Abstract
Previous studies in an acid-sulfate-chloride spring in Yellowstone National Park found that microbial arsenite [As(III)] oxidation is absent in regions of the spring outflow channel where H2S exceeds ∼5 μM and served as a backdrop for continued efforts in the present study. Ex situ assays with microbial mat samples demonstrated immediate As(III) oxidation activity when H2S was absent or at low concentrations, suggesting the presence of As(III) oxidase enzymes that could be reactivated if H2S is removed. Cultivation experiments initiated with mat samples taken from along the H2S gradient in the outflow channel resulted in the isolation of an As(III)-oxidizing chemolithotroph from the low-H2S region of the gradient. The isolate was phylogenetically related to Acidicaldus and was characterized in vitro for spring-relevant properties, which were then compared to its distribution pattern in the spring as determined by denaturing gradient gel electrophoresis and quantitative PCR. While neither temperature nor oxygen requirements appeared to be related to the occurrence of this organism within the outflow channel, H2S concentration appeared to be an important constraint. This was verified by in vitro pure-culture modeling and kinetic experiments, which suggested that H2S inhibition of As(III) oxidation is uncompetitive in nature. In summary, the studies reported herein illustrate that H2S is a potent inhibitor of As(III) oxidation and will influence the niche opportunities and population distribution of As(III) chemolithotrophs.
Microbial arsenite [As(III)] chemolithotrophy was first reported by Ilyaletdinov and Abdrashitova (18) and then more recently by Santini et al. (39), Oremland et al. (34), and Rhine et al. (38). Progress toward understanding the genetics and physiology of As(III) oxidation is at an early stage, being limited to a few definitive papers that describe the biochemical and structural features of one of the two identified As(III) oxidases (4, 11, 37) and recent studies that have identified the structural genes that encode these As(III) oxidases (31, 40).
Even less is known about the ecology of As(III) chemolithotrophs. Drainage waters originating from commercial mining operations often contain appreciable amounts of As(III) and thus are potential habitats for organisms capable of using As(III) as an energy source. Such was the case for the As(III) chemolithotrophs isolated from gold mines (39, 40). Also, waters originating from geothermal sources often carry significant As(III) (5, 28, 45). The facultative anaerobic As(III) chemolithotroph isolated by Oremland et al. (34) was obtained from anaerobic bottom waters of Mono Lake, a meromictic lake containing high concentrations of As(III) derived from geothermal inputs (33). Thermophiles capable of oxidizing As(III) have been isolated (10, 14), and a recent PCR-based survey documented the presence and expression of As(III) oxidase structural genes in geothermal springs in Yellowstone National Park (YNP), WY (21). However, to date, the lone documentation of a thermophile capable of As(III) chemolithotrophy is a brief notation that Sulfurihydrogenibium azorense can use As(III) as an electron donor (1).
Our previous studies in the Norris Geyser Basin in YNP have focused on an acidic geothermal feature referred to as Dragon Spring (22, 28). Chemical analysis of Dragon Spring found the As(III) concentration in the source waters to be 33 μM (28), which we have documented on subsequent occasions to be nearly invariable (unpublished data). At a constant flux, this level of As(III) should be adequate for supporting growth of As(III) chemolithotrophs. And, while biological As(III) oxidation has been documented in this spring, it was only found in specific regions of the outflow channel where H2S was absent or was present at low concentrations (28). In subsequent in vitro experiments with a pure culture of an As(III)-oxidizing Hydrogenobaculum-like organism isolated from this spring, we showed that H2S inhibited As(III) oxidation (10). However, other environmental factors may also contribute to the specific localization of As(III) oxidation in this spring. In the present study, As(III) oxidation in Dragon Spring was reexamined to assess the potential for As(III) chemolithotrophs to inhabit this geothermal feature and to gain a more thorough understanding of the H2S inhibition phenomenon with respect to how it may relate to microbial function and population distribution in the environment.
MATERIALS AND METHODS
Study site and chemical analysis.
All field experiments were conducted at Dragon Spring (44°43′54.8″N, 110°42′39.9"W, spring number NHSP106 in the YNP thermal inventory), located in the Norris Geyser Basin, YNP. Water temperature and pH were routinely monitored throughout the 2-year study. At the point of discharge, the pH was a nearly constant 3.1 and the temperature ranged from 68 to 72°C during the course of the study, with changes in the latter being unpredictable. Dissolved H2S was measured by the amine-sulfuric acid methodology previously described (28), except that the spring water was assayed within seconds of sampling and without prior filter sterilization. Dissolved oxygen was assayed with a Hach OX-2P test kit (Hach Company, Loveland, CO).
As(III) oxidation assays.
Mat samples were collected with autoclaved tools or sterile pipette tips and placed into sterile 15-ml conical tubes, homogenized by mixing, and then split into two subsamples. One subsample was used to measure As(III) oxidation, while the other was boiled for 20 min to serve as a killed control. Mat samples were aseptically transferred into 150-ml serum bottles containing 40 ml of filter-sterilized spring water that was amended to 60 μM As(III) (added as NaH2AsO3). Assay bottles were closed with butyl rubber septa, pressure sealed with an aluminum ring, and then incubated at in situ temperatures by placing the bottles in the spring at the same location from which the mat material was sampled. Water samples were withdrawn at 10-min intervals to assay for As(III) oxidation by sodium borohydride speciation as described previously (10). Samples were stored at 4°C until analysis by hydride generation atomic absorption spectrometry (24). As(V) formation and H2S disappearance data were normalized on the basis of mat sample dry weight, which was determined by collection of all of the mat material from each serum bottle onto preweighed 0.2-μm-pore-size filters, overnight drying at 65°C, and then weighing to determine the mat dry weight contained in each serum bottle. The maximum mat dry weight variation between samples was 7.6%.
Cultivation work.
Approximately 2 g of mat material was aseptically collected with wide-bore pipette tips along the five transect sites shown in Fig. 1. Mat material was stored for transportation to the laboratory in 15-ml conical tubes and maintained at 55°C in a thermos bottle filled with site water. Mat samples were resuspended in 10 ml spring water collected directly above the mat sampling site, and 100 μl of this suspension was used to inoculate 16-ml serum bottles containing 4.9 ml of filter-sterilized spring water amended with 1 mM As(III) (supplied as NaH2AsO3) and degassed to remove the H2S. The bottles were sealed as described above, with the headspace made 50% CO2 and 50% air with filter-sterilized gasses. Enrichments were incubated at the same temperature as recorded for each sample in the field, and the headspace gasses were replaced every third day via syringe. Every 30 days, 100 μl of culture was transferred to similarly prepared serum bottles. In the latter stages of the enrichment process, the addition of a dilute synthetic defined medium, pH 3.1 (see reference 15), was found to enhance growth yields, and thus a 1:1 mixture of filter-sterilized spring water and synthetic medium was the composition of late-stage enrichment medium. Throughout, enrichment cultures were monitored for growth by phase-contrast microscopy. After 120 days, cultures were serially diluted and used as inocula for spread plates containing the same medium solidified with 1.8% Gelrite (Research Products International, Mt. Prospect, IL). A pure culture was established by repeated subculturing of isolated colonies and is referred to herein as strain AO5.
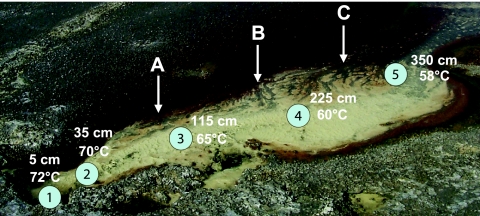
FIG. 1.
Color image of Dragon Spring. Almost all sampling occurred in the yellow zone, where the solid-phase material is composed of S0. The main transect points are numbered 1 to 5. Sampling for some experiments occurred at many locations not always corresponding exactly to the primary transect points. The water temperatures shown were measured on 15 June 2003, were the typical upper boundary temperatures observed at each transect point during this study, and illustrate the temperature gradient in this portion of the outflow channel. Vertical arrows indicate the sampling sites for DNA used for PCR-DGGE analysis and correspond to lanes A, B, and C in Fig. 7. This color image is modified (with permission of the publisher) from a photograph taken by T.R.M. and used previously (36).
Further experiments with the pure culture found that the organism would grow on the defined medium without spring water and thus the undiluted defined medium was used in all subsequent experiments. Cardinal temperatures were determined in water bath shakers (containing polyethylene glycol 400) set at various temperatures. The sealed serum bottles contained 20 ml of medium and 80 ml of headspace gasses (50% CO2 and 50% air, both filter sterilized) that were replaced daily to maintain optimum levels of CO2 and O2. The same culture conditions were used for As(III) oxidation and sulfide inhibition kinetics studies. For anaerobic culture work, the liquid medium was amended with sulfate (as FeSO4), nitrate (NaNO3−), or ferric iron (FeCl3) as representative dominant spring-relevant electron acceptors previously documented in this spring (28), and all at 1 mM. The medium and headspace in each autoclaved sealed serum bottle were then purged for 2 h with filter-sterilized certified O2-free N2 gas, being vented via a sterile syringe needle. All gassing treatments were conducted aseptically in a Labconco biosafety cabinet (Labconco Corp., Kansas City, MO).
As(III) oxidation kinetics.
Late-log-phase AO5 cells (1.0 × 106 total) were added to sealed serum bottles containing 20 ml sterile medium amended with 500 μM citric acid (pH 3.0) as a pH buffer and 30 μM As(III). Na2S was added to a final concentration of 0, 3, 15, 30, or 60 μM, and then 100-μl samples were removed at 10-min intervals starting immediately upon the addition of the Na2S (time = 0) and continuing for 60 min. Samples were immediately frozen in liquid nitrogen and stored at −80°C until speciation, with the analysis carried out as described above for the ex situ assays. To model the mode of enzyme inhibition, As(III) oxidation rates were calculated from AO5 cultures incubated in the same fashion, except that As(III) was added to final concentrations ranging from 3.75 to 30 μM in medium amended with 0, 2.91, or 5.82 μM Na2S.
PCR and phylogenetic analysis.
DNA was extracted from the pure culture as previously described (6). The nearly full-length 16S rRNA gene was amplified with Bacteria-specific primer 8F and the universal primer 1492R (3). The PCR mixture contained 2.5 mM MgCl2, 0.2 mM deoxynucleoside triphosphates (dNTPs), 0.5 μM each primer, approximately 10 ng template DNA, 0.5 mg ml−1 bovine serum albumin, 2.5 U Taq polymerase, and 1× buffer (Promega, Madison, WI) in a total volume of 50 μl. The amplification program consisted of 2 min at 95°C and then 30 cycles of 1 min of denaturation at 95°C, 1 min of annealing at 45°C, and 1 min of extension at 72°C, followed by a final extension of 7 min at 72°C. Sequencing was carried out at the Ohio State University Plant Microbe Genomics Facility with primers 8F, 533F, 533R, and 1492R. Sequences were aligned with ClustalX (47) and then edited and trimmed with Se-Al v2.0a11 (http://evolve.zoo.ox.ac.uk). Phylogenetic analysis of the aligned sequences was performed by the maximum-likelihood algorithm, and bootstrap values for 100 pseudoreplicates were generated with the PAUP* v4.0b10 software package (46). Hydrogenobaculum acidophilum (GenBank accession AJ320225), a member of the deeply branching family Aquificaceae, was used as an outgroup for these analyses.
Denaturing gradient gel electrophoresis (DGGE).
The PCR primers and conditions and techniques used for DGGE analysis were the same as previously described (32). Briefly, the primers used were Bacteria-specific 1070F (13) and the universal reverse primer 1392R (3) containing a GC clamp (32). DGGE was performed with a Bio-Rad DCode system (Bio-Rad, Hercules, CA) and an 8% polyacrylamide gel containing a 40 to 70% denaturant concentration (where 100% denaturant contains 7 M urea and 40% [vol/vol] formamide). DNA was extracted from DGGE gels by excising bands from the gel and crushing them in a 1.5-ml microcentrifuge tube. Extraction solution (150 μl containing 500 mM ammonium acetate, 0.1% [wt/vol] sodium dodecyl sulfate, and 0.1 mM EDTA) was added to the crushed gel slice, and the mixture was incubated at 75°C for 2 h. The tube was centrifuged at 14,000 × g for 10 min, and the supernatant was removed to a separate tube. DNA was precipitated by adding 0.1 volume of 3 M sodium acetate and 2.5 volumes of 95% ethanol.
Q-PCR.
Quantitative PCR (Q-PCR) was used to study the in situ distribution pattern of the As(III) chemolithotroph isolated and characterized in this study. Nucleic acids were extracted from the mat material by the method of Botero et al. (7). Each Q-PCR mixture contained a total of 100 ng DNA. Standards contained 0, 0.001, 0.01, 0.1, 1, or 10 ng DNA extracted from isolate AO5 in a background of sheared herring sperm DNA. Each 25-μl reaction mixture contained 2.5 μl Mg-free buffer, 2.5 μl 25 mM MgCl2, 2 μl dNTP mixture (2.5 mM each dNTP), 0.1 μl Taq DNA polymerase (Promega, Madison, WI), 1 μl 20× SYBR green (Invitrogen, Carlsbad, CA), and 1 μl each of primers AO5-965F (5′-ATCGGTCGTCCCGCAAC-3′) and AO5-1181R (5′-CTGTCACCGCCATTGTAGCA-3′). The Q-PCR cycling conditions used were 95°C for 10 min and 35 cycles of 60 s at 95°C, 60 s at 53.5°C, and 45 s at 72°C, followed by a final melting step gradually ramping the temperature up to 105°C. Cycling and fluorescence monitoring were carried out in a Rotor-Gene RG-3000 (Corbett Research, Mortlake, New South Wales, Australia). Results were analyzed with the Rotor Gene software package version 6.0.
Primer specificity was evaluated first by in silico methods with both the probe match functions at the Ribosomal Database Project II (http://rdp.cme.msu.edu) and the BLAST (2) search for short, nearly identical matches at the National Center for Biotechnology Information (http://ncbi.nlm.nih.gov) (43). In addition, the Q-PCR primer design took into account all of the 16S sequences cloned with two different primer sets from a previous molecular analysis of Dragon Spring (22) and sequences derived from the DGGE analysis (above) conducted in the present study. The closest identity of any PCR clones derived from this spring deviated by at least six nucleotides in the 18-mer primers. As expected, testing the primers against various Hydrogenobaculum PCR clones that dominate the clone libraries derived from the yellow S0 zone this spring (22) did not result in an amplicon (data not shown). However, these primers could not differentiate AO5 from clone YNPFFP86 (accession no, AF391980; a single nucleotide mismatch in AO5-965F), which we originally obtained by reverse transcriptase PCR from RNA extracted from a geothermally heated soil at a geographically distant (∼50 km) YNP location (7) but has thus far not been detected in Dragon Spring.
Nucleotide sequence accession number.
The 16S rRNA gene sequence for isolate AO5 is available as GenBank accession number EF151282.
RESULTS
Mapping of spring H2S gradients.
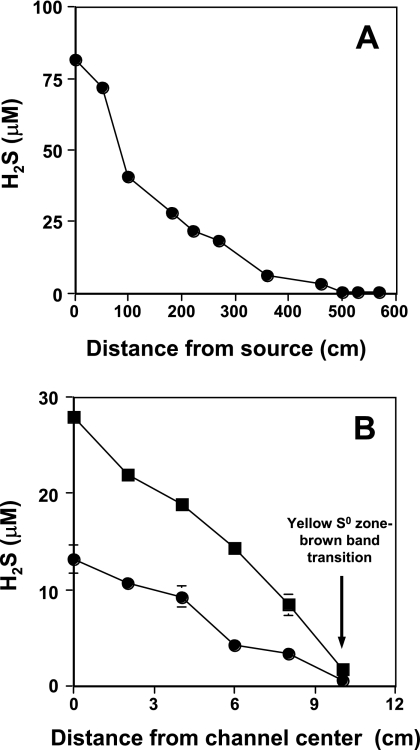
H2S gradients were defined in two dimensions, with all work focused in the yellow S0 zone (Fig. 1). A spatially intensive sampling transect of the S0 zone demonstrated an H2S gradient consistent with that observed by Langner et al. (28), showing that the aqueous H2S concentration decreases as a function of distance from the spring source (Fig. 2A) and that such gradients in this spring are stable over time (years). H2S levels decreased to roughly 5 μM at sampling transect site 5 in the yellow S0 zone (Fig. 1 and 2A) and then to undetectable levels at a distance of approximately 5 m and beyond (Fig. 2A). Additional measurements quantified H2S levels across the width of the yellow S0 zone to assess H2S concentrations in a second dimension. The H2S concentration was highest at mid-channel, coinciding with the highest flow rates, but sharply declined to below detection (∼1 μM) at the interface between the yellow S0 mat area and the brown band zone (Fig. 1 and 2B).
FIG. 2.
Quantifying Dragon Spring H2S gradients. (A) H2S concentration decreasing with increasing distance from the spring source along the main flow channel. (B) H2S concentration as a function of distance perpendicular to stream flow beginning at the center of the outflow channel and corresponding to transect site 4 (▪) and transect site 5 (•). For both panels, data points represent the mean of three samples, with error bars (where visible) representing 1 standard deviation of the mean. Samples for panel B were taken on a separate sampling trip, and thus maximum values at the channel center do not exactly correspond to those plotted in panel A.
As(III) oxidation potential in the H2S zone.
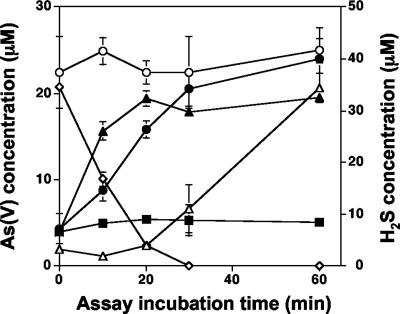
In previous work with a Hydrogenobaculum sp. isolate obtained from this spring (10), we observed that As(III) oxidase activity was constitutively expressed; i.e., As(III) oxidation activity profiles were the same for cultures preexposed to As(III) or for As(III)-naive cells. As previous molecular analyses also demonstrated that the yellow S0 mat area is heavily dominated by Hydrogenobaculum-like populations (19, 22; unpublished data), additional experiments were undertaken to determine whether the presence of As(III) oxidase activity could be demonstrated in this region of the spring if H2S concentrations were manipulated. In experiments where microbial mat material sampled from transect site 2 (Fig. 1) was suspended in filter-sterilized spring water from the same location, As(III) oxidation was not observed until aqueous H2S concentrations decreased to below 5 μM (Fig. 3). In contrast, As(III) oxidation was observed to occur within 10 min (the first sampling time point) in incubations where mat material from site 2 was incubated with filter-sterilized spring water taken at 5 m from the source (i.e., H2S was below detection, Fig. 2A) (Fig. 3). Similarly, As(III) oxidation was detected at the first sampling point with site 5 mat samples incubated in spring water taken from that same location. As(III) oxidation was not observed in the heat-killed samples (Fig. 3).
FIG. 3.
Ex situ assays of As(III) oxidation by a microbial mat from the yellow S0 zone in Dragon Spring. Symbols: ○, total dissolved As in transect site 2 mat assays showing no appreciable change in total dissolved As concentrations during these assays; ⋄, dissolved H2S measured in assays containing transect site 2 filter-sterilized water with mat material collected from transect site 2; ▵, As(V) formation in assays containing mat material from transect site 2 suspended in spring water from transect site 2; •, As(V) formation in assays containing mat material from transect site 2 suspended in filter-sterilized spring water collected from transect site 5; ▴, As(V) formation in assays containing mat material from transect site 5 in filter-sterilized spring water collected from transect site 5; ▪, As(V) concentration in heat-killed mat material from transect site 2 suspended in filter-sterilized spring water from transect site 5. Data are from one representative field trip experiment, with symbols representing the average of duplicate assays at each time point for each treatment and error bars (where visible) representing the range of the duplicate assays. H2S concentrations were lower than those shown in Fig. 2A because volatile losses occurred during the filter sterilization of spring water.
Isolation of an As(III) chemolithotroph.
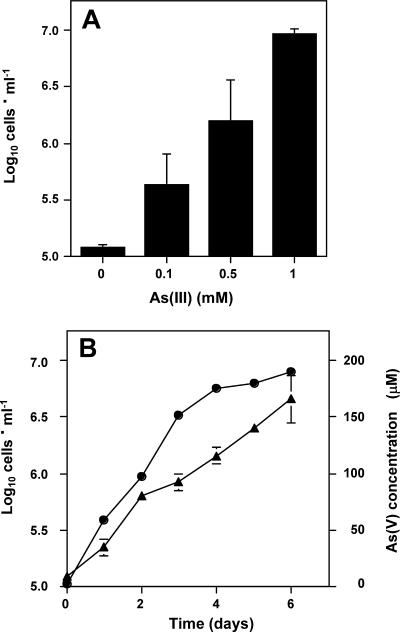
Given the apparent potential for As(III) oxidation activity in the yellow S0 zone, subsequent experiments sought to determine whether As(III) chemolithotrophs could be cultivated from this spring. Only enrichments inoculated with mat material from transect site 5 (Fig. 1) and corresponding to ∼5 μM H2S (Fig. 2A) generated a positive enrichment, which yielded pure culture strain AO5. In liquid defined medium, AO5 oxidized As(III) at a rate of approximately 1 nmol · min−1 · 106 cells−1. No growth was observed in the absence of As(III) (Fig. 4A), and although it was somewhat variable, the final culture density was positively correlated with incremental increases in As(III) in the medium (Fig. 4A). Growth rates declined as As(V) accumulated in the culture fluids (Fig. 4B), perhaps reflecting As(V) toxicity as As(V)/phosphate ratios increased or the exhaustion of another of the defined nutrients in this medium. With extended incubation (14 days), As(III) was nearly quantitatively oxidized (results not shown).
FIG. 4.
As(III) chemolithoautotrophic growth by isolate AO5. (A) Culture cell density after 10 days of growth at 55°C with various amounts of As(III) (shown as initial concentrations). The cell number recorded for 0 mM As(III) reflects the starting inoculum cell density. (B) Growth of isolate AO5 (•) with 1 mM As(III). (▴), As(V) accumulating in culture medium during growth of isolate AO5. For both panels, data represent the mean of three replicate cultures, with error bars (where visible) representing 1 standard deviation.
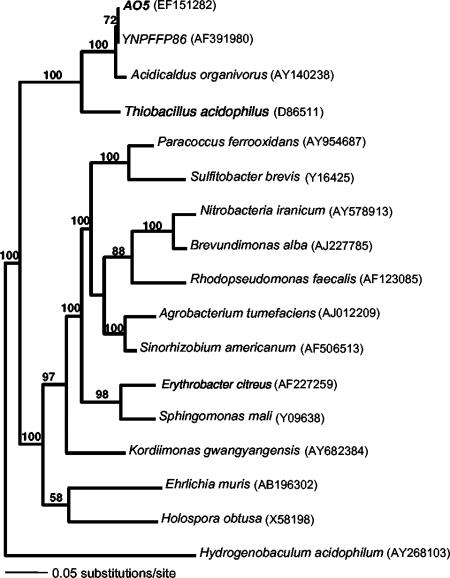
Nearly full-length (1,320 bp) sequence analysis of the 16S rRNA gene showed that strain AO5 was 97.8% identical to Acidicaldus organivorus strain Y008T, an acidothermophilic heterotrophic alphaproteobacterium also isolated from YNP (23). AO5 was 99.2% identical to above-mentioned clone YNPFFP86 (accession no. AF391980) and 98.9 to 99.2% identical to nine distinct 16S rRNA gene environmental clones (GenBank accession no. AY882679, AY882680, AY882682, AY882695, AY882794, AY882795, AY882809, DQ834208, and DQ834209) retrieved from two other geographically distant YNP springs, Joseph's Coat (∼32 km) and Rainbow Springs (∼48 km), where As(III) oxidation has also been documented (20). Phylogenetic comparison with other alphaproteobacteria showed that AO5 and A. organivorus have in common a node distinct from other characterized alphaproteobacteria (Fig. 5).
FIG. 5.
Maximum-likelihood tree showing the phylogenetic placement of isolate AO5. H. acidophilum, a deep-branching bacterium of the family Aquificaceae, was used as an outgroup. Numbers at nodes are bootstrap values for 100 pseudoreplicates, and NCBI accession numbers for each sequence are in parentheses.
Given the relatedness of AO5 to Acidicaldus organivorus strain Y008T, AO5 was examined for growth on various carbon sources and with spring-relevant alternative electron acceptors. The best growth was observed with xylose [doubling time of ∼18 h, compared to 23.4 h with As(III)], with slower growth on fructose and ethanol and very weak growth with glucose (doubling time, >7 days). AO5 would not grow anaerobically with spring-relevant electron acceptors such as SO4−2, NO3−, or Fe(III), regardless of whether As(III) or xylose was used as the electron donor. AO5 also failed to demonstrate growth when incubated in filter-sterilized spring water without the addition of As(III), showing that any organic carbon present in the spring water was incapable of supporting measurable growth. Previous work found the dissolved organic carbon concentration in the spring source water to be 80 μM (28).
Reaction of strain AO5 to H2S.
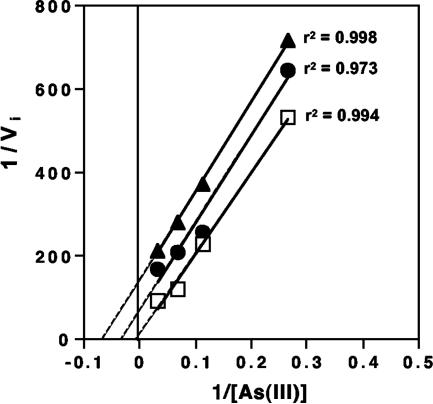
In liquid cultures with a starting As(III) concentration of 30 μM and with 0 to 60 μM H2S, half-maximal inhibition of As(III) oxidation was modeled to occur at approximately 5.8 μM H2S (data not shown). Additional kinetic analysis demonstrated that the H2S inhibition was uncompetitive in nature (Fig. 6). However, inhibition was determined to be unique to growth on As(III), as H2S (added at 0 to 60 μM) had no inhibitory effect on strain AO5 when it was cultured heterotrophically on xylose (results not shown).
FIG. 6.
Kinetic analysis of H2S inhibition of As(III) oxidation in isolate AO5. Lineweaver-Burk plot of H2S inhibition of As(III) oxidation by whole cells of strain AO5. Symbols: □, no Na2S added to assay samples; •, 2.01 μM Na2S added; ▴, 5.82 μM Na2S added. Solid lines indicate best-fit linear functions of measured data, and dashed portions indicate extrapolated data. r2 values for each line are given. Initial velocity (Vi) = 0.0287 μmol As(V) produced per min with 30 μM initial As(III) and no added Na2S.
Localization of strain AO5 within Dragon Spring.
The localization of strain AO5 in the spring was established by two different culture-independent approaches. First, DGGE was used to assess the microbial diversity present at outflow channel locations where chemical analysis indicated that H2S was absent (Fig. 2B). The DGGE profiles of three separate sites (A, B, and C) along the interface between the yellow S0 and brown band zones (Fig. 1) were essentially identical (Fig. 7). Band purification and sequencing identified strain AO5 (100% sequence match) at each location (Fig. 7).
FIG. 7.
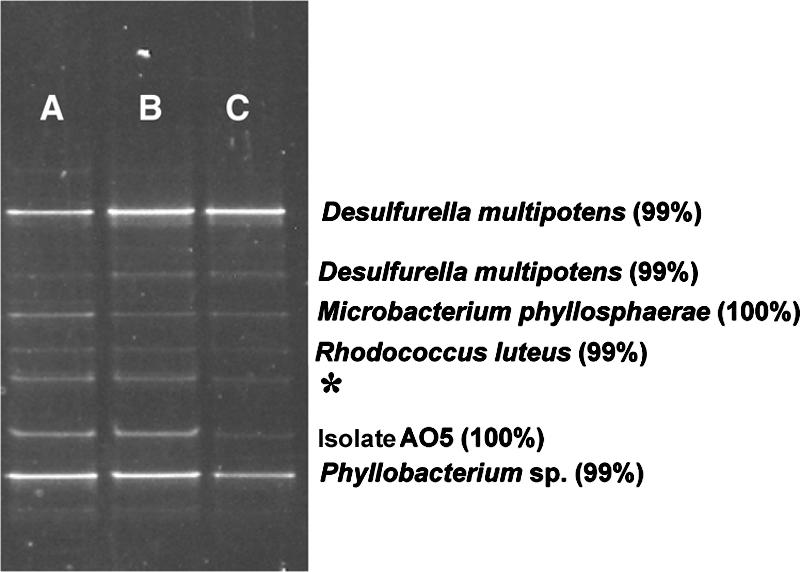
DGGE analysis of microbial diversity found at the interface between the yellow S0 zone and the brown band zone. Lanes A, B, and C correspond to sites A, B, and C, respectively, in Fig. 1. Blast identification of band sequences is shown with percent identity scores. We were unable to purify and sequence the band marked with an asterisk.
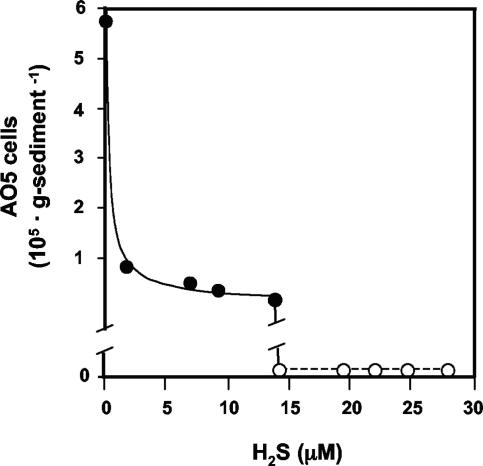
DNA sequences representing other organisms identified in the DGGE profiles provided information that was also important for designing primers for Q-PCR, which was the second approach taken to locate AO5 in the spring. Of particular interest was the distribution of AO5 in relation to H2S concentrations. Mat samples were taken for DNA extraction from sampling points along the outflow channel, as well as perpendicular to the water flow in the channel center (5 to 10 cm apart). Water samples were taken at the same mat sample locations and immediately measured for H2S content. The Q-PCR detection limit for the AO5 primers was determined to be 0.001 ng AO5 DNA in 100 ng total DNA from either sheared herring sperm DNA or from total DNA extracted from the microbial mat in the center of the yellow S0 zone. This corresponds to approximately three cells, assuming 0.39 pg DNA per cell, as was estimated by the method of Hermansson and Lindgren (17), and assuming one copy of a 3.5-Mb genome per cell (average for all genome-sequenced alphaproteobacteria) and a single rRNA operon per genome. In addition to determining the sensitivity limits of the primers, these experiments also demonstrated that lack of an AO5 Q-PCR signal in the yellow S0 zone was not due to technical problems with the Q-PCRs or to template bias. As approximated by Q-PCR, AO5 cell numbers were ∼2.5 × 106 cells · g−1 dry mat (Fig. 8) where the yellow S0 zone transitioned to the brown band zone (Fig. 1), which corresponded to low or undetectable levels of H2S (Fig. 2B). On the basis of the threshold cycle number and as derived from standard curves, the AO5 DNA was calculated to represent roughly 18% (wt/wt basis; 100 ng total DNA in each Q-PCR) of the total template population at this location. The AO5 template concentration was significantly reduced (∼2.5% of the total) where H2S concentrations increased above approximately 2 μM (Fig. 8) and fell below the detection limit at roughly 15 μM H2S.
FIG. 8.
Q-PCR estimate of AO5 (or AO5-like organisms) in mat samples in the yellow S0 zone in relation to dissolved H2S. For locations where AO5 DNA was not detectable, the counts are shown as zero.
To determine whether other environmental factors might also be important in establishing AO5 distribution patterns, AO5 temperature limits were examined. AO5 was capable of chemolithotrophic growth at 50 to 70°C, with the optimal temperature (Topt) determined to be 55 to 60°C. The Topt of AO5 corresponded to temperatures typically observed at transect sites 4 and 5 (Fig. 1) and at the S0-brown zone interface, where the water temperature measured during several sampling trips was consistently 58°C at sites A, B, and C (Fig. 1). Thus, it was concluded that while the temperature at the yellow S0-brown zone interface area was nearly optimum, water temperatures well into the yellow S0 zone should have also allowed for nearly optimum growth and thus would not limit AO5 distribution.
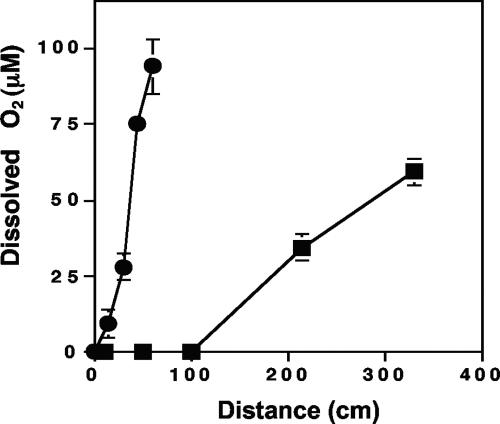
Since AO5 characterization experiments suggested that this organism would not grow without oxygen (above), we also measured O2 availability in the spring outflow channel to determine whether O2 would limit AO5 distribution. O2 was undetectable in the initial 100 cm of the channel center flow region (Fig. 9), corresponding to transect sites 1, 2, and 3 (Fig. 1). However, O2 content increased rapidly in the more distal reaches of the yellow S0 zone center channel and O2 gradients perpendicular to the water flow in the channel center were quite steep, suggesting that adequate electron acceptor would be available in much of the yellow S0 zone where AO5 was not detectable by Q-PCR and from where we were unable to isolate AO5 [or any As(III) chemolithotroph].
FIG. 9.
Dissolved oxygen in the Dragon Spring yellow S0 zone. Symbols: ▪, as a function of distance from the spring source; •, as a function of distance from the center of the flow channel with sampling increments perpendicular to the direction of spring flow at transect site 3. Symbols represent the average of three replicate samples, with error bars (where visible) representing 1 standard deviation.
DISCUSSION
The study summarized herein is one of a continuing series of investigations of a geothermal spring in YNP and the microbial populations that occur along its geochemical and thermal gradients. In this study, we were able to isolate an organism that is contributing to a prominent feature of the spring biogeochemical profile and that could also be detected by culture-independent techniques. These features, along with its sensitivity to H2S and the apparent uniqueness of its 16S rRNA gene (relative to the balance of the microbial community) suggested this organism would be an interesting model for exploring fundamental questions in microbial ecology, namely, the relationship between a microbe's functional attributes and its environment. Dragon Spring is ideally suited for such autecological studies as it is characterized by superimposable chemical and physical gradients that are measurable and definable (Fig. 1, 2, 8, and 9) and that correspond to distinct color changes (Fig. 1). These gradients define a continuum of potential niche opportunities, with the H2S gradient and its effect on As(III) oxidation being the focal point in the present study. Presumably due primarily to off-gassing, aqueous H2S concentrations decline rapidly as a function of distance from the spring source (Fig. 2A) and even more rapidly in relation to distance from the center of the spring channel (Fig. 2B). In the latter case, the spring waters are shallow and quiescent and thus allow rapid gas equilibration with the air.
While experimental evidence clearly demonstrated that H2S inhibits As(III) oxidation (Fig. 3 and 6), the high flux of As(III) and extremely high concentrations of CO2 in the source water of this spring (28; unpublished data) nevertheless provide otherwise ideal conditions for the proliferation of As(III) chemolithoautotrophs. The Acidicaldus-like organism characterized in the present study constitutes the first detailed description of a thermophilic As(III) chemolithoautotroph. Phylogenetically, AO5 is closely related to the heterotrophic alphaproteobacterium A. organivorus and could also grow on some of the same carbon compounds. However, A. organivorus Y008T was reported to be capable of anaerobic growth with Fe(III) as an electron acceptor but incapable of As(III) oxidation (23). Along with differences in the 16S rRNA gene sequence, these physiologic features distinguish isolate AO5 from A. organivorus Y008T. Additional characterization would be required to assess whether AO5 represents a new species of Acidicaldus.
The environmental features of the spring correlate well with the properties of AO5 and its distribution within the spring outflow channel. DGGE analysis (Fig. 7), Q-PCR determinations (Fig. 8), and enrichment and isolation success document the presence and numerical dominance of AO5 at the interface of the yellow and brown band zones (Fig. 1). H2S is depleted at this location (Fig. 2B), which agrees with the H2S sensitivity of As(III) oxidation in this organism and suggests that H2S is a major niche determinant for AO5. Conversely, the Topt and O2 requirement of this organism were less predictive as the temperature gradients (Fig. 1) and aqueous O2 measurements (Fig. 9) implied that AO5 could thrive in a significant portion of the yellow S0 zone. The microgeographical boundaries of the AO5 niche along the gradients may be very focused. The 16S rRNA gene signature of this organism was not encountered in a previous molecular survey of this spring (22) that sampled regions of the yellow S0 zone and the middle of the brown band zone but not the interface between these two major gradient zones.
Sulfide inhibition of other microbial processes has also been documented. This includes nitrate reduction by Desulfovibrio desulfuricans (8), nitric and nitrous oxide reduction by Pseudomonas fluorescens (44), oxygenic photosynthesis in cyanobacteria (35), and nitrification in estuarine sediments (25). Sulfide has also been shown to apparently influence microbial species composition in hot springs in Iceland (42), in marine coastal systems (16), and in salt marsh sediments (9). Miller and Bebout (30) also showed that sulfide tolerance among different cyanobacteria was positively correlated with sulfide levels in the environments from which they were isolated. The present autecology study extends beyond these general observations to more specifically quantify sulfide influence on a microbial process in nature, combining in situ and in vitro experiments to demonstrate the importance of such environmental constraints on function and how sulfide apparently correlates very closely with the distribution of the microorganism studied.
The H2S sensitivity of As(III) oxidation in isolate AO5 was similar to our observations with a Hydrogenobaculum-like isolate (10) and prompted us to examine an Agrobacterium tumefaciens soil isolate (29) we have been using as a model organism for studying the genetics and physiology underlying As(III) oxidation (26, 27). Sulfide inhibited As(III) oxidation in this bacterium as well (results not shown), suggesting that sulfide inhibition of As(III) oxidation may be common among As(III)-oxidizing microorganisms. However, there may be exceptions. For example, the facultative As(III) chemolithotroph isolated by Oremland et al. (34) was enriched from the anaerobic bottom waters of Mono Lake, where sulfide levels exceed 1 mM (33). Thus, either sulfide is not an inhibitor of As(III) oxidation in that organism or it may use As(III) as an energy source in regions of the Mono Lake water column where sulfide is absent or at least present at lower concentrations. Another example comes from YNP itself, where Inskeep et al. (20) reported increasing As(V)/As(III) ratios occurring in circumneutral geothermal springs containing 30 to 50 μM sulfide.
While previous chemical analysis of solid phases associated with the yellow S0 zone mat material failed to detect As-S solid phases (28), the studies of Wilkin et al. (48) suggest the potential for thioarsenite complexes to occur in Dragon Spring. The consequences for enzymatic As(III) oxidation are unknown, and therefore we cannot rule out As-S complexation as a factor influencing or controlling microbial As(III) oxidation in situ. However, the results of the kinetic analyses suggest that such As-S complexes may be of little significance. The periplasmic location of the As(III) oxidase enzyme (4, 41) provides the opportunity to examine the impact of inhibitors with whole cells, as opposed to purified enzyme preparations. The precision of enzyme catalysis rates would be influenced by the requirement for the substrate [As(III)] and product [As(V)] to diffuse in or out of the periplasm; however, assays with whole cells would nevertheless still provide an opportunity to gather evidence about the mechanism of inhibition. As(III) oxidation was reduced by roughly 50% at an As(III)/H2S ratio of approximately 5:1 and nearly completely inhibited at an As(III)/H2S ratio of 2.0 (Fig. 6A). The Km for As(III) under these conditions was estimated to be roughly 0.109 μM (extracted from the Lineweaver-Burk plots), and thus in at least the in vitro assays, H2S/As(III) complexing should be of little consequence as there still should have been As(III) available for oxidation at nearly maximum rates. Therefore, we suggest that the outcomes of the kinetic analysis (i.e., uncompetitive inhibition, Fig. 6) are more indicative of the true basis of H2S inhibition. The complete lack of H2S inhibition of AO5 heterotrophic growth is also probably informative in that it implies that if AO5 were growing heterotrophically on organic carbon in situ, then its distribution would not be expected to correspond so closely to the H2S concentration. Furthermore, it would also imply that the inhibitory effect does not derive from H2S binding to iron centers of cytochromes in electron transport (12).
Finally, the ex situ in-field assays yielded important information on two separate issues. First, by extending the As(III) oxidation assay incubations beyond the time frame used by Langner et al. (28), we were able to observe As(V) formation coinciding with the disappearance of H2S (Fig. 3) and thus illustrated agreement between in situ- and in vitro-based observations. Second, these assays also generated data that suggested constitutive expression of the As(III) oxidase enzyme(s) in situ. Microbial mat As(III) oxidation was observed in the earliest measurement (10 min) with mat samples incubated in spring water that did not contain detectable H2S (Fig. 3). The occurrence of measurable As(V) within such short time frames suggests that As(III) oxidation observed with mat organisms inhabiting the high-sulfide zone was not due to gene induction and de novo As(III) oxidase enzyme synthesis. Rather, the As(III) oxidase(s) appears to be present in the yellow S0 zone mat, with the removal of sulfide resulting in rapid reactivation.
Acknowledgments
This study was supported primarily by the National Science Foundation Microbial Observatories Program (MCB-0132022). Additional support was provided by the National Aeronautics and Space Administration (NAG 5-8807) and the Montana Agricultural Experiment Station (911310).
We thank John Varley and Christie Hendrix at the Yellowstone Center for Resources, YNP, WY, for assistance in permitting.
Footnotes
Published ahead of print on 7 September 2007.
REFERENCES
- 1.Aguiar, P., T. J. Beveridge, and A.-L. Reysenbach. 2004. Sulfurihydrogenibium azorense, sp. nov., a thermophilic hydrogen-oxidizing microaerophile from terrestrial hot springs in the Azores. Int. J. Syst. Evol. Microbiol. 54:33-39. [DOI] [PubMed] [Google Scholar]
- 2.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. H. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amann, R. I., W. Ludwig, and K. H. Schleifer. 1995. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 59:143-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anderson, G. L., J. Williams, and R. Hille. 1992. The purification and characterization of arsenite oxidase from Alcaligenes faecalis, a molybdenum-containing hydroxylase. J. Biol. Chem. 267:23674-23682. [PubMed] [Google Scholar]
- 5.Ball, J. W., R. B. McCleskey, D. K. Nordstrom, J. M. Holloway, and P. L. Verplanck. 2002. Water-chemistry data for selected springs, geysers, and streams in Yellowstone National Park, Wyoming 1999-2000. United States Geological Survey open file report 02-382. U.S. Geological Survey, Boulder, CO.
- 6.Botero, L. M., K. B. Brown, S. Brumefield, M. Burr, R. W. Castenholz, M. Young, and T. R. McDermott. 2004. Thermobaculum terrenum gen. nov., sp. nov.: a non-phototrophic gram-positive thermophile representing an environmental clone group related to the Chloroflexi (green non-sulfur bacteria) and Thermomicrobia. Arch. Microbiol. 181:269-277. [DOI] [PubMed] [Google Scholar]
- 7.Botero, L. M., S. D'Imperio, M. Burr, T. R. McDermott, M. Young, and D. J. Hassett. 2005. Poly(A) polymerase modification and reverse transcriptase PCR amplification of environmental RNA. Appl. Environ. Microbiol. 71:1267-1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dalsgaard, T., and F. Bak. 1994. Nitrate reduction in a sulfate-reducing bacterium, Desulfovibrio desulfuricans, isolated from rice paddy soil: sulfide inhibition, kinetics, and regulation. Appl. Environ. Microbiol. 60:291-297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dollhopf, S. L., J. H. Hyun, A. C. Smith, H. J. Adams, S. O'Brien, and J. E. Kostka. 2005. Quantification of ammonia-oxidizing bacteria and factors controlling nitrification in salt marsh sediments. Appl. Environ. Microbiol. 71:240-246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Donahoe-Christiansen, J., S. D'Imperio, C. R. Jackson, W. P. Inskeep, and T. R. McDermott. 2004. Arsenite-oxidizing Hydrogenobaculum strain isolated from an acid-sulfate-chloride geothermal spring in Yellowstone National Park. Appl. Environ. Microbiol. 70:1865-1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ellis, P. J., T. Conrads, R. Hille, and P. Kuhn. 2001. Crystal structure of the 100kDa arsenite oxidase from Alcaligenes faecalis in two crystal forms at 1.64 Å and 2.03 Å. Structure 9:125-132. [DOI] [PubMed] [Google Scholar]
- 12.Evans, C. L. 1967. The toxicity of hydrogen sulphide and other sulphides. Q. J. Exp. Physiol. 52:231-248. [DOI] [PubMed] [Google Scholar]
- 13.Ferris, M. J., G. Muyzer, and D. M. Ward. 1996. Denaturing gradient gel electrophoresis profiles of 16S rRNA-defined populations inhabiting a hot spring microbial mat community. Appl. Environ. Microbiol. 62:340-346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gihring, T. M., G. K. Druschel, R. B. McCleskey, R. J. Hamers, and J. F. Banfield. 2001. Rapid arsenite oxidation by Thermus aquaticus and Thermus thermophilus: field and laboratory investigations. Environ. Sci. Technol. 35:3857-3862. [DOI] [PubMed] [Google Scholar]
- 15.Hallberg, K. B., M. Dopson, and E. B. Lindstrom. 1996. Reduced sulfur compound oxidation by Thiobacillus caldus. J. Bacteriol. 178:6-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Herbert, R. A. 1999. Nitrogen cycling in coastal marine ecosystems. FEMS Microbiol. Rev. 23:563-590. [DOI] [PubMed] [Google Scholar]
- 17.Hermansson, A., and P. E. Lindgren. 2001. Quantification of ammonia-oxidizing bacteria in arable soil by real-time PCR. Appl. Environ. Microbiol. 67:972-976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ilyaletdinov, A. N., and S. A. Abdrashitova. 1981. Autotrophic oxidation of arsenic by Pseudomonas arsenitoxidans. Mikrobiologiya 50:197-205. [PubMed] [Google Scholar]
- 19.Inskeep, W. P., and T. R. McDermott. 2005. Geomicrobiology of acid-sulfate-chloride springs in Yellowstone National Park, p. 143-162. In W. P. Inskeep and T. R. McDermott (ed.), Geothermal biology and geochemistry in Yellowstone National Park. Thermal Biology Institute, Montana State University, Bozeman.
- 20.Inskeep, W. P., G. G. Ackerman, W. P. Taylor, M. Kozubal, S. Korf, and R. E. Macur. 2005. On the energetics of chemolithotrophy in nonequilibrium systems: case studies of geothermal springs in Yellowstone National Park. Geobiology 3:297-317. [Google Scholar]
- 21.Inskeep, W. P., R. E. Macur, N. Hamamura, T. P. Warelow, S. A. Ward, and J. M. Santini. 2007. Detection, diversity and expression of aerobic bacterial arsenite oxidase genes. Environ. Microbiol. 9:934-943. [DOI] [PubMed] [Google Scholar]
- 22.Jackson, C. R., H. W. Langner, J. Donahoe-Christiansen, W. P. Inskeep, and T. R. McDermott. 2001. Molecular analysis of microbial community structure in an arsenite-oxidizing acidic thermal spring. Environ. Microbiol. 3:532-542. [DOI] [PubMed] [Google Scholar]
- 23.Johnson, D. B., B. Stallwood, S. Kimura, and K. B. Hallberg. 2006. Isolation and characterization of Acidicaldus organivorus, gen. nov., sp nov.: a novel sulfur-oxidizing, ferric iron-reducing thermo-acidophilic heterotrophic Proteobacterium. Arch. Microbiol. 185:212-221. [DOI] [PubMed] [Google Scholar]
- 24.Jones, C. A., H. W. Langner, K. Anderson, T. R. McDermott, and W. P. Inskeep. 2000. Rates of microbially mediated arsenate reduction and solubilization. Soil Sci. Soc. Am. J. 64:600-608. [Google Scholar]
- 25.Joye, S. B., and J. T. Hollibaugh. 1995. Influence of sulfide inhibition of nitrification on nitrogen regeneration in sediments. Science 270:623-625. [Google Scholar]
- 26.Kashyap, D. R., L. M. Botero, C. Lehr, D. J. Hassett, and T. R. McDermott. 2006. A Na+:H+ antiporter and a molybdate transporter are essential for arsenite oxidation in Agrobacterium tumefaciens. J. Bacteriol. 188:1577-1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kashyap, D. R., L. M. Botero, W. L. Franck, D. J. Hassett, and T. R. McDermott. 2006. Complex regulation of arsenite oxidation in Agrobacterium tumefaciens. J. Bacteriol. 188:1081-1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Langner, H. W., C. R. Jackson, T. R. McDermott, and W. P. Inskeep. 2001. Rapid oxidation of arsenite in a hot spring ecosystem, Yellowstone National Park. Environ. Sci. Technol. 35:3302-3309. [DOI] [PubMed] [Google Scholar]
- 29.Macur, R. E., C. R. Jackson, L. M. Botero, T. R. McDermott, and W. P. Inskeep. 2004. Bacterial populations associated with the oxidation and reduction of arsenic in an unsaturated soil. Environ. Sci. Technol. 38:104-111. [DOI] [PubMed] [Google Scholar]
- 30.Miller, S. R., and B. M. Bebout. 2004. Variation in sulfide tolerance of photosystem II in phylogenetically diverse cyanobacteria from sulfidic habitats. Appl. Environ. Microbiol. 70:736-744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Muller, D., D. Lievremont, D. D. Simeonova, J.-C. Hubert, and M.-C. Lett. 2003. Arsenite oxidase aox genes from a metal-resistant β-proteobacterium. J. Bacteriol. 185:135-141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Norris, T. B., J. Wraith, R. C. Castenholz, and T. R. McDermott. 2002. Soil microbial community structure across a thermal gradient following a recent geothermal heating event. Appl. Environ. Microbiol. 68:6300-6309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Oremland, R. S., P. R. Dowdle, S. Hoeft, J. O. Sharp, J. K. Schaefer, L. G. Miller, J. Switzer Blum, R. L. Smith, N. S. Bloom, and D. Wallschlaeger. 2000. Bacterial dissimilatory reduction of arsenate and sulfate in meromictic Mono Lake, California. Geochim. Cosmochim. Acta 64:3073-3084. [Google Scholar]
- 34.Oremland, R. S., S. E. Hoeft, J. M. Santini, N. Bano, R. A. Hollibaugh, and J. T. Hollibaugh. 2002. Anaerobic oxidation of arsenite in Mono Lake water and by a facultative, arsenite-oxidizing chemoautotroph, strain MLHE-1. Appl. Environ. Microbiol. 68:4795-4802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Oren, A., E. Padan, and S. Malkin. 1979. Sulfide inhibition of photosystem-II in cyanobacteria (blue-green-algae) and tobacco chloroplasts. Biochim. Biophys. Acta 546:270-279. [DOI] [PubMed] [Google Scholar]
- 36.Peters, J., B. Bothner, and S. Kelly. 2007. Unfolding the mystery of protein thermostability. Yellowstone Sci. 15:5-14. [Google Scholar]
- 37.Phillips, S. E., and M. L. Taylor. 1976. Oxidation of arsenite to arsenate by Alcaligenes faecalis. Appl. Environ. Microbiol. 32:392-399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rhine, E. D., C. D. Phelps, and L. Y. Young. 2006. Anaerobic arsenite oxidation by novel denitrifying isolates. Environ. Microbiol. 8:899-908. [DOI] [PubMed] [Google Scholar]
- 39.Santini, J. M., L. I. Sly, R. D. Schnagl, and J. M. Macy. 2000. A new chemolithoautotrophic arsenite-oxidizing bacterium isolated from a gold mine: phylogenetic, physiological, and preliminary biochemical studies. Appl. Environ. Microbiol. 66:92-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Santini, J. M., L. I. Sly, A. Wen, D. Combrie, P. De Wulf-Durand, and J. M. Macy. 2002. New arsenite-oxidizing bacteria isolated from Australian gold mining environments—phylogenetic relationships. Geomicrobiol. J. 19:67-76. [Google Scholar]
- 41.Santini, J. M., and R. N. vanden Hoven. 2004. Molybdenum-containing arsenite oxidase of the chemolithoautotrophic arsenite oxidizer NT-26. J. Bacteriol. 186:1614-1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Skirnisdottir, S., G. O. Hreggvidsson, S. Hjorleifsdottir, V. T. Marteinsson, S. K. Petursdottir, O. Holst, and J. K. Kristjansson. 2000. Influence of sulfide and temperature on species composition and community structure of hot spring microbial mats. Appl. Environ. Microbiol. 66:2835-2841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Skovhus, T. L., N. B. Ramsing, C. Holmstrom, S. Kjelleberg, and I. Dahllof. 2004. Real-time quantitative PCR for assessment of abundance of Pseudoalteromonas species in marine samples. Appl. Environ. Microbiol. 70:2373-2382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sorensen, J., J. M. Tiedje, and R. B. Firestone. 1980. Inhibition by sulfide of nitric and nitrous-oxide reduction by denitrifying Pseudomonas fluorescens. Appl. Environ. Microbiol. 39:105-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stauffer, R. E., E. A. Jenne, and J. W. Ball. 1980. Chemical studies of selected trace elements in hot-spring drainages of Yellowstone National Park. U.S. Geological Survey professional paper 1044-F. U.S. Government Printing Office, Washington, DC.
- 46.Swofford, D. L. 2002. PAUP*. Phylogenetic analysis using parsimony (*and other methods). Version 4.0b10. Sinauer Associates, Inc., Sunderland, MA.
- 47.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL_X Windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wilkin, R. T., D. Wallschlager, and R. G. Ford. 2003. Speciation of arsenic in sulfidic waters. Geochem. Trans. 4:1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]