
Expression analysis of geoA1 and geoA2.
The Phormidium sp. culture produced geosmin in large amounts at 20°C and a light-dark-cycle of 12 h (data not shown). To test if geoA1 and geoA2 are expressed under the same conditions, RNA was isolated at the end of the light phase. RNA was transcribed into cDNA using random primers. As positive control we used rbcL-specific primers. rbcL encodes the large subunit of ribulose-1,5-bisphosphate carboxylase and was expressed under the tested growth conditions in the light phase. PCR with specific primers for geoA1 and geoA2 yielded the expected fragments (Fig. 4), indicating that geoA1 and geoA2 are expressed. No geoA1-specific fragments were obtained with RNA alone.
FIG. 4.
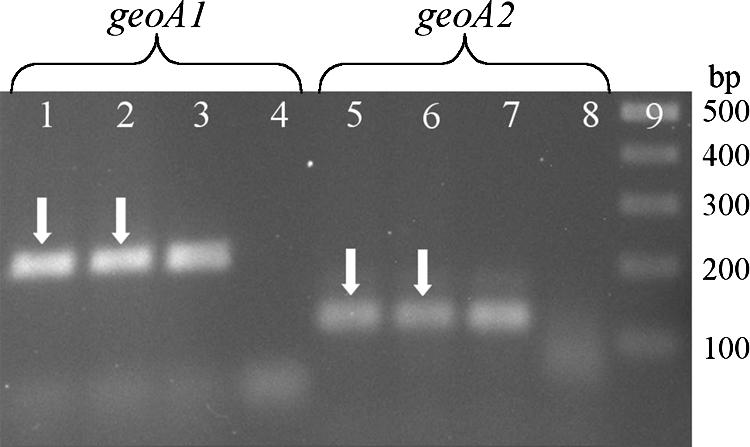
Separation of RT-PCR products (arrows) obtained with RNA isolated from cultures grown at 20°C and at the end of a 12-h light period. Lanes 1, 2, 5, and 6, amplification products from two independent experiments with specific primers for geoA1 and geoA2, respectively; lanes 3 and 7, positive controls using genomic DNA of Phormidium sp. as the template for PCR; lanes 4 and 8, negative control without addition of template; lane 9, size marker.
To test if low temperature affects expression, cultures were incubated at 10°C for 24 h with the normal light-dark-cycle of 12 h. RNA was isolated at the end of the light phase. RT-PCR revealed that geoA1, geoA2, and rbcL were expressed (data not shown). In contrast, if we isolated RNA after incubation of the cultures for 24 h in the dark (20°C), geoA1 and geoA2 expression could not be found. However, rbcL expression was still detectable (data not shown).
Putative sesquiterpene synthase genes in environmental samples.
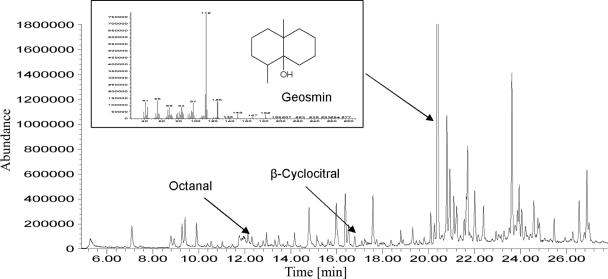
In the Klingenberg drinking water reservoir, TO episodes were monitored for many decades. At a water depth of 3 to 5 meters, a belt of epipelic phytobenthos (mainly cyanobacteria and diatoms) was found. For several years samples were taken from this belt. In every biomass sample large amounts of geosmin were detected (Fig. 5). By using GC/MS in scan mode, other odor compounds as 2-methylisoborneol, β- ionone and some aldehydes were identified, but their concentrations were significantly lower than those of geosmin.
FIG. 5.
Chromatogram (headspace-SPME and GC/MS scan) from a suspension of biomass of benthic cyanobacteria from the Klingenberg drinking water reservoir.
To test for the prevalence of sesquiterpene synthase genes, total DNA was extracted. PCR fragments were generated using the degenerate primers CycFW and CycRW. After cloning, the nucleotide sequences of the fragments were established. Fifty-five different sequences have been obtained. All derived amino acid sequences showed similarity to sesquiterpene synthases. The diversity suggests that they were derived from taxonomically very different organisms. This is also reflected by the divergent GC content, which ranged from 42% to 67%.
DISCUSSION
In this study, the geoA1 and geoA2 genes were identified from a cyanobacterial culture that produced geosmin in a very high quantity. Microscopic analysis suggests that the isolate belongs to Phormidium sp. Although we could not eliminate heterotrophic bacteria in our culture, several results support the conclusion that geoA1 and geoA2 are involved in geosmin production. Odor production was observed only if Phormidium sp. was present in the cultures. PCR fragments with geoA1- and geoA2-specific primers could be obtained only if the sample contained DNA of Phormidium sp. Furthermore, expression studies showed that the genes are expressed under light but not after 24 h in the dark, indicating that the geosmin-producing organism is a phototrophic bacterium. Interestingly, rbcL mRNA was still detectable under the same conditions. Chow and Tabita (4), using Northern analysis, reported that the rbcL gene was still expressed at a low level during a 12-h dark period. We also found expression of geoA1 and geoA2 at 10°C, which is more similar to the temperature under environmental conditions. Geosmin production at low temperature is a well-established fact (20, 26). Our experimental data together with the high similarity of GeoA1 and GeoA2 to Cyc2 and GeoA of Streptomyces (Fig. 1) suggest that these genes are responsible for geosmin synthesis in Phormidium sp. Although we found Phormidium sp. only at low abundance (Limnothrix sp. was dominant), it seems to be a main producer of geosmin.
geoA1 and geoA2 are likely to form an operon. The two ORFs are only a few base pairs apart. At 13 nucleotides upstream of geoA1 is an AG-rich region, which might be involved in translation initiation. For Synechocystis sp., Yada et al. (31) calculated an average distance of 9.7 bp between the Shine-Dalgarno sequence and the translational start codon.
To test for the prevalence of sequences similar to geoA in environmental samples, we used the same degenerate primer pair that was used for the identification of geoA1 and geoA2. The GC content of the obtained partial sesquiterpene synthase sequences varies from 42% to 67%, indicating that the primers are suitable for amplification of sesquiterpene synthase genes with very different base compositions. The differences in GC content could mean that the fragments originated from taxonomically very different organisms. However, cyanobacteria are also very diverse, and their GC content ranges from 35% to 71% (8). Therefore, the obtained sequences might be of cyanobacterial origin.
Further studies are required to identify the corresponding microorganisms and their contribution to geosmin production. The question of under what environmental conditions geosmin is released into the water is still open.
Acknowledgments
This study was partly financed by Landestalsperrenverwaltung Sachsen (LTV Sachsen) in cooperation with DREWAG Dresden GmbH.
We thank Claudia Häder for cloning and sequencing of environmental sesquiterpene synthase sequences. We are grateful to Dietrich Uhlmann for valuable comments on the manuscript.
Footnotes
Published ahead of print on 7 September 2007.
REFERENCES
- 1.Bentley, R., and R. Meganathan. 1981. Geosmin and methylisoborneol biosynthesis in streptomycetes. Evidence for an isoprenoid pathway and its absence in non-differentiating isolates. FEBS Lett. 125:220-222. [DOI] [PubMed] [Google Scholar]
- 2.Cane, D. E., and R. M. Watt. 2003. Expression and mechanistic analysis of a germacradienol synthase from Streptomyces coelicolor implicated in geosmin biosynthesis. Proc. Natl. Acad. Sci. USA 100:1547-1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cane, D. E., X. He, S. Kobayashi, S. Omura, and H. Ikeda. 2006. Geosmin biosynthesis in Streptomyces avermitilis. Molecular cloning, expression, and mechanistic study of the germacradienol/geosmin synthase. J. Antibiot. (Tokyo) 59:471-479. [DOI] [PubMed] [Google Scholar]
- 4.Chow, T.-J., and F. R. Tabita. 1994. Reciprocal light-dark transcriptional control of nif and rbc expression and light-dependent posttranslational control of nitrogenase activity in Synechococcus sp. strain RF-1. J. Bacteriol. 176:6281-6285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ferris, M. J., and C. F. Hirsch. 1991. Method for isolation and purification of cyanobacteria. Appl. Environ. Microbiol. 57:1448-1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gerber, N. N., and H. A. Lechevalier. 1965. Geosmin, an earthy-smelling substance isolated from actinomycetes. Appl. Microbiol. 13:935-938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gust, B., G. L. Challis, K. Fowler, T. Kieser, and K. F. Chater. 2003. PCR-targeted Streptomyces gene replacement identifies a protein domain needed for biosynthesis of the sesquiterpene soil odor geosmin. Proc. Natl. Acad. Sci. USA 100:1541-1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Herdman, M., M. Janvier, J. B. Waterbury, R. Rippka, R. Y. Stanier, and M. Mandel. 1979. Deoxyribonucleic acid base composition of cyanobacteria. J. Gen. Microbiol. 111:63-71. [Google Scholar]
- 9.Izaguirre, G., C. J. Hwang, S. W. Krasner, and M. J. McGuire. 1982. Geosmin and 2-methylisoborneol from cyanobacteria in three water supply systems. Appl. Environ. Microbiol. 43:708-714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Izaguirre, G., C. J. Hwang, S. W. Krasner, and M. J. McGuire. 1983. Production of 2-methylisoborneol by two benthic cyanophyta. Water Sci. Technol. 15:211-220. [Google Scholar]
- 11.Izaguirre, G., and W. D. Taylor. 1995. Geosmin and 2-methylisoborneol production in a major aqueduct system. Water Sci. Technol. 31:41-48. [Google Scholar]
- 12.Izaguirre, G., and W. D. Taylor. 2004. A guide to geosmin- and MIB-producing cyanobacteria in the United States. Water Sci. Technol. 49:19-24. [PubMed] [Google Scholar]
- 13.Jiang, J., X. He, and D. E. Cane. 2006. Geosmin biosynthesis. Streptomyces coelicolor germacradienol/germacrene D synthase converts farnesyl diphosphate to geosmin. J. Am. Chem. Soc. 128:8128-8129. [DOI] [PubMed] [Google Scholar]
- 14.Jüttner, F. 1984. Dynamics of the volatile organic substances associated with cyanobacteria and algae in a eutrophic shallow lake. Appl. Environ. Microbiol. 47:814-820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kikuchi, T., S. Kadota, H. Suehara, A. Nishi, and K. Tsubaki. 1981. Odorous metabolites of a fungus, Chaetomium globosum KINZE ex FR.; identification of geosmin, a musty-smelling compound. Chem. Pharm. Bull. (Tokyo) 29:1782-1784. [Google Scholar]
- 16.Ko, W. Y., R. M. David, and H. Akashi. 2003. Molecular phylogeny of the Drosophila melanogaster species subgroup. J. Mol. Evol. 57:562-573. [DOI] [PubMed] [Google Scholar]
- 17.Komárek, J., and K. Anagnostides. 2005. Cyanoprokaryota. 2. Teil: Oscillatoriales, p. 340-343, 390-483. In B. Büdel, L. Krienitz, G. Gärtner, and M. Schagerl (ed.), Süsswasserflora von Mitteleuropa, band 19(2). Elsevier, Munich, Germany.
- 18.Oikawa, E., and Y. Ishibashi. 2004. Species specificity of musty odor producing Phormidium tenue in Lake Kamafusa. Water Sci. Technol. 49:41-46. [PubMed] [Google Scholar]
- 19.Rozen, S., and H. Skaletsky. 2000. Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 132:365-386. [DOI] [PubMed] [Google Scholar]
- 20.Sabater, S., E. Vilalta, A. Gaudes, H. Guasch, I. Muñoz, and A. Romaní. 2003. Ecological implications of mass growth of benthic cyanobacteria in rivers. Aquat. Microb. Ecol. 32:175-184. [Google Scholar]
- 21.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 22.Schrader, K. K., and M. E. Dennis. 2005. Cyanobacteria and earthy/musty compounds found in commercial catfish (Ictalurus punctatus) ponds in the Mississippi Delta and Mississippi-Alabama Blackland Prairie. Water Res. 39:2807-2814. [DOI] [PubMed] [Google Scholar]
- 23.Skjevrak, I., V. Lund, K. Ormerod, and H. Herikstad. 2005. Volatile organic compounds in natural biofilm in polyethylene pipes supplied with lake water and treated water from the distribution network. Water Res. 39:4133-4141. [DOI] [PubMed] [Google Scholar]
- 24.Stanier, R. Y., R. Kunisawa, M. Mandel, and G. Cohen-Bazire. 1971. Purification and properties of unicellular blue-green algae (order Chroococcales). Bacteriol. Rev. 35:171-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Trowitzsch, W., L. Witte, and H. Reichenbach. 1981. Geosmin from earthy smelling cultures of Nannocystis exedens (Myxobacterales). FEMS Microbiol. Lett. 12:257-260. [Google Scholar]
- 26.Vilalta, E., H. Guasch, I. Muñoz, A. Romaní, F. Valero, J. J. Rodriguez, R. Alcaraz, and S. Sabater. 2004. Nuisance odours produced by benthic cyanobacteria in a Mediterranean river. Water Sci. Technol. 49:25-31. [PubMed] [Google Scholar]
- 27.Watson, S. B., and J. Ridal. 2004. Periphyton: a primary source of widespread and severe taste and odour. Water Sci. Technol. 49:33-39. [PubMed] [Google Scholar]
- 28.Watson, S. B., J. Ridal, B. Zaitlin, and A. Lo. 2003. Odours from pulp mill effluent treatment ponds: the origin of significant levels of geosmin and 2-methylisoborneol (MIB). Chemosphere 51:765-773. [DOI] [PubMed] [Google Scholar]
- 29.Westerhoff, P., M. Rodriguez-Hernandez, L. Baker, and M. Sommerfeld. 2005. Seasonal occurrence and degradation of 2-methylisoborneol in water supply reservoirs. Water Res. 39:4899-4912. [DOI] [PubMed] [Google Scholar]
- 30.Wood, S., S. T. Williams, and W. R. White. 2001. Microbes as a source of earthy flavours in potable water—a review. Int. Biodet. Biodeg. 48:26-40. [Google Scholar]
- 31.Yada, T., T. Sazuka, and M. Hirosawa. 1997. Analysis of sequence patterns surrounding the translation initiation sites on cyanobacterium genome using the hidden Markov model. DNA Res. 4:1-7. [DOI] [PubMed] [Google Scholar]
- 32.Yagi, M., M. Kajino, U. Matsuo, K. Ashitani, T. Kita, and T. Nakamura. 1983. Odor problems in Lake Biwa. Water Sci. Technol. 15:311-321. [Google Scholar]
- 33.Young, W. F., H. Horth, R. Crane, T. Ogden, and M. Arnott. 1996. Taste and odour threshold concentrations of potential potable water contaminants. Water Res. 30:331-340. [Google Scholar]