Abstract
Anoxia occurs in bottom waters of stratified estuaries when respiratory consumption of oxygen, primarily by bacteria, outpaces atmospheric and photosynthetic reoxygenation. Once water becomes anoxic, bacterioplankton must change their metabolism to some form of anaerobic respiration. Analysis of redox chemistry in water samples spanning the oxycline of Chesapeake Bay during the summer of 2004 suggested that there was a succession of respiratory metabolism following the loss of oxygen. Bacterial community doubling time, calculated from bacterial abundance (direct counts) and production (anaerobic leucine incorporation), ranged from 0.36 to 0.75 day and was always much shorter than estimates of the time that the bottom water was anoxic (18 to 44 days), indicating that there was adequate time for bacterial community composition to shift in response to changing redox conditions. However, community composition (as determined by PCR-denaturing gradient gel electrophoresis analysis of 16S rRNA genes) in anoxic waters was very similar to that in surface waters in June when nitrate respiration was apparent in the water column and only partially shifted away from the composition of the surface community after nitrate was depleted. Anoxic water communities did not change dramatically until August, when sulfate respiration appeared to dominate. Surface water populations that remained dominant in anoxic waters were Synechococcus sp., Gammaproteobacteria in the SAR86 clade, and Alphaproteobacteria relatives of Pelagibacter ubique, including a putative estuarine-specific Pelagibacter cluster. Populations that developed in anoxic water were most similar (<92% similarity) to uncultivated Firmicutes, uncultivated Bacteroidetes, Gammaproteobacteria in the genus Thioalcalovibrio, and the uncultivated SAR406 cluster. These results indicate that typical estuarine bacterioplankton switch to anaerobic metabolism under anoxic conditions but are ultimately replaced by different organisms under sulfidic conditions.
Every summer in many stratified estuaries, eutrophication-elevated phytoplankton production drives rapid bacterial respiration, creating zones in which dissolved oxygen is greatly reduced (hypoxic, <3 mg/liter dissolved oxygen) or eliminated (anoxic). These so-called “dead zones” exclude fish, kill benthic organisms, and eliminate habitat for commercially and recreationally important marine species (48). The management community considers hypoxia/anoxia to be a bellwether of eutrophication in coastal waters. This phenomenon is extensive in the United States, occurring in the waters of 16 of the 21 coastal continental states and in 66 of 137 estuaries (7). In some systems, like the Chesapeake Bay, hypoxic/anoxic bottom water covers large areas and may have a significant impact on food webs and on carbon and nutrient cycles.
Despite their popular name, hypoxic/anoxic zones are not really dead but rather are populated with living and very active communities of bacteria and protists. In fact, bacterial production in anoxic marine and fresh waters is often similar to or even greater than that in overlying oxic waters (2, 12, 58).
Two factors act in concert to produce deep-water anoxia in estuaries: stratification and aerobic respiration. First, vertical stratification traps high-density water away from the atmosphere and prevents reoxygenation (3, 4, 42). Second, aerobic respiration removes oxygen from the water column while consuming organic matter from several potential sources, including phytoplankton production, river inputs, and detritus from salt marshes and mangroves. In many eutrophied estuaries, elevated phytoplankton production is thought to be responsible for expansion of bottom water anoxia (50). For example, in Chesapeake Bay there is a strong relationship between chlorophyll a (phytoplankton) deposition and the loss of dissolved oxygen in the spring (6).
Chesapeake Bay is a vertically stratified estuary that supports an extensive seasonal anoxic zone that forms every year. The magnitude of hypoxia/anoxia in this system is somewhat predictable based on measurements of spring nutrient and water inputs to the estuary (28). Depending on river flow, bottom water anoxia occurs in May or June as temperatures rise and the pycnocline strengthens, and hydrogen sulfide is evident in bottom waters by July (25). Hydrogen sulfide helps maintain anoxic conditions throughout the summer by chemically consuming oxygen (34). Anoxia remains through August and begins to shift back to hypoxia by mid-September. By November to December the bottom water oxygen concentrations are high.
We investigated the activity and phylogenetic composition of bacterioplankton communities across the oxycline of the Chesapeake Bay during four cruises in the summer of 2004. We hypothesized that bacterial communities in anoxic water would shift away from the aerobic surface water community and that this shift would be related to redox conditions in the anoxic zone.
MATERIALS AND METHODS
Depth profile samples were collected in the mesohaline reach of the Chesapeake Bay north of the mouth of the Patuxent River (38o34.1′N, 76o26.6′W; Chesapeake Bay Program site CB4.3C) on four dates during the summer of 2004. Samples were collected with an on-board diaphragm pump (10 June), with an impeller-driven submersible pump (1 July and 4 August) (21), and with a rosette of Niskin bottles (23 July). Temperature, salinity, depth, and oxygen concentration were measured with a conductivity-temperature-depth sensor deployed either with the impeller pump or with the Niskin bottles. On 10 June the measurements were obtained with a hand-held sensor by submersing its probe in a beaker that was continually flooded with water from the sampling depth.
Samples collected for chemical analysis were filtered immediately using a syringe filter (which was tested for contamination) and then frozen (nutrients) or preserved with acid (metals). Samples for gas analyses were collected in 8-ml, gas-tight vials, which were overflowed three times, preserved with 10 μl of 50% saturated mercuric chloride, capped, and stored under water until they were analyzed. Measurements were obtained for dissolved nitrogen and phosphorus species (NO3−, NO2−, NH4+, soluble reactive phosphorus) and silica using standard colorimetric analyses (51), for iron using ferrozine (64), and for hydrogen sulfide by using the method of Cline (11). Dissolved oxygen was measured by membrane inlet mass spectrometry (38). Dissolved organic carbon and total nitrogen were measured with a Control Instrument CHN analyzer.
