Abstract
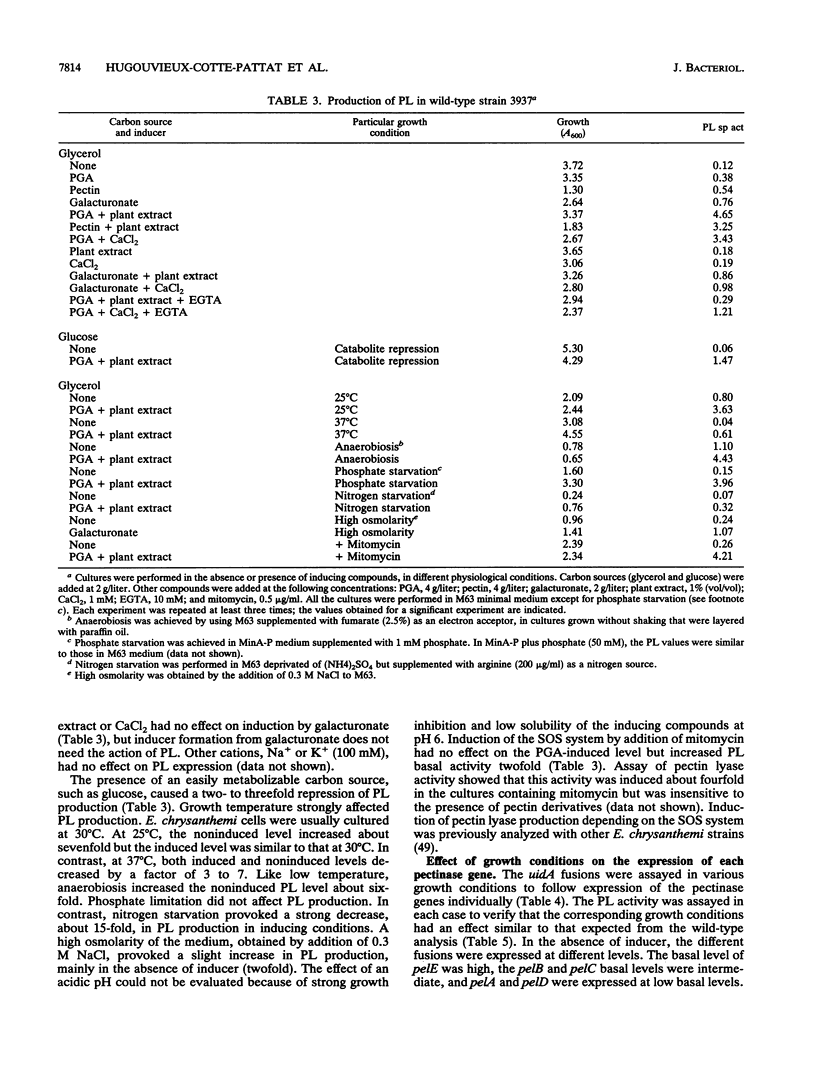
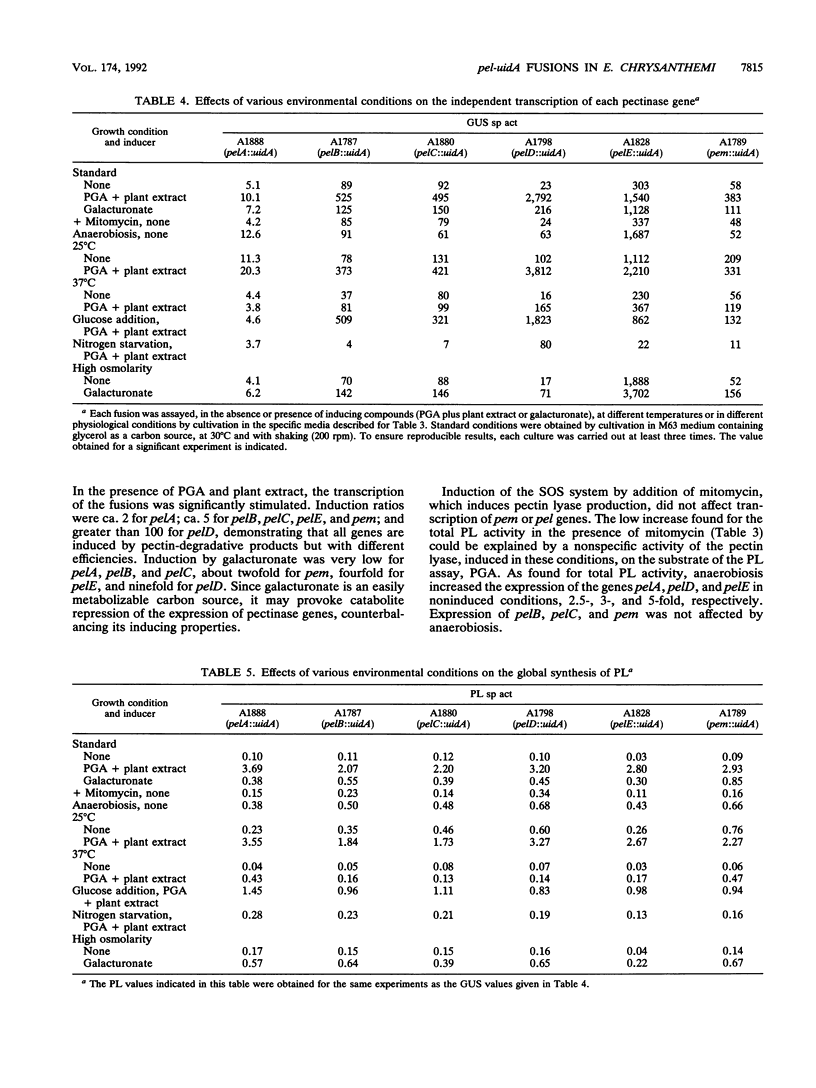
To depolymerize plant pectin, the phytopathogenic enterobacterium Erwinia chrysanthemi produces a series of enzymes which include a pectin-methyl-esterase encoded by the pem gene and five isoenzymes of pectate lyases encoded by the five genes pelA, pelB, pelC, pelD, and pelE. We have constructed transcriptional fusions between the pectinase gene promoters and the uidA gene, encoding beta-glucuronidase, to study the regulation of these E. chrysanthemi pectinase genes individually. The transcription of the pectinase genes is dependent on many environmental conditions. All the fusions were induced by pectic catabolic products and responded, to different degrees, to growth phase, catabolite repression, temperature, and nitrogen starvation. Transcription of pelA, pelD, and pelE was also increased in anaerobic growth conditions. High osmolarity of the culture medium increased expression of pelE but decreased that of pelD; the other pectinase genes were not affected. The level of expression of each gene was different. Transcription of pelA was very low under all growth conditions. The expression of the pelB, pelC, and pem genes was intermediate. The pelE gene had a high basal level of expression. Expression of pelD was generally the most affected by changes in culture conditions and showed a low basal level but very high induced levels. These differences in the expression of the pectinase genes of E. chrysanthemi 3937 presumably reflect their role during infection of plants, because the degradation of pectic polymers of the plant cell walls is the main determinant of tissue maceration caused by soft rot erwiniae.
Full text
PDF





Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bachmann B. J. Linkage map of Escherichia coli K-12, edition 8. Microbiol Rev. 1990 Jun;54(2):130–197. doi: 10.1128/mr.54.2.130-197.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardonnet N., Blanco C. 'uidA-antibiotic-resistance cassettes for insertion mutagenesis, gene fusions and genetic constructions. FEMS Microbiol Lett. 1992 Jun 15;72(3):243–247. doi: 10.1016/0378-1097(92)90469-5. [DOI] [PubMed] [Google Scholar]
- Bertheau Y., Madgidi-Hervan E., Kotoujansky A., Nguyen-The C., Andro T., Coleno A. Detection of depolymerase isoenzymes after electrophoresis or electrofocusing, or in titration curves. Anal Biochem. 1984 Jun;139(2):383–389. doi: 10.1016/0003-2697(84)90022-8. [DOI] [PubMed] [Google Scholar]
- Boccara M., Chatain V. Regulation and role in pathogenicity of Erwinia chrysanthemi 3937 pectin methylesterase. J Bacteriol. 1989 Jul;171(7):4085–4087. doi: 10.1128/jb.171.7.4085-4087.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castilho B. A., Olfson P., Casadaban M. J. Plasmid insertion mutagenesis and lac gene fusion with mini-mu bacteriophage transposons. J Bacteriol. 1984 May;158(2):488–495. doi: 10.1128/jb.158.2.488-495.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Condemine G., Hugouvieux-Cotte-Pattat N., Robert-Baudouy J. Isolation of Erwinia chrysanthemi kduD mutants altered in pectin degradation. J Bacteriol. 1986 Mar;165(3):937–941. doi: 10.1128/jb.165.3.937-941.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Condemine G., Robert-Baudouy J. 2-keto-3-deoxygluconate transport system in Erwinia chrysanthemi. J Bacteriol. 1987 May;169(5):1972–1978. doi: 10.1128/jb.169.5.1972-1978.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diolez A., Coleno A. Mu-lac insertion-directed mutagenesis in a pectate lyase gene of Erwinia chrysanthemi. J Bacteriol. 1985 Sep;163(3):913–917. doi: 10.1128/jb.163.3.913-917.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diolez A., Richaud F., Coleno A. Pectate lyase gene regulatory mutants of Erwinia chrysanthemi. J Bacteriol. 1986 Jul;167(1):400–403. doi: 10.1128/jb.167.1.400-403.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favey S., Bourson C., Bertheau Y., Kotoujansky A., Boccara M. Purification of the acidic pectate lyase and nucleotide sequence of the corresponding gene (pelA) of Erwinia chrysanthemi strain 3937. J Gen Microbiol. 1992 Mar;138(3):499–508. doi: 10.1099/00221287-138-3-499. [DOI] [PubMed] [Google Scholar]
- Gold S., Nishio S., Tsuyumu S., Keen N. T. Analysis of the pelE promoter in Erwinia chrysanthemi EC16. Mol Plant Microbe Interact. 1992 Mar-Apr;5(2):170–178. [PubMed] [Google Scholar]
- Hinton J. C., Sidebotham J. M., Gill D. R., Salmond G. P. Extracellular and periplasmic isoenzymes of pectate lyase from Erwinia carotovora subspecies carotovora belong to different gene families. Mol Microbiol. 1989 Dec;3(12):1785–1795. doi: 10.1111/j.1365-2958.1989.tb00164.x. [DOI] [PubMed] [Google Scholar]
- Hugouvieux-Cotte-Pattat N., Reverchon S., Robert-Baudouy J. Expanded linkage map of Erwinia chrysanthemi strain 3937. Mol Microbiol. 1989 May;3(5):573–581. doi: 10.1111/j.1365-2958.1989.tb00204.x. [DOI] [PubMed] [Google Scholar]
- Hugouvieux-Cotte-Pattat N., Robert-Baudouy J. Analysis of the regulation of the pelBC genes in Erwinia chrysanthemi 3937. Mol Microbiol. 1992 Aug;6(16):2363–2376. doi: 10.1111/j.1365-2958.1992.tb01411.x. [DOI] [PubMed] [Google Scholar]
- Hugouvieux-Cotte-Pattat N., Robert-Baudouy J. Hexuronate catabolism in Erwinia chrysanthemi. J Bacteriol. 1987 Mar;169(3):1223–1231. doi: 10.1128/jb.169.3.1223-1231.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hugouvieux-Cotte-Pattat N., Robert-Baudouy J. Isolation of Erwinia chrysanthemi mutants altered in pectinolytic enzyme production. Mol Microbiol. 1989 Nov;3(11):1587–1597. doi: 10.1111/j.1365-2958.1989.tb00144.x. [DOI] [PubMed] [Google Scholar]
- Hugouvieux-Cotte-Pattat N., Robert-Baudouy J. Lactose metabolism in Erwinia chrysanthemi. J Bacteriol. 1985 Apr;162(1):248–255. doi: 10.1128/jb.162.1.248-255.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotoujansky A., Lemattre M., Boistard P. Utilization of a thermosensitive episome bearing transposon TN10 to isolate Hfr donor strains of Erwinia carotovora subsp. chrysanthemi. J Bacteriol. 1982 Apr;150(1):122–131. doi: 10.1128/jb.150.1.122-131.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange R., Hengge-Aronis R. Growth phase-regulated expression of bolA and morphology of stationary-phase Escherichia coli cells are controlled by the novel sigma factor sigma S. J Bacteriol. 1991 Jul;173(14):4474–4481. doi: 10.1128/jb.173.14.4474-4481.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mekalanos J. J. Environmental signals controlling expression of virulence determinants in bacteria. J Bacteriol. 1992 Jan;174(1):1–7. doi: 10.1128/jb.174.1.1-7.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran F., Nasuno S., Starr M. P. Extracellular and intracellular polygllacturonic acid trans-eliminases of Erwinia carotovora. Arch Biochem Biophys. 1968 Feb;123(2):298–306. doi: 10.1016/0003-9861(68)90138-0. [DOI] [PubMed] [Google Scholar]
- Nasser W., Condemine G., Plantier R., Anker D., Robert-Baudouy J. Inducing properties of analogs of 2-keto-3-deoxygluconate on the expression of pectinase genes of Erwinia chrysanthemi. FEMS Microbiol Lett. 1991 Jun 1;65(1):73–78. doi: 10.1016/0378-1097(91)90474-o. [DOI] [PubMed] [Google Scholar]
- Nasser W., Reverchon S., Robert-Baudouy J. Purification and functional characterization of the KdgR protein, a major repressor of pectinolysis genes of Erwinia chrysanthemi. Mol Microbiol. 1992 Jan;6(2):257–265. doi: 10.1111/j.1365-2958.1992.tb02007.x. [DOI] [PubMed] [Google Scholar]
- Plastow G. S. Molecular cloning and nucleotide sequence of the pectin methyl esterase gene of Erwinia chrysanthemi B374. Mol Microbiol. 1988 Mar;2(2):247–254. doi: 10.1111/j.1365-2958.1988.tb00026.x. [DOI] [PubMed] [Google Scholar]
- Preston J. F., 3rd, Rice J. D., Ingram L. O., Keen N. T. Differential depolymerization mechanisms of pectate lyases secreted by Erwinia chrysanthemi EC16. J Bacteriol. 1992 Mar;174(6):2039–2042. doi: 10.1128/jb.174.6.2039-2042.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reverchon S., Huang Y., Bourson C., Robert-Baudouy J. Nucleotide sequences of the Erwinia chrysanthemi ogl and pelE genes negatively regulated by the kdgR gene product. Gene. 1989 Dec 21;85(1):125–134. doi: 10.1016/0378-1119(89)90472-1. [DOI] [PubMed] [Google Scholar]
- Reverchon S., Nasser W., Robert-Baudouy J. Characterization of kdgR, a gene of Erwinia chrysanthemi that regulates pectin degradation. Mol Microbiol. 1991 Sep;5(9):2203–2216. doi: 10.1111/j.1365-2958.1991.tb02150.x. [DOI] [PubMed] [Google Scholar]
- Reverchon S., Van Gijsegem F., Rouve M., Kotoujansky A., Robert-Baudouy J. Organization of a pectate lyase gene family in Erwinia chrysanthemi. Gene. 1986;49(2):215–224. doi: 10.1016/0378-1119(86)90282-9. [DOI] [PubMed] [Google Scholar]
- Roeder D. L., Collmer A. Marker-exchange mutagenesis of a pectate lyase isozyme gene in Erwinia chrysanthemi. J Bacteriol. 1985 Oct;164(1):51–56. doi: 10.1128/jb.164.1.51-56.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiro S., Guest J. R. FNR and its role in oxygen-regulated gene expression in Escherichia coli. FEMS Microbiol Rev. 1990 Aug;6(4):399–428. doi: 10.1111/j.1574-6968.1990.tb04109.x. [DOI] [PubMed] [Google Scholar]
- Tamaki S. J., Gold S., Robeson M., Manulis S., Keen N. T. Structure and organization of the pel genes from Erwinia chrysanthemi EC16. J Bacteriol. 1988 Aug;170(8):3468–3478. doi: 10.1128/jb.170.8.3468-3478.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Gijsegem F. Relationship between the pel genes of the pelADE cluster in Erwinia chrysanthemi strain B374. Mol Microbiol. 1989 Oct;3(10):1415–1424. doi: 10.1111/j.1365-2958.1989.tb00124.x. [DOI] [PubMed] [Google Scholar]



