Abstract
The cotton bollworm Helicoverpa armigera is the major insect pest targeted by cotton genetically engineered to produce the Bacillus thuringiensis toxin (transgenic Bt cotton) in the Old World. The evolution of this pest's resistance to B. thuringiensis toxins is the main threat to the long-term effectiveness of transgenic Bt cotton. A deletion mutation allele (r1) of a cadherin gene (Ha_BtR) was previously identified as genetically linked with Cry1Ac resistance in a laboratory-selected strain of H. armigera. Using a biphasic screen strategy, we successfully trapped two new cadherin alleles (r2 and r3) associated with Cry1Ac resistance from a field population of H. armigera collected from the Yellow River cotton area of China in 2005. The r2 and r3 alleles, respectively, were created by inserting the long terminal repeat of a retrotransposon (designated HaRT1) and the intact HaRT1 retrotransposon at the same position in exon 8 of Ha_BtR, which results in a truncated cadherin containing only two ectodomain repeats in the N terminus of Ha_BtR. This is the first time that the B. thuringiensis resistance alleles of a target insect of Bt crops have been successfully detected in the open field. This study also demonstrated that bollworm larvae carrying two resistance alleles can complete development on Bt cotton. The cadherin locus should be an important target for intensive DNA-based screening of field populations of H. armigera.
The global area of cotton (Gossypium hirsutum L.) genetically engineered to produce the Bacillus thuringiensis toxin (transgenic Bt cotton) planted in 2006 was 12.1 million hectares (12). Transgenic Bt cotton has been adopted extensively to reduce reliance on insecticides and save labor and costs in many countries. The rapid evolution of resistance by target pests could completely undermine the advantages of this new technology. Despite the expectations of many scientists and the predictions from models, resistance to Bt crops in the field has not yet been reported after a decade of adoption of Bt crops (2, 10, 19). However, we should not underestimate the ability of target insects to adapt to Bt crops, because resistance to Bt crops by target insects remains a question not of “if” but of “when.”
Invertebrates have demonstrated a range of resistance mechanisms to B. thuringiensis toxins (11). The diamondback moth, Plutella xylostella, has already evolved resistance to Bacillus thuringiensis subspecies in the field (16, 18). The cotton bollworm Helicoverpa armigera (Hübner) is the most important target pest of transgenic Bt cotton in China, India, Australia, and South Africa. Laboratory selection experiments from several countries demonstrate that this pest has the capacity to evolve resistance to the B. thuringiensis toxin Cry1Ac protein contained in Bt cotton (1, 14, 24).
Understanding the molecular mechanisms of B. thuringiensis resistance is critical for developing efficient DNA-based monitoring of the resistance frequency for early resistance warning and is key to informed and proactive resistance management. Mutations in a cadherin gene were identified as the causes for the recessive resistance to Cry1Ac in three major lepidopteran cotton pests, Heliothis virescens, Pectinophora gossypiella, and H. armigera (7, 15, 24). However, the mutant alleles found in laboratory-selected strains may not be identical to those present in field populations (3). In fact, two recent publications reported that no resistance alleles of cadherin of P. gossypiella and H. virescens were detected from field populations after large-scale screening (8, 20). Because the cadherin genes are not essential to the survival of P. gossypiella, H. virescens, and H. armigera (at least under laboratory conditions) (7, 15, 24), any mutation that nullifies the cadherin protein as a key functional receptor could confer Cry1Ac resistance. It is therefore very important to establish whether any such mutant cadherin alleles are present in the field, leading to the development of an appropriate DNA-based resistance surveillance program.
As suggested by Yang et al. (26), it is both feasible and necessary to screen for resistance-associated cadherin alleles from field populations of H. armigera by using the single-pair mating technique (or F1 screen). Field-derived moths of H. armigera can be crossed individually with moths from the laboratory-selected GYBT strain (r1r1), which shows recessive resistance to Cry1Ac (24). The mutated cadherin genotype status of field-derived moths will be revealed if their F1 offspring show significant Cry1Ac resistance. Here, we report the isolation of two new cadherin alleles conferring Cry1Ac resistance from a field population of H. armigera by using this strategy. Our results provide an important foundation for a DNA-based detection method for monitoring the frequency of mutant cadherin alleles in field populations of H. armigera.
MATERIALS AND METHODS
Insect strains.
A resistant strain (GYBT) of H. armigera was derived from the original GY strain (collected from the Gaoyang district of Hebei Province of China) by 28 generations of selection with activated Cry1Ac by surface contamination bioassay. The GYBT strain has 564-fold resistance to Cry1Ac compared with that of the GY strain (24) and is homozygous for the mutant cadherin r1 allele (26). Detailed information for the GYBT strain is presented in the report by Xu et al. (24). More than 20,000 eggs were collected from transgenic Bt cotton plants in Anyang (36°06′N, 114°21′E) of Henan Province, China, in early July of 2005. Bt cotton has been planted in Anyang since 1997 and has exceeded 90% of the total cotton area since 2001. Field-collected insect eggs were moved from Bt cotton leaves to an artificial diet, hatched, and reared to the second-instar larval stage for screening bioassays.
Larvae of H. armigera were reared on an artificial diet based on wheat germ and soybean powder (17) at 27 ± 1°C with a 16:8 light/dark (L:D, respectively) photoperiod. Adults were held under the same temperature and light conditions at an rH of 60% and were supplied with a 10% sugar solution.
Bioassay.
A surface contamination bioassay was used in this study. The toxin suspensions of activated Cry1Ac toxin were diluted with a 0.01 M, pH 7.4, phosphate buffer solution (PBS). PBS was used as a control. A disc of artificial diet (1.6-cm diameter) was put into a 24-well plate and made to fit into the inner wall and bottom of the plate by gentle pressure. The toxin solution (100 μl) was applied to the diet surface and allowed to air dry. One second-instar larva, starved for 4 h, was placed in each well of the plate, which was covered with two layers of nylon net to prevent escape. The mortality rates (for dead larvae or those with a body mass of less than 5 mg) and the body masses of survivors were measured after being maintained for 5 days at 26 ± 1°C, with a 16:8 L:D photoperiod and a 60% rH.
Biphasic screen.
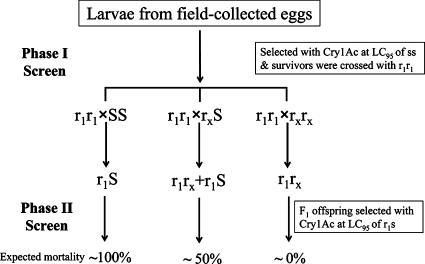
A schematic diagram of the experimental design for the biphasic screen of mutant cadherin alleles in field populations of H. armigera is shown in Fig. 1. In the first phase, second-instar larvae hatched from field-collected eggs were screened with a discriminating dose of activated Cry1Ac (1 μg/cm2 of diet surface, the 95% lethal concentration [LC95] of the susceptible homozygotes), which killed most of the susceptible larvae. In the second phase, survivors from the first-phase screen were individually crossed to homozygous-resistant moths (r1r1) from the GYBT strain, and then F1 offspring (48 second-instar larvae) from each single-pair family were screened with a second discriminating dose of activated Cry1Ac (2.5 μg/cm2 of diet surface, the LC95 of the r1S heterozygotes). If a field-derived moth was a susceptible homozygote (SS), then all of the F1 offspring would be heterozygous (r1S) and would have only a few survivors following the second discriminating dose. If a field-derived moth was a heterozygote (rxS) at the Ha_BtR locus, then 50% of the F1 offspring would be resistant homozygotes (r1rx), which would survive the second discriminating dose. If a field-derived moth was homozygous (rxrx) at the Ha_BtR locus, then 100% of the F1 offspring would be resistant homozygotes (r1rx).
FIG. 1.
Schematic diagram of the experimental design for a biphasic screening for cadherin mutations in field populations of Helicoverpa armigera. The laboratory-selected GYBT strain is homozygous for the cadherin allele r1. The cadherin allele rx is putatively carried by field-collected insects.
cDNA cloning of Ha_BtR alleles of H. armigera.
Total RNA from the midguts of final-instar larvae was extracted with an SV total RNA isolation system (Promega, Madison, WI), according to the manufacturer's instructions, and reverse transcribed with Moloney murine leukemia virus reverse transcriptase (Promega). Two pairs of primers (1-F/7-R and 7-F/9-R) were used to amplify the full-length cDNA of the r1 allele originating from the GYBT strain. Three primer pairs (1-F/4-R, 4-F/7-R, and 7-F/9-R) and 3′ rapid amplification of cDNA ends were used to amplify cDNA fragments of other Ha_BtR alleles derived from field moths. The sequences of primers can be found in the supplementary material of the article by Yang et al. (26).
PCR amplification of the genomic DNA fragments of the r2 and r3 alleles of Ha_BtR.
Genomic DNA of individual adults was prepared according to the method detailed by Yang et al. (26). The genomic sequences flanking the mutation sites of the r2 and r3 alleles (between exon 1 and exon 11) were determined by sequencing overlapping PCR products amplified from genomic DNA of the field-derived moths of the single-pair families 15 and 17.
PCR products of the expected size were excised and purified using a Wizard DNA purification system (Promega) and cloned using pGEM-T easy vector (Promega). All clones were sequenced by Invitrogen (Shanghai, China).
DNA-based detection of the r1, r2, and r3 alleles of Ha_BtR.
Allele-specific PCR for the r1 allele was performed as described by Yang et al. (26). A pair of specific primers (the forward cadherin primer for family 15 [15Cad-F], 5′-AGACAGGGACACTCTTGAGAAG-3′; and 15Cad-R, 5′-GGCTCGTTCGTTACACTCAGTA-3′) was used to detect both the r2 and the r3 alleles of Ha_BtR, and the expected length of the target genomic DNA fragment was 180 bp.
The amplification-reactive mixture (25 μl) contained 100 ng of genomic DNA, 1 μM of each primer, 150 μM of each deoxynucleoside triphosphate, 2 mM of MgCl2, 1 U of rTaq DNA polymerase, and 2.5 μl of 10× PCR buffer. PCR was performed for 30 cycles of 30 s at 94°C, 1 min at 55°C, and 2 min at 72°C. PCR products were analyzed by 1.5% agarose gel electrophoresis and ethidium bromide staining.
Survival test on a Cry1Ac-coated diet and on Bt cotton for larvae carrying cadherin mutations.
The 15Sel strain was established from survivors of the family 15 after the second screen. In the seventh generation of the 15Sel strain, 48 second-instar larvae were exposed to an activated Cry1Ac-coated diet (2.5 μg/cm2 of diet surface), which killed all of the susceptible and most of the heterozygous larvae. Five days later, survivors (those with a body mass of more than 5 mg) were collected, and their genomic DNA was extracted for cadherin genotyping.
Transgenic Bt cotton plants (Bollgard DP410B, Monsanto, China) were grown in a plant growth chamber (25°C to 30°C). Five plants were planted in each pot (20-cm diameter). Ten first-instar larvae (1 day old) from the seventh generation of the 15Sel strain were placed on each of 50 transgenic Bt cotton plants with five to six fully expanded leaves.
Nucleotide sequence accession numbers.
Genomic DNA sequences of the r2 and r3 alleles of Ha_BtR have been deposited in the GenBank database under the accession numbers EU016078 and EU016079, respectively. cDNA sequences of the r2 and r3 alleles of Ha_BtR are available in the GenBank database under accession numbers DQ973282 and EU016080, respectively.
RESULTS
Biphasic screen for cadherin mutants from field populations of H. armigera.
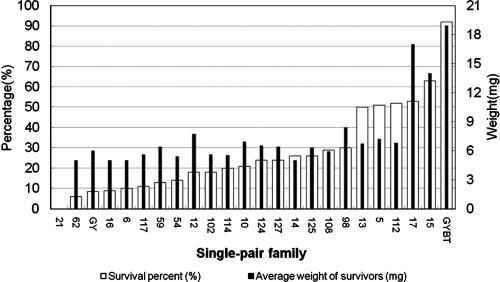
Second-instar larvae (9,984) of H. armigera from a population from Anyang district in Henan Province were individually treated with 1 μg of activated Cry1Ac/cm2 of diet surface. Of the 123 surviving insects, 70 larvae completed development. The resulting 70 moths were single-pair crossed with adults from the laboratory-selected GYBT strain. Twenty-two single-pair crosses produced fertile offspring. F1 offspring from these 22 single-pair families were subjected to a secondary screen (activated Cry1Ac, 2.5 μg/cm2 of diet surface). The survival rates and average body weights of the survivors of each single-pair family are shown in Fig. 2. Of 22 single-pair families, 5 single-pair families had about 50% survival (family 5, 51%; family 13, 50%; family 15, 63%; family 17, 53%; and family 112, 52%); others were less than 30%. The average body weights of survivors from the second screen varied significantly among these five single-pair families. The average body weights of survivors from families 15 (14.0 ± 1.2 mg) and 17 (17.0 ± 1.4 mg) were most similar to that of survivors from the GYBT strain (18.9 ± 1.2 mg), with the dosage of 2.5 μg/cm2 of diet surface, but all of them were significantly heavier than survivors from families 5 (7.2 ± 0.7 mg), 13 (6.7 ± 0.4 mg), and 112 (7.7 ± 0.8 mg) (analysis of variance, α = 0.05). This suggested that the field-derived parents of families 15 and 17 were possibly heterozygous for cadherin mutations.
FIG. 2.
Survival percentage and average body weight of survivors of the second screen with Cry1Ac (2.5 μg/cm2 of diet surface) with F1 offspring of single-pair matings between survivors from the primary screening and moths of the GYBT strain of H. armigera.
Cloning and sequencing of cDNA of cadherin alleles from field populations of H. armigera.
cDNA of F1 survivors (three final-instar larvae) from the second screen of the aforementioned five single-pair lines was prepared for cadherin genotyping. cDNA of Ha_BtR from the three surviving F1 larvae from each of the five single-pair families (5, 13, 15, 17, and 112) was individually cloned and sequenced. Each F1 survivor carried two alleles of Ha_BtR, one from the GYBT strain (r1) and the other from the field-derived parent. As expected, the r1 allele was recovered from all F1 survivors (GenBank accession no. DQ973284, DQ973285, DQ523168, DQ973286, and DQ973289 for families 5, 13, 15, 17, and 112, respectively). In the F1 survivors of family 15, a new transcript of Ha_BtR (GenBank accession no. DQ973282), putatively encoding a truncated cadherin protein (with 371 amino acids relative to 1,730 amino acids of the wild type), was cloned and sequenced. In the F1 survivors of family 17, another new transcript of Ha_BtR (GenBank accession no. EU016080, with the difference of only one nucleotide in the coding region compared with that in DQ973282) was cloned and sequenced from each of the three larvae. In the F1 survivors of families 5, 13, and 112, besides the r1 allele from the GYBT strain, we found only the full-length wild-type transcript of Ha_BtR, which was passed from their field-derived parents.
Identification of mutations in the genomic DNA of the r2 and r3 alleles of Ha_BtR.
To further confirm whether the two new transcripts originated from field-derived parents, genomic DNA sequences of Ha_BtR of the field-derived moths were checked. From the field-derived parent of family 15, a 5,813-bp genomic DNA fragment (GenBank accession no. EU016078, named the r2 allele of Ha_BtR) encoding the new transcript of DQ973282 was successfully amplified and sequenced. The insertion of a 1,007-bp fragment was identified between bp 111 and 112 of exon 8 of Ha_BtR. From the field-derived parent of family 17, a 10,605-bp genomic DNA fragment (GenBank accession no. EU016079, designated the r3 allele of Ha_BtR) encoding the new transcript of EU016080 was obtained through a set of PCR amplifications. The insertion of a 5,800-bp fragment was also identified between bp 111 and 112 of exon 8 of Ha_BtR. The wild-type allele of Ha_BtR could also be detected with allele-specific PCR in the field-derived parents of both family 15 and family 17. It confirmed that these two field-derived moths are heterozygous for the Ha_BtR mutations, which is consistent with the results from the F1 progeny bioassay and cDNA sequencing.
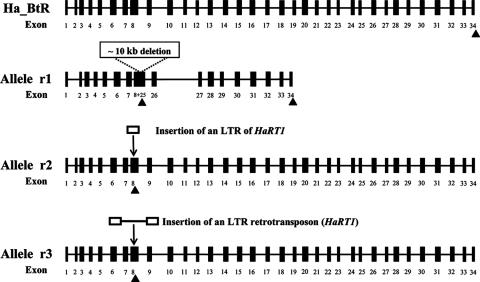
The 5,800-bp insertion in the r3 allele was identified as a long terminal repeat retrotransposon (LTR-RT), which was named HaRT1 (DNA sequences are included in GenBank accession no. EU016079). It revealed the common characteristics of an LTR-RT: LTRs of 1,007 bp, gag and protease domains (in open reading frame 1 [ORF1] of HaRT1), retrotranscriptase, and an RNase H domain (in ORF2) and an integrase domain (in ORF3). Interestingly, the sequence of the insertion in the r2 allele was almost identical to that of the LTR of HaRT1 (only 2 bases out of 1,007 bp were different). At a position 9 bp downstream of the 5′ end of the inserted LTR, there was a premature stop codon (TGA), which caused the truncation of the r2 and r3 alleles. The genomic structures of the wild-type allele and the r1, r2, and r3 alleles of the Ha_BtR gene from H. armigera are shown in Fig. 3. The truncated transcripts of the r2 and r3 alleles putatively encode only 2 of the 11 ectodomain repeats in the N terminus of Ha_BtR, which lacks transmembrane and toxin-binding domains.
FIG. 3.
Genomic structures of the wild-type allele and the r1, r2, and r3 alleles of the Ha_BtR gene from H. armigera. Exons are shown as vertical bars and introns as thin horizontal lines. The stop codon and poly(A) sites are marked with ▴.
Survival of larvae carrying cadherin mutations on a Cry1Ac-coated diet and on Bt cotton.
Survivors of family 15 following the second screen were maintained in the laboratory as the founders of the 15Sel strain for further investigation. In the seventh generation of the 15Sel strain, survival of larvae on a Cry1Ac-coated diet (2.5 μg/cm2 of diet surface) and on Bt cotton was tested, and the results are detailed in Table 1.
TABLE 1.
Genotypes of survival of the 15Sel strain of H. armigera on Cry1Ac-coated diet and on Bt cotton
| Treatment | No. of insects genotyped | No. of survivors of cadherin genotypes:
|
|||||
|---|---|---|---|---|---|---|---|
| r1r1 | r2r2 | r1r2 | r1S | r2S | SS | ||
| Artificial diet (without B. thuringiensis toxin) | 50 | 4 | 2 | 7 | 11 | 12 | 14 |
| Artificial diet with Cry1Ac (2.5 μg/cm2) | 15a | 2 | 1 | 10 | 1 | 1 | 0 |
| Bt cotton (DP410B) | 2b | 1 | 0 | 1 | 0 | 0 | 0 |
Survivors of 48 larvae reared on a Cry1Ac-coated diet (2.5 μg/cm2 of diet surface).
Number of pupae that survived from Bt cotton plants infested with 500 1-day-old larvae of the 15Sel strain.
For the untreated larvae from the 15Sel family, the ratio among homozygous-resistant individuals (carrying two r alleles, i.e., r1r1, r2r2, or r1r2), heterozygotes (r1S or r2S), and homozygous susceptible individuals (SS) is nearly 1:2:1 (∑χ2 = 0.36 < χ2 [P = 0.05]; df = 2). Following treatment with 2.5 μg Cry1Ac/cm2 of diet surface, 87% of survivors were homozygous individuals, and the remaining 13% were heterozygotes. This indicates that H. armigera carrying two mutated cadherin alleles can survive but that most heterozygotes carrying one mutated cadherin allele and all susceptible homozygotes cannot survive on this discriminating dose, exemplifying the recessive nature of the gene.
Treated with transgenic Bt cotton, only 2 out of 500 larvae (presumably containing about 25% larvae with two resistance alleles, i.e., around 125 larvae) completed their larval development. These two survivors (pupae) were determined to be r1r1 and r1r2. This demonstrates that H. armigera carrying two mutated cadherin alleles can survive on transgenic Bt cotton, and the survival is about 1 to 2%. Heterozygotes carrying one mutated cadherin allele cannot survive on transgenic Bt cotton. Under the conditions of selection by transgenic Bt cotton in this study, resistance conferred by the disruption of Ha_BtR is completely recessive.
DISCUSSION
The F1 screen method was first developed to estimate the frequency of major B. thuringiensis resistance alleles in field populations of H. virescens (9). Field-trapped males and males that developed from field-collected eggs of H. virescens were used in single-pair crosses to laboratory-selected homozygous-resistant YHD2 strain virgin females, and their progeny were screened with a discriminating dose of Cry1Ac. Three out of 1,025 field-derived males were tested heterozygous for a major resistance allele of BtR-4, and the field frequency of the BtR-4 allele in field populations of H. virescens was estimated to be 0.0015 (9). BtR-4 was identified later as a cadherin gene disrupted by the insertion of a retrotransposon (Hel-1) (7). However, the Hel-1 insertion was not detected from the three field-caught males thought to carry BtR-4 in a recent study (8). There are two possible explanations for this discrepancy: (i) the three field-caught males have other kinds of cadherin mutation than the Hel-1 insertion or (ii) the BtR-4 frequency could be overestimated because the F1 screen procedure may detect dominant resistance alleles at other loci. In the present study, the progeny tests indicated that the field-derived parents of families 5, 13, and 112 may have been heterozygous for a Ha_BtR resistance allele, but no mutation was found other than that from the GYBT strain. This may be explained by the second-screen dosage of Cry1Ac (2.5 μg/cm2 of diet surface, LC95 of the susceptible strain) not being high enough to kill 100% of the Ha_BtR heterozygotes. This is supported by results from the survival tests (Table 1). In future, the second-screen dosage should be increased to an LC99 (5 μg/cm2 of diet surface) of the susceptible GY strain of H. armigera, which is expected to reduce ambiguity when choosing candidate families with resistance mutations from the field.
The biphasic F1 screen employed in this study is very effective and promising for screening diverse resistance alleles in field populations. The primary screen was designed with the assumption of incomplete recessiveness of Cry1Ac resistance in H. armigera. This step is intended to keep most resistance alleles in the screen and to limit the scale of the secondary screen, providing huge labor and cost savings. It should be a standard practice before the initiation of any DNA-based detection program.
Pink bollworm (P. gossypiella) resistance to Cry1Ac was monitored for 8 years with laboratory bioassays of strains collected annually from 10 to 17 cotton fields in Arizona, and bioassay results showed no net increase from 1997 to 2004 in the mean frequency of pink bollworm resistance to B. thuringiensis toxin (19, 21, 22). A recent DNA-based screening of pink bollworm resistance to B. thuringiensis toxin across Arizona, California, and Texas from 2001 to 2005 detected no resistance alleles (20). Delayed resistance of target pests to Bt crops compared with the rapid evolution of resistance with laboratory selection could result from one or more of the following factors: refuges of non-Bt crops that enable survival of susceptible insects, recessive inheritance of resistance, incomplete resistance, and field fitness costs associated with resistance (5, 19).
Bt cotton has been commercialized in China since 1997. In 2006, China's Bt cotton area increased to 3.5 million ha (about two-thirds of the total cotton area) (12). In the Yellow River cotton area, Bt cotton has comprised nearly 100% of the crop for several years. The high-dose/refuge strategy is not mandated for resistance management in China. The susceptibilities of Chinese field populations of H. armigera to the Cry1Ac toxin were monitored using bioassays from 1997 to 2006, and no resistance was detected (23). In the current study, providing that the cadherin mutation frequency in the 70 survivors from the second screen is similar to that in the 22 survivors producing fertile progeny, the resistance mutation frequency in the Anyang field population in 2005 can be estimated to be around 3.5 × 10−4. After nearly a decade of Bt cotton planting, the frequency of mutant cadherin alleles in the Anyang population of H. armigera is still low. Fitness cost, incomplete resistance, and alternative host crops as a natural refuge could be key factors for delayed resistance of H. armigera to Bt cotton in the small-scale cotton system of China. Ongoing work in this area will address these questions.
The mode of action of Cry1Ac toxins is a multistep process that involves interactions with several receptor molecules leading to membrane insertion and cell lysis (4). Cadherin appears to be one key functional receptor in the multistep intoxication process, at least in H. virescens, P. gossypiella, and H. armigera (25). It is possible that a large number of mutations in the cadherin gene can cause B. thuringiensis resistance, complicating DNA-based B. thuringiensis resistance-monitoring strategies. Together with previous results (24, 26), three resistance alleles (r1, r2, and r3) of Ha_BtR of H. armigera have now been detected. All three alleles are associated with a mutation in exon 8 of Ha_BtR, which may be a regional hot spot for mutations. If other mutations are also uncovered in this region, it will simplify DNA-based monitoring methods.
As far as we know, this is the first time that B. thuringiensis resistance alleles of a target insect of a Bt crop have been successfully detected in the field. The biphasic F1 screen method could be used to trap resistance alleles of other populations of H. armigera or other pest insects. Although the presence of additional resistance alleles at other loci cannot be excluded, the cadherin locus of H. armigera (Ha_BtR) should be a major resistance locus for molecular monitoring of resistance to Bt cotton, given that bollworm larvae with two mutant cadherin alleles can complete development on Bt cotton, albeit at a low frequency.
Transposable elements are mobile sequences that are abundant within eukaryotic genomes and that are endogenous mutators with the potential to produce a wide array of changes in the genomes of their hosts (13). Transposable elements can make insects become resistant to insecticides either by altering the metabolic enzymes that detoxify insecticides or by altering the targets of insecticides (6). In laboratory-selected Heliothis virescens, disruption of a cadherin (HevCLP) mediated by a defective LTR-RT (Hel-1) insertion is responsible for high levels of resistance to the B. thuringiensis toxin Cry1Ac (7). In the current study, an insertion of an LTR-RT (HaRT1) in the r3 allele of Ha_BtR resulted in a truncated cadherin, thereby conferring resistance to the B. thuringiensis toxin Cry1Ac in H. armigera. The r2 allele of Ha_BtR was disrupted by an insertion of a remnant LTR of HaRT1. Insertions in the r2 and r3 alleles occurred at exactly the same site in the Ha_BtR gene. This suggests that the r2 allele originated from an earlier insertion event of HaRT1 and that the r3 allele of Ha_BtR came from a more recent insertion event of HaRT1.
Acknowledgments
This research was supported by grants from the National Basic Research Program of China (2006CB102003), the National Natural Science Foundation of China (30571233), the National High Technology Research and Development Program of China (2007AA10Z420), and the 111 project (B07030).
Footnotes
Published ahead of print on 7 September 2007.
REFERENCES
- 1.Akhurst, R. J., W. J. James, L. J. Bird, and C. Beard. 2003. Resistance to the Cry1Ac δ-endotoxin of Bacillus thuringiensis in the cotton bollworm, Helicoverpa armigera (Lepidoptera: Noctuide). J. Econ. Entomol. 96:1290-1299. [DOI] [PubMed] [Google Scholar]
- 2.Bates, S. L., J.-Z. Zhao, R. T. Roush, and A. M. Shelton. 2005. Insect resistance management in GM crops: past, present and future. Nat. Biotechnol. 23:57-62. [DOI] [PubMed] [Google Scholar]
- 3.Bravo, A., M. Soberón, and S. S. Gill. 2005. Bacillus thuringiensis: mechanism and use, p. 175-205. In L. I. Gilbert, K. Iatrou, and S. S. Gill (ed.), Comprehensive molecular insect science, vol. 6. Elsevier Pergamon, Oxford, United Kingdom. [Google Scholar]
- 4.Bravo, A., S. S. Gill, and M. Soberon. 2007. Mode of action of Bacillus thuringiensis Cry and Cyt toxins and their potential for insect control. Toxicon 49:423-435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carrière, Y., T. J. Dennehy, C. Ellers-Kirk, D. Holley, Y. B. Liu, M. A. Simms, and B. E. Tabashnik. 2002. Fitness costs, incomplete resistance, and management of resistance to Bt crops, p. 82-91. In R. J. Akhurst, C. E. Beard, and P. Hughes (ed.), Biotechnology of Bacillus thuringiensis and its environmental impact. Proceedings of the 4th Pacific Rim Conference, CSIRO, Canberra, Australia.
- 6.Ffrench-Constant, R., P. Daborn, and R. Feyereisen. 2006. Resistance and the jumping gene. Bioessays 28:6-8. [DOI] [PubMed] [Google Scholar]
- 7.Gahan, L. J., F. Gould, and D. G. Heckel. 2001. Identification of a gene associated with Bt resistance in Heliothis virescens. Science 293:857-860. [DOI] [PubMed] [Google Scholar]
- 8.Gahan, L. J., F. Gould, J. D. López, Jr., S. Micinski, and D. G. Heckel. 2007. A polymerase chain reaction screen of field populations of Heliothis virescens for a retrotransposon insertion conferring resistance to Bacillus thuringiensis toxin. J. Econ. Entomol. 100:187-194. [DOI] [PubMed] [Google Scholar]
- 9.Gould, F., A. Anderson, A. Jones, D. Sumerford, D. G. Heckel, J. Lopez, S. Micinski, R. Leonard, and M. Laster. 1997. Initial frequency of alleles for resistance to Bacillus thuringiensis toxins in field populations of Heliothis virescens. Proc. Natl. Acad. Sci. USA 94:3519-3523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gressel, J. 2005. Problems in qualifying and quantifying assumptions in plant protection models: resultant simulations can be mistaken by a factor of million. Crop Prot. 24:1007-1015. [Google Scholar]
- 11.Griffitts, J. S., and R. V. Aroian. 2005. Many roads to resistance: how invertebrates adapt to Bt toxins. Bioessays 27:614-624. [DOI] [PubMed] [Google Scholar]
- 12.James, C. 2006. ISAAA Brief 35-2006: global status of commercialized Biotech/GM crops. International Service for the Acquisition of Agri-biotech Applications, Ithaca, NY.
- 13.Kidwell, M. G., and D. R. Lisch. 2000. Transposable elements and host genome evolution. Trends Ecol. Evol. 15:95-99. [DOI] [PubMed] [Google Scholar]
- 14.Kranthi, K. R., S. Kranthi, S. Ali, and S. K. Banerjee. 2000. Resistance to Cry1Ac δ-endotoxin of Bacillus thuringiensis in a laboratory selected strain of Helicoverpa armigera (Hübner). Curr. Sci. 78:1001-1004. [Google Scholar]
- 15.Morin, S., R. W. Biggs, M. S. Sisterson, L. Shriver, C. Ellers-Kirk, D. Higginson, D. Holley, L. J. Gahan, D. G. Heckel, Y. Carrière, T. J. Dennehy, J. K. Brown, and B. E. Tabashnik. 2003. Three cadherin alleles associated with resistance to Bacillus thuringiensis in pink bollworm. Proc. Natl. Acad. Sci. USA 100:5004-5009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shelton, A. M., J. L. Robertson, J. D. Tang, C. Perez, S. D. Eigenbrode, H. K. Prcisler, W. T. Wilsey, and R. J. Cooley. 1993. Resistance of diamondback moth (Lepidoptera: Plutellidae) to Bacillus thuringiensis subspecies in the field. J. Econ. Entomol. 86:697-705. [Google Scholar]
- 17.Shen, J., and Y. Wu. 1995. Resistance to Helicoverpa armigera to insecticides and its management, p. 92-94. China Agricultural Press, Beijing, China.
- 18.Tabashnik, B. E., N. L. Cushing, N. Finson, and M. W. Johnson. 1990. Field development of resistance to Bacillus thuringiensis in diamondback moth (Lepidoptera: Plutellidae). J. Econ. Entomol. 83:1671-1676. [Google Scholar]
- 19.Tabashnik, B. E., T. J. Dennehy, and Y. Carrière. 2005. Delayed resistance to transgenic cotton in pink bollworm. Proc. Natl. Acad. Sci. USA 102:15389-15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tabashnik, B. E., J. A. Fabrick, S. Henderson, R. W. Biggs, C. M. Yafuso, M. E. Nyboer, N. M. Manhardt, L. A. Coughlin, J. Sollome, Y. Carrière, T. J. Dennehy, and S. Morin. 2006. DNA screening reveals pink bollworm resistance to Bt cotton remains rare after a decade of exposure. J. Econ. Entomol. 99:1525-1530. [DOI] [PubMed] [Google Scholar]
- 21.Tabashnik, B. E., A. L. Patin, T. J. Dennehy, Y. B. Liu, Y. Carrière, M. A. Sims, and L. Antilla. 2000. Frequency of resistance to Bacillus thuringiensis in field populations of pink bollworm. Proc. Natl. Acad. Sci. USA 97:12980-12984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tabashnik, B. E., Y. Carrière, T. J. Dennehy, S. Morin, M. S. Sisterson, R. T. Roush, A. M. Shelton, and J. Z. Zhao. 2003. Insect resistance to transgenic Bt crops: lessons from the laboratory and field. J. Econ. Entomol. 96:1031-1038. [DOI] [PubMed] [Google Scholar]
- 23.Wu, K. 2007. Monitoring and management strategy for Helicoverpa armigera resistance to Bt cotton in China. J. Invertebr. Pathol. 95:220-223. [DOI] [PubMed] [Google Scholar]
- 24.Xu, X., L. Yu, and Y. Wu. 2005. Disruption of a cadherin gene associated with resistance to Cry1Ac δ-endotoxin of Bacillus thuringiensis in Helicoverpa armigera. Appl. Environ. Microbiol. 71:948-954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xu, X., and Y. Wu. 2007. Disruption of Ha_BtR alters binding of Bacillus thuringiensis δ-endotoxin Cry1Ac to midgut BBMVs of Helicoverpa armigera. J. Invertebr. Pathol. [Epub ahead of print.] doi: 10.1016/j.jip.2007.04.009. [DOI] [PubMed]
- 26.Yang, Y., H. Chen, S. Wu, Y. Yang, X. Xu, and Y. Wu. 2006. Identification and molecular detection of a deletion mutation responsible for a truncated cadherin of Helicoverpa armigera. Insect Biochem. Mol. Biol. 36:735-740. [DOI] [PubMed] [Google Scholar]