The water samples used for biological measurements were collected by following precautions described by Pedrós-Alió et al. (52) for handling anaerobic samples. Water was pumped directly into the bottom of gas-tight biological oxygen demand bottles and allowed to overflow three to five times before capping, making sure the bottles contained no visible bubbles. These bottles were stored under water at in situ temperature in the dark for up to 5 h before processing in the lab.
Heterotrophic bacterial production was measured by determining the rate of incorporation of [3H]leucine into cold trichloroacetic acid-insoluble macromolecules (40) during 1-h incubations using the methods of Bastviken and Tranvik (2) to avoid oxygen contamination of anoxic and hypoxic samples. Incubation mixtures were prepared in an anaerobic glove bag, and incubation was carried out in sealed plastic syringes with the headspace removed. All plastic was stored in an oxygen-free environment for 24 h prior to use. Bacterial carbon production was calculated from leucine incorporation using a ratio of cellular carbon to protein of 0.86, a fraction of leucine in protein of 0.073, and an intracellular leucine isotope dilution of two (40).
Bacteria were enumerated by direct microscopic counting of glutaraldehyde-fixed cells after labeling with the fluorescent DNA stain SYBR gold (47). Bacterial community doubling times were calculated for samples collected on 1 July and 4 August from leucine incorporation rates and cell counts. Bacterial biomass (BB) was assumed to be 25 fg carbon per cell. The equation μ = ln[(BB + BP)/BB] (where BP is bacterial carbon production) was used to estimate cell-specific exponential growth (μ), and the equation DT = ln2/μ was used to calculate the doubling time (DT) (1).
DNA samples were collected on 0.2-μm-pore-size Sterivex filter capsules (Millipore) with a peristaltic pump (∼500 ml/filter) and were extracted and purified with standard techniques (17). Bacterioplankton community compositions were compared based on the presence/absence, relative peak heights, and relative densities of bands in banding patterns created by denaturing gradient gel electrophoresis (DGGE) separation of PCR-amplified 16S rRNA genes (17) using GelcomparII v3.5 software (Bionumerics). Banding patterns were subjected to rolling-ball background subtraction (disk width, 50), and bands were considered present when the peak height exceeded 5% of the peak height of the darkest band in the sample. Relative band density was determined by dividing the “relative band surface” (i.e., the area under each densitometric curve) by the total band area in the sample. Relative peak height was calculated in a similar manner.
Pairwise similarity matrices were calculated using the Dice equation for presence/absence data (SPSS v11.0) and the Bray-Curtis equation for relative peak height and relative density (Primer v5.0). Relationships among samples were visualized using the ordination technique multidimensional scaling (MDS) calculated with the PROXSCAL algorithm (SPSS v11.0). Analysis of similarity (ANOSIM) (Primer v5.0), as described by Clark and Green (10), was used to test the hypothesis that communities in anoxic waters were more similar to each other than to communities in oxic waters. Canonical correspondence analysis (CANOCO v.4.5; Microcomputer Power) was used to determine the extent to which environmental variables explained patterns of similarity in community composition (59). The variables included in the analysis (salinity, temperature, depth, dissolved oxygen, N2-N, NO3−, NO2−, NH4+, sulfides) were transformed when necessary to correct for deviations from normality. Variables were added by forward selection until model improvement failed (P < 0.05). The DNA in select DGGE bands was isolated, reamplified, and sequenced using standard techniques (16) to identify dominant populations in oxic and anoxic waters.
Clone libraries of nearly full-length 16S rRNA genes were prepared for one oxic sample (depth, 5 m) and one anoxic sample (depth, 15 m) collected on 4 August using previously described methods (17), with the following modifications. The primers used were 27f (5′-AGAGTTTGATCCTGGCTCAG-3′) and 1492r (5′-GGCTACCTTGTTACGACTT-3′). PCR conditions were modified to include a “reconditioning PCR” step; after initial amplification with 27 PCR cycles, products were diluted 1:10 in fresh PCR cocktail and subjected to three more PCR cycles (60). DNA sequences were determined by High-Throughput Sequencing Solutions (www.htseq.org), aligned using the ARB sequence alignment program (www.arb-home.de), and screened for chimera formation using BELLEROPHON (32), CHIMERA_CHECK (13), and visual inspection of secondary structure base pair matches. Fourteen chimeras were identified and excluded from further analysis.
The diversity of bacteria represented by clone library sequences was estimated as operational taxonomic unit richness using rarefaction analysis (56) and the Chao 1 nonparametric species richness estimator (9) for 97% sequence similarity groups. Aligned sequences were subjected to a 50% base pair frequency filter prior to analysis to eliminate regions that were difficult to align. Plastid sequences were excluded from the analysis.
The two clone libraries were compared statistically using the LIBSHUFF computer program (57), which calculates coverage (26) at various levels of evolutionary distance using the Jukes-Cantor model for nucleic acid substitution as described previously (15). Aligned sequences were subjected to a 50% base pair frequency filter prior to analysis to eliminate regions that were difficult to align.
Nucleotide sequence accession numbers.
DNA sequences for clone libraries and DGGE bands are available in the GenBank database under accession numbers EF395606 to EF395789.
RESULTS
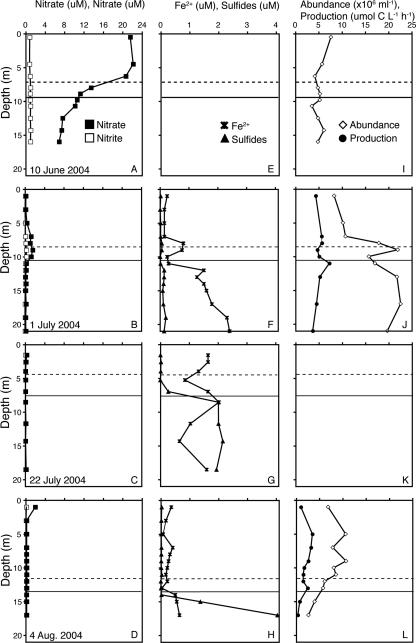
The oxycline on all sampling dates mirrored the pycnocline in depth and thickness, and the depth of the hypoxic/anoxic boundary (O2 concentration, <0.75 mg liter−1) varied from 7 to 14 m (Fig. 1). Nitrate and nitrite concentrations were high throughout the water column on 10 June but by 1 July were elevated only near the oxycline (Fig. 1). After this date, the nitrate and nitrate concentrations were low (<0.45 μM NO3− and <0.16 μM NO2−). The concentration of reduced iron (Fe2+; not measured on 10 June) was elevated in anoxic and hypoxic waters on 1 July and 22 July but was somewhat reduced on 4 August. The concentration of sulfide (also not measured on 10 June) was low on 1 July but was elevated in anoxic waters on 22 July and 4 August.
FIG. 1.
Depth profiles of the nitrate and nitrite concentrations (A to D), reduced iron and sulfide concentrations (E to H), and bacterial abundance and carbon production (I to L) on each sampling date. The dashed horizontal lines indicate the upper depth of the hypoxic layer (<3 mg liter−1 oxygen). The solid horizontal lines indicate the upper depth of the anoxic layer (oxygen concentration below the detection limit, 0.5 mg liter−1). An absence of data indicates that measurements were not obtained.
Bacterial cell abundance ranged from 2.6 × 106 to 22.6 × 106 cells ml−1 and varied with sampling date and depth (Fig. 1). In oxic waters the bacterial cell abundance was fairly constant throughout the summer (average, 9.0 × 106 ± 3.3 × 106 cells ml−1), but in anoxic waters it changed dramatically, varying from about the same as that in oxic waters on 10 June, to much higher than that in oxic waters on 1 July, to much lower than that in oxic waters on 4 August. This pattern matches those found previously for anoxic waters of Chesapeake Bay (29, 62).
Bacterial production on 1 July was similar in oxic and anoxic waters and showed a small peak just below the hypoxic/anoxic interface (Fig. 1). On 4 August, bacterial production was reduced in anoxic waters.
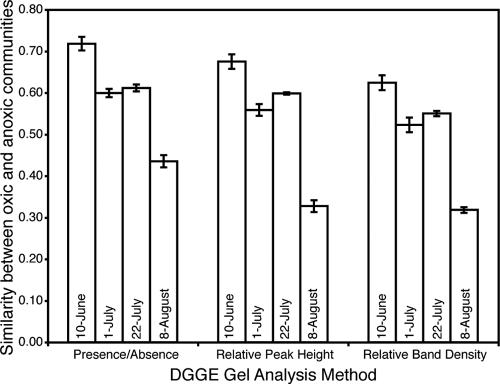
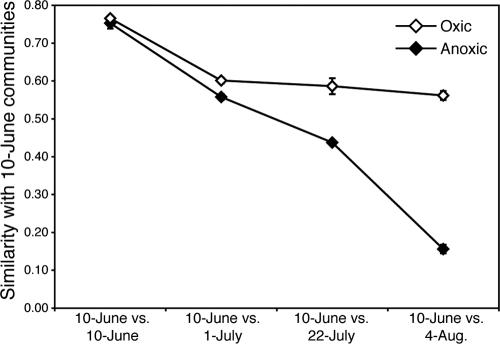
The compositions of bacterioplankton communities in oxic and anoxic waters were similar at the beginning of this study, but as the season progressed, communities in anoxic bottom waters shifted away from those in oxic surface waters (Fig. 2). The results were the same regardless of the method used to analyze DGGE banding patterns. The bacterioplankton communities changed over time in both oxic and anoxic waters, but this change was much more pronounced in anoxic waters (Fig. 3). Note that MDS diagrams of DGGE banding patterns were similar for all analysis methods (presence/absence, peak height, band density), so only one diagram is shown. The bacterial communities in oxic waters showed moderate changes during the summer, as indicated by the relatively high DGGE similarity values between communities on 10 June and 4 August (Fig. 4). In contrast, the bacterial communities in anoxic waters changed dramatically during the 8 weeks of this study (Fig. 4).
FIG. 2.
Average DGGE similarity values (± standard errors) for bacterioplankton communities in oxic and anoxic samples for each sampling date, calculated for pairwise data based on band presence/absence (Dice equation), relative peak height (Bray-Curtis equation), and relative band density (Bray-Curtis equation).
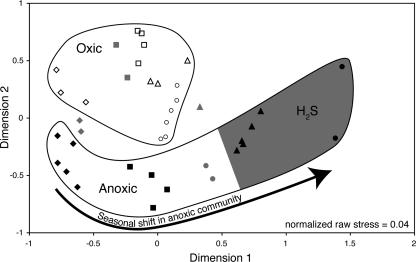
FIG. 3.
Multidimensional scaling ordination of pairwise estimates of similarity (Bray-Curtis equation) of bacterioplankton communities calculated from DGGE banding patterns based on relative band density for DNA samples collected on 10 June (diamonds), 1 July (squares), 22 July (triangles), and 4 August (circles) of 2004. Symbol shading indicates oxic (open), hypoxic (gray), and anoxic (black) samples. Oxic and anoxic samples are circled. The gray region indicates samples containing sulfides.
FIG. 4.
Average DGGE similarity values (± standard errors) for comparisons between bacterioplankton communities on 10 June and bacterioplankton communities on the other sampling dates for oxic and anoxic waters based on relative band density (Bray-Curtis equation).
Despite these temporal shifts, ANOSIM analyses showed that the communities in anoxic waters were more similar to each other than to those in oxic waters on all sampling dates (Table 1). Hypoxic water communities also grouped separately from oxic water communities but could not be distinguished from anoxic water communities. Communities in anoxic waters that contained measurable concentrations of sulfide grouped together on MDS plots (Fig. 3). This grouping was confirmed by ANOSIM (Table 1). Canonical correspondence analysis demonstrated that 56% of the variability in community composition (Σλ = 1.32, Σλcanonical = 0.74) was described by a combination of variables (temperature, depth, dissolved oxygen, NO3−, NO2−, NH4+, sulfides).
TABLE 1.
ANOSIM statistics for comparisons of communities using DGGE similarity values derived from band presence/absence data, relative peak height, and relative band densities
| Comparison | Presence/absence
|
Relative peak ht
|
Relative band density
|
|||
|---|---|---|---|---|---|---|
| R-statistic | P value | R-statistic | P value | R-statistic | P value | |
| Anoxic vs oxic | 0.41 | 0.001 | 0.38 | 0.001 | 0.41 | 0.001 |
| Hypoxic vs oxic | 0.26 | 0.012 | 0.22 | 0.023 | 0.26 | 0.01 |
| Anoxic vs hypoxic | −0.05 | NSa | −0.01 | NS | −0.04 | NS |
| Sulfides vs no sulfides | 0.74 | 0.001 | 0.58 | 0.001 | 0.63 | 0.001 |
NS, not significant.
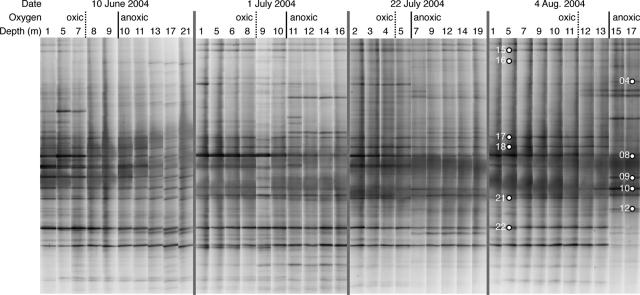
Most DGGE bands identified with DNA sequencing matched longer DNA sequences from clone libraries, which were used for phylogenetic identification of these bands (Table 2). DGGE bands in oxic surface waters contained DNA from typical marine populations of Gammaproteobacteria in the SAR86 cluster and Alphaproteobacteria in the SAR11 cluster (Fig. 5). One very strong band contained DNA from a SAR11 relative (band 21) that clustered with several other clones from estuaries and coastal waters, forming a putative estuarine-specific clade of the cultivated SAR11 organism Pelagibacter ubique (Table 2), here named after clone OM155 (accession no. U70686).
TABLE 2.
Information on DGGE band sequences, including oxygen conditions under which bands were strongest, phylogenetic grouping, and information on top BLAST matches for PCR clone library sequences matching DGGE bands
| Band no. | Designation | Depth | Phylum, genus, and/or cluster | Clone library match | Difference from DGGE band | BLAST match accession no. | DNA sequence similarity (%) | Source |
|---|---|---|---|---|---|---|---|---|
| 01 | CBM01Z01 | Anoxic | Firmicutes | CBM01B08 | Exact | AB013835 | 91 | Marine sediments |
| 04 | CBM01Z04 | Anoxic | Bacteroidetes | CBM01B03 | Exact | AB100009 | 91 | Inactive hydrothermal vent chimney |
| 08 | CBM01Z08 | Oxic | Synechococcus | CBM01G09 | Exact | BX569694 | 98 | Marine waters |
| 09 | CBM01Z09 | Anoxic | SAR406 | CBM01D05 | Exact | DQ009452 | 85 | Marine waters |
| 10 | CBM01Z10 | Anoxic | Gammaproteobacteria | CBM01A03 | Exact | AF126545 | 89 | Soda lakes |
| 12 | CBM01Z12 | Anoxic | Gammaproteobacteria | CBM01B07 | Exact | AF126545 | 89 | Soda lakes |
| 13 | CBM02Z13 | Oxic | Gammaproteobacteria | CBM02E10 | 1 bp | DQ300651 | 95 | Marine waters |
| 15a | CBM02Z15a | Oxic/anoxic | Bacteroidetes | CBM02C10 | 1 bp | AJ697707 | 86 | Mesotrophic lake |
| 15b | CBM02Z15b | Oxic/anoxic | Gammaproteobacteria, SAR86 | CBM02A07 | 2 bp | DQ071142 | 99 | Marine waters |
| 16 | CBM02Z16 | Oxic | Gammaproteobacteria, SAR86 | CBM02A07 | 2 bp | DQ071142 | 99 | Marine waters |
| 17 | CBM02Z17 | Oxic | Alphaproteobacteria, SAR11 | No match | AF245620a | 98 | Marine water | |
| 18 | CBM02Z18 | Oxic | Alphaproteobacteria, SAR11 | No match | AF245620a | 98 | Marine water | |
| 21 | CBM02Z21 | Oxic | Alphaproteobacteria, OM155b | CBM02B03 | 2 bp | AF406547 | 92 | Marine waters |
| 22 | CBM02Z22 | Oxic | Alphaproteobacteria, SAR11 | CBM02B02 | Exact | AB193892 | 97 | Hydrothermal vent water |
BLAST match for DGGE sequence.
“Estuarine” SAR11.
FIG. 5.
DGGE banding patterns for each depth on each sampling date. The oxygen conditions at each depth are indicated. The dashed vertical lines indicate the upper depth of the hypoxic layer (<3 mg liter−1 oxygen). The solid black vertical lines indicate the upper depth of the anoxic layer (oxygen concentration below the detection limit, 0.5 mg liter−1). DGGE bands analyzed with DNA sequencing are indicated. Bands 1 and 13 were off the image for the 17- and 5-m samples collected on 4 August, respectively.
DGGE bands from anoxic waters contained DNA sequences most similar to those of uncultivated Gammaproteobacteria, uncultivated Bacteroidetes from marine sediments, cultivated members of the Firmicutes, cultivated Synechococcus sp., and the uncultivated SAR406 cluster of marine bacterioplankton. Two bands (bands 10 and 12) contained DNA that exactly matched DNA of a large number of anoxic water clones that were most similar (∼89% sequence similarity) to the genus Thioalcalovibrio of the Gammaproteobacteria, which includes several obligately autotrophic sulfur-oxidizing and denitrifying organisms. Band 9 contained DNA most similar to DNA of uncultivated marine organisms in the SAR406 cluster, which is distantly related to Chlorobium and Fibrobacter (27). Band 8 exactly matched clones from both oxic and anoxic waters that were similar to Synechococcus sp. strain WH8102 (accession no. BX569694).
Clone libraries from oxic surface and anoxic bottom waters revealed diverse bacterial communities (Table 3). Oxic waters were dominated by typical coastal marine bacteria in the Alphaproteobacteria, Cyanobacteria, and Planctomycetes, whereas anoxic waters were dominated by Gammaproteobacteria, Bacteroidetes, Verrucomicrobia, and Deltaproteobacteria. Anoxic waters contained many of the same classes of organisms found in surface waters but also contained organisms in the Chloroflexi and candidate divisions OD1, OP11, and WS1. Chao-1 estimators of taxonomic diversity calculated from clone library sequence data indicated that the diversity in anoxic waters (124 taxa at 97% sequence similarity) was much higher than that in oxic waters (87 taxa at 97% sequence similarity).
TABLE 3.
Classification of bacterial taxa identified in environmental clone libraries
| Phylum or subphylym | No. of clones
|
|
|---|---|---|
| Oxic | Anoxic | |
| Actinobacteria | 3 | 4 |
| Alphaproteobacteria | 17 | 7 |
| Gammaproteobacteria | 8 | 27 |
| Deltaproteobacteria | 5 | 7 |
| Bacteroidetes | 8 | 10 |
| Planctomycetes | 12 | 5 |
| Cyanobacteria | 16 | 5 |
| Verrucomicrobia | 3 | 8 |
| Chloroflexi | 0 | 3 |
| Candidate division OD1 | 0 | 1 |
| Candidate division OP11 | 0 | 1 |
| Candidate division WS3 | 0 | 1 |
| Plastid | 14 | 1 |
| Unknown | 1 | 2 |
Several bacterial populations were identified in both clone libraries, including three members of the Bacteroidetes (two of which match DGGE bands 04 and 15a), two members of the Planctomycetes, two members of the Cyanobacteria (including one match to DGGE band 08), one member of the Gammaproteobacteria, and three members of the Alphaproteobacteria (including matches to DGGE bands 21 and 22). However, statistical comparison of homologous and heterologous coverage curves for these libraries using LIBSHUFF suggested that these two bacterioplankton communities are significantly different (P < 0.001).
DISCUSSION
The plasticity of bacterioplankton community composition is well documented across environmental gradients and over time in a broad array of environments. In fact, the simple act of capturing bacterioplankton in a bottle can drive dramatic changes in community composition (45). Moreover, many phylogenetically distinct bacteria appear to be capable of filling the same ecological niche (i.e., functional redundancy), suggesting that components of natural communities may be replaced at random without altering their ecological roles (18). Thus, when environmental controls on bacterioplankton community composition are investigated, it is often more informative when a bacterial community does not change in response to changing environmental conditions. Recent work demonstrated striking consistency in bacterioplankton community composition across nearly 250 km and a 20-fold change in salinity in Chesapeake Bay surface waters (36). Also, bacterioplankton communities in several systems have been shown to reassemble year after year (15, 24, 36), suggesting that functional redundancy may be limited. These patterns indicate that aquatic environments host native communities composed of persistent and well-adapted bacterial populations.
Respiratory and phylogenetic succession.
Bacterioplankton communities in anoxic bottom waters of the Chesapeake Bay were very similar to those in oxic surface waters on 10 June, suggesting that aerobic bacteria in Chesapeake Bay continue to function under anoxic conditions. However, the community in anoxic bottom water became increasingly different than the surface community over the following 8.5 weeks. During this time environmental conditions in the bottom waters changed, reflecting a gradual succession in respiratory processes in response to depletion of electron acceptors, such as oxygen and nitrate. On 10 June, elevated nitrate concentrations and detectable nitrite suggested that bacteria were using nitrate respiration, an ability common in aerobic bacteria. On 1 July nitrate was depleted except in hypoxic waters at the pycnocline. On this date the sulfide concentration was still very low, so bacteria were probably using electron acceptors other than sulfate. Midwater peaks in reduced iron on 1 July and 22 July suggest that there was iron respiration by bacteria in the water column, but they could also indicate that there was deep water precipitation of iron by sediment-produced hydrogen sulfide (14). On 22 July and 4 August the sulfide concentration was elevated, indicating that there was persistent sulfate respiration in the sediments and possibly in the water column.
In contrast to systems such as the Black Sea and the Cariaco Basin, which experience long-term anoxia, the biogeochemical properties of Chesapeake Bay subpycnocline waters change rapidly. The disappearance of oxidants (e.g., O2) and the appearance of reduced species, such as Mn(II), Fe(II), and H2S (25), indicate that there is a succession in terminal electron acceptors for microbial respiration. In the Chesapeake Bay, the mass of bottom water moves northward from the southern bay, where it originates as surface water. Along this traverse, oxygen is depleted and reduced products, such as N2 and NH4+, accumulate (37). Although bottom water transect data are unavailable for Mn(II), Fe(II), and H2S, seasonal changes in the concentrations of these species (Fig. 1) are consistent with their sequential importance as electron acceptors. Thus, much like the typical vertical sequence of electron acceptors found in aquatic sediments (8, 22), the bottom waters of the Chesapeake Bay may be viewed as having a horizontal sequence of redox processes, with modest vertical exchange across the pycnocline (39). The temporal succession that we observed at our sampling station likely resulted from sampling this horizontal sequence at different points during anoxic zone expansion, but it may have also reflected seasonal changes in the supply of oxidants from southern bay source waters and changes in the flux of reduced species from sediments.
In relatively shallow coastal ecosystems like the Chesapeake Bay, sediment microbial processes can greatly influence the chemistry of overlying waters. Sediments contain the largest pools of solid-phase oxidants (Mn and Fe oxides), as well as high concentrations of labile organic matter, and it seems likely that water column accumulation of Fe (Fig. 1) and Mn (20) reflects mostly sedimentary microbiological and chemical processes. Also, measured fluxes of sulfide from sediment to bottom water (54) and a comparison of sulfate reduction rates in sediments and overlying water (62) indicate that most hydrogen sulfide in the water column is produced by sediment sulfate respiration. However, Kemp et al. (39) suggested that aerobic respiration in subpycnocline water of this region generally exceeds sediment aerobic respiration. Moreover, we documented high bacterial production in anoxic waters under conditions that require some form of anaerobic respiration. It is likely that both water column and sediment processes shape spatial and temporal gradients in redox conditions that feed back to control the forms of respiration used by bacterioplankton and, ultimately, control the phylogenetic composition of bacterioplankton communities.
The time available for the bacterial community composition to change in estuarine anoxic zones depends on the amount of time that the water is anoxic. Unlike meromictic lakes and permanent anoxic marine basins like the Black Sea, estuarine anoxic zones contain water that is transitory and is constantly replaced as estuarine circulation moves bottom water up-estuary, as described above. In Chesapeake Bay, the tidally averaged residual flow of bottom water varies seasonally and with wind speed and direction, but a reasonable summer average is 2 to 5 cm s−1 (43). Our sampling site is approximately 76 km from a shoal at the mouth of the Potomac River, which was the down-estuary limit of anoxia during our sampling period (Chesapeake Bay Program [www.chesapeakebay.net]). Thus, after becoming oxygen depleted, bottom waters were anoxic for 18 to 44 days before they reached our sampling site.
Bacterial community doubling times were much shorter than this anoxic period, ranging from 0.36 day in surface waters in July to 0.75 day in anoxic waters in August. Therefore, adequate time was available for the community composition to shift in response to oxygen depletion. However, DGGE results indicate that a dramatic shift in bacterioplankton community composition did not occur until 4 August, when sulfide concentrations were very high. In fact, prior to 4 August more than one-half of the DGGE bands in surface waters were also found in deep anoxic waters (Fig. 5), suggesting that the bacterial populations continued to grow after oxygen was depleted.
Bacterioplankton diversity.
Most of what we know about bacterioplankton diversity in anoxic waters comes from studies of semipermanently anoxic marine basins and meromictic lakes (30, 53). One large-scale study of bacterioplankton across the oxycline of the Black Sea demonstrated that there were very different oxic and anoxic communities (63). Another study in anoxic waters of the Cariaco Basin off the coast of Venezuela described a bacterioplankton community that resembled deep-sea sediment communities and included members of the Fibrobacteres, Deltaproteobacteria, and Epsilonproteobacteria (44). Meromictic lakes with anoxic bottom waters have essentially the same pattern of diversity (5, 31), although in some systems there is a some overlap between oxic and anoxic bacterioplankton (41).
Unlike most systems described above, seasonally anoxic zones in estuaries, coastal waters, and monomictic lakes are dynamic and are periodically refreshed with oxygen and other electron acceptors. Bacterioplankton communities in these systems are probably in a constant state of succession, shifting respiratory processes and phylogenetic composition as chemical conditions change over time. In one study, Hollibaugh et al. (31) observed gradual reestablishment of distinct oxic and anoxic communities over the several months following a rare water column mixing event in meromictic Mono Lake in California. This likely reflects events that occur during the establishment of seasonal anoxic zones in estuaries.
Most bacterioplankton populations in oxic surface waters of Chesapeake Bay were also found in anoxic bottom waters during part of the summer. Several of these apparently anoxia-tolerant populations were very similar to common marine and estuarine bacterioplankton in the SAR11 and SAR86 clusters. These organisms may be capable of shifting from aerobic to anaerobic respiration using nitrate, iron, or some other electron acceptor. Also, one Synechococcus sp. identified in a DGGE band was found in both oxic and anoxic waters, and several Synechococcus spp. were found in both oxic and anoxic clone libraries. In culture, some Synechococcus sp. strains grow under anoxic conditions (33), and others seem to be capable of heterotrophic growth (49, 55). One study found elevated abundance and rapid growth of Synechococcus-like bacteria in anoxic waters below a depth of 120 m in the Baltic Sea (19). It is possible that Synechococcus shifts metabolism and grows heterotrophically in dark, anoxic waters of the Chesapeake Bay.
Community succession in anoxic waters produced a mixture of surface water populations and new populations that developed after the onset of anoxia. This caused an increase in the bacterial taxonomic diversity above that in oxic surface waters. New populations that developed in anoxic waters were not similar to any cultivated or uncultivated organisms, including those identified in molecular surveys of anoxic waters in the Black Sea (63), Cariaco Basin (44), Baltic Sea (GenBank accession no. DQ385015 to DQ385056), and meromictic lakes (Lake Tanganika [accession no. DQ463691 to DQ463742] and Lake Pavin [accession no. DQ642315 to DQ642430]). However, these new populations were most similar to marine sediment organisms (bands 1 and 4), chemoautotrophic Gammaproteobacteria (bands 10 and 12), and the SAR406 cluster (band 9), which is abundant in the oxygen minimum zone of the Arabian Sea (23). Clones from anoxic waters also included sequences similar to those of organisms found in low-oxygen environments, such as members of the Deltaproteobacteria, members of the Gammaproteobacteria related to sulfur-oxidizing endosymbionts, and members of candidate divisions OP11, OD1, and WS3.
Bacterial production and respiration.
One striking result of this study is the high rate of bacterial production in anoxic waters. Heterotrophic bacterial production is often calculated from the rate of incorporation of radioactive tracers (leucine and thymidine) into newly produced macromolecules (protein and DNA). Thymidine works well with oxic water but gives reduced estimates of production with anoxic water (46, 61) because many anaerobes do not take up and incorporate thymidine into DNA. Leucine works well for both aerobes and anaerobes (2) and has been used in several studies of bacterial production in anoxic waters (12, 46).
The bacterial production in anoxic waters on 1 July was similar to the production in surface waters and showed a peak just below the oxycline in anoxic water, a pattern typical of other oxygen-stratified systems (2, 12, 46, 58). But later in the season on 4 August bacterial production was reduced in anoxic bottom water (Fig. 1). These results contrast with earlier studies of the Chesapeake Bay using the thymidine method which, predictably, found very low rates of production in anoxic waters (35, 62).
Acknowledgments
We thank V. Kelly, D. Kimmel, and M. S. Owens for assistance in the field, L. A. Codispoti for the impeller-driven submersible pump, and E. Kiss for assistance in the laboratory.
This work was supported by the Maryland Sea Grant's NSF-funded REU program, by UMCES Horn Point Laboratory, and by the National Science Foundation (grant OCE-9981617).
Footnotes
Published ahead of print on 31 August 2007.
REFERENCES
- 1.Barbosa, A. B., H. M. Galvao, P. A. Mendes, X. A. Alvarez-Salgado, F. G. Figueiras, and I. Joint. 2001. Short-term variability of heterotrophic bacterioplankton during upwelling off the NW Iberian margin. Prog. Oceanogr. 51:339-359. [Google Scholar]
- 2.Bastviken, D., and L. Tranvik. 2001. The leucine incorporation method estimates bacterial growth equally well in both oxic and anoxic lake waters. Appl. Environ. Microbiol. 67:2916-2921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boicourt, W. C. 1992. Influences of circulation processes on dissolved oxygen in the Chesapeake Bay, p. 7-59. In D. E. Smith, M. Leffler, and G. Mackiernan (ed.), Oxygen dynamics in the Chesapeake Bay. Maryland Sea Grant, College Park.
- 4.Borsuk, M. E., C. A. Stow, R. A. Luettich, H. W. Paerl, and J. L. Pinckney. 2001. Modelling oxygen dynamics in an intermittently stratified estuary: estimation of process rates using field data. Estuar. Coast. Shelf Sci. 52:33-49. [Google Scholar]
- 5.Bosshard, P. P., R. Stettler, and R. Bachofen. 2000. Seasonal and spatial community dynamics in the meromictic Lake Cadagno. Arch. Microbiol. 174:168-174. [DOI] [PubMed] [Google Scholar]
- 6.Boynton, W. R., and W. M. Kemp. 2000. Influence of river flow and nutrient loads on selected ecosystem processes, p. 269-298. In J. E. Hobbie (ed.), Estuarine science, a synthetic approach to research and practice. Island Press, Washington, DC.
- 7.Bricker, S. B., C. G. Clement, D. E. Pirhalla, S. P. Orlando, and D. R. G. Farrow. 1999. National estuarine eutrophication assessment: effects of nutrient enrichment in the nation's estuaries. NOAA National Ocean Service Special Projects Office and the National Centers for Coastal Ocean Science, Silver Spring, MD.
- 8.Burdige, D. J. 2006. Geochemistry of marine sediments. Princeton University Press, Princeton, NJ.
- 9.Chao, A. 1984. Nonparametric-estimation of the number of classes in a population. Scand. J. Stat. 11:265-270. [Google Scholar]
- 10.Clarke, K. R., and R. H. Green. 1988. Statistical design and analysis for a biological effects study. Mar. Ecol. Prog. Ser. 46:213-226. [Google Scholar]
- 11.Cline, J. D. 1969. Spectrophotometric determination of hydrogen sulfide in natural waters. Limnol. Oceanogr. 14:454-458. [Google Scholar]
- 12.Cole, J. J., and M. L. Pace. 1995. Bacterial secondary production in oxic and anoxic fresh-waters. Limnol. Oceanogr. 40:1019-1027. [Google Scholar]
- 13.Cole, J. R., B. Chai, R. J. Farris, Q. Wang, S. A. Kulam, D. M. McGarrell, G. M. Garrity, and J. M. Tiedje. 2005. The Ribosomal Database Project (RDP-II): sequences and tools for high-throughput rRNA analysis. Nucleic Acids Res. 33:D294-D296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cornwell, J. C., and P. A. Sampou. 1995. Environmental controls on iron sulfide mineral formation in a coastal plain estuary, p. 224-242. In Geochemical transformations of sedimentary sulfur, vol. 612. American Chemical Society, Washington, DC. [Google Scholar]
- 15.Crump, B. C., and J. E. Hobbie. 2005. Synchrony and seasonality of bacterioplankton communities in two temperate rivers. Limnol. Oceanogr. 50:1718-1729. [Google Scholar]
- 16.Crump, B. C., C. S. Hopkinson, M. L. Sogin, and J. E. Hobbie. 2004. Microbial biogeography along an estuarine salinity gradient: combined influences of bacterial growth and residence time. Appl. Environ. Microbiol. 70:1494-1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Crump, B. C., G. W. Kling, M. Bahr, and J. E. Hobbie. 2003. Bacterioplankton community shifts in an arctic lake correlate with seasonal changes in organic matter source. Appl. Environ. Microbiol. 69:2253-2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Curtis, T. P., and W. T. Sloan. 2004. Prokaryotic diversity and its limits: microbial community structure in nature and implications for microbial ecology. Curr. Opin. Microbiol. 7:221-226. [DOI] [PubMed] [Google Scholar]
- 19.Detmer, A. E., H. C. Giesenhagen, V. M. Trenkel, H. A. D. Venne, and F. J. Jochem. 1993. Phototrophic and heterotrophic pico-plankton and nanoplankton in anoxic depths of the central baltic sea. Mar. Ecol. Prog. Ser. 99:197-203. [Google Scholar]
- 20.Eaton, A. 1979. Impact of anoxia on Mn fluxes in the Chesapeake Bay. Geochim. Cosmochim. Acta 43:429-432. [Google Scholar]
- 21.Friederich, G. E., and L. A. Codispoti. 1987. An analysis of continuous vertical nutrient profiles taken during a cold-anomaly off Peru. Deep-Sea Res. Part A 34:1049-1065. [Google Scholar]
- 22.Froelich, P. N., G. P. Klinkhammer, M. L. Bender, N. A. Luedtke, G. R. Heath, C. Cullen, P. Dauphin, and D. Hammond. 1979. Early oxidation of organic matter in pelagic sediments of the eastern equatorial Atlantic: suboxic diagenesis. Geochim. Cosmochim. Acta 43:1075-1090. [Google Scholar]
- 23.Fuchs, B. M., D. Woebken, M. V. Zubkov, P. Burkill, and R. Amann. 2005. Molecular identification of picoplankton populations in contrasting waters of the Arabian Sea. Aquat. Microb. Ecol. 39:145-157. [Google Scholar]
- 24.Fuhrman, J. A., I. Hewson, M. S. Schwalbach, J. A. Steele, M. V. Brown, and S. Naeem. 2006. Annually reoccurring bacterial communities are predictable from ocean conditions. Proc. Natl. Acad. Sci. USA 103:13104-13109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gavis, J., and V. Grant. 1986. Sulfide, iron, manganese, and phosphate in the deep water of the Chesapeake Bay during anoxia. Estuar. Coast. Shelf Sci. 23:451-463. [Google Scholar]
- 26.Good, I. J. 1953. The population frequencies of species and the estimation of population parameters. Biometrika 40:237-264. [Google Scholar]
- 27.Gordon, D. A., and S. J. Giovannoni. 1996. Detection of stratified microbial populations related to Chlorobium and Fibrobacter species in the Atlantic and Pacific Oceans. Appl. Environ. Microbiol. 62:1171-1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hagy, J. D., W. R. Boynton, C. W. Keefe, and K. V. Wood. 2004. Hypoxia in Chesapeake Bay, 1950-2001: long-term change in relation to nutrient loading and river flow. Estuaries 27:634-658. [Google Scholar]
- 29.Hamdan, L. J., and R. B. Jonas. 2006. Seasonal and interannual dynamics of free-living bacterioplankton and microbially labile organic carbon along the salinity gradient of the Potomac River. Estuar. Coasts 29:40-53. [Google Scholar]
- 30.Hofle, M. G., and I. Brettar. 1995. Taxonomic diversity and metabolic activity of microbial communities in the water column of the central Baltic Sea. Limnol. Oceanogr. 40:868-874. [Google Scholar]
- 31.Hollibaugh, J. T., P. S. Wong, N. Bano, S. K. Pak, E. M. Prager, and C. Orrego. 2001. Stratification of microbial assemblages in Mono Lake, California, and response to a mixing event. Hydrobiologia 466:45-60. [Google Scholar]
- 32.Huber, T., G. Faulkner, and P. Hugenholtz. 2004. Bellerophon: a program to detect chimeric sequences in multiple sequence alignments. Bioinformatics 20:2317-2319. [DOI] [PubMed] [Google Scholar]
- 33.Ikemoto, H., and A. Mitsui. 1994. Diazotrophic synchronous growth of a marine unicellular cyanobacterium, Synechococcus sp. strain Miami-Bg-043511, under aerobic and microaerobic/anaerobic conditions. Microbiology 140:2153-2158. [Google Scholar]
- 34.Jonas, R. B. 1997. Bacteria, dissolved organics and oxygen consumption in salinity stratified Chesapeake Bay, an anoxia paradigm. Am. Zool. 37:612-620. [Google Scholar]
- 35.Jonas, R. B., and J. H. Tuttle. 1990. Bacterioplankton and organic-carbon dynamics in the lower mesohaline Chesapeake Bay. Appl. Environ. Microbiol. 56:747-757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kan, J. J., B. C. Crump, K. Wang, and F. Chen. 2006. Bacterioplankton community in Chesapeake Bay: predictable or random assemblages. Limnol. Oceanogr. 51:2157-2169. [Google Scholar]
- 37.Kana, T. M., J. C. Cornwell, and L. J. Zhong. 2006. Determination of denitrification in the Chesapeake Bay from measurements of N2 accumulation in bottom water. Estuar. Coasts 29:222-231. [Google Scholar]
- 38.Kana, T. M., C. Darkangelo, M. D. Hunt, J. B. Oldham, G. E. Bennett, and J. C. Cornwell. 1994. Membrane inlet mass-spectrometer for rapid high-precision determination of N2, O2, and Ar in environmental water samples. Anal. Chem. 66:4166-4170. [Google Scholar]
- 39.Kemp, W. M., P. A. Sampou, J. Garber, J. Tuttle, and W. R. Boynton. 1992. Seasonal depletion of oxygen from bottom waters of Chesapeake Bay—roles of benthic and planktonic respiration and physical exchange processes. Mar. Ecol. Prog. Ser. 85:137-152. [Google Scholar]
- 40.Kirchman, D. L. 1993. Leucine incorporation as a measure of biomass production by heterotrophic bacteria, p. 509-512. In P. F. Kemp, B. F. Sherr, E. B. Sherr, and J. J. Cole (ed.), Handbook of methods in aquatic microbial ecology. Lewis Publishers, Boca Raton, FL.
- 41.Konopka, A., T. Bercot, and C. Nakatsu. 1999. Bacterioplankton community diversity in a series of thermally stratified lakes. Microb. Ecol. 38:126-135. [DOI] [PubMed] [Google Scholar]
- 42.Kuo, A. Y., K. Park, and M. Z. Moustafa. 1991. Spatial and temporal variabilities of hypoxia in the Rappahannock River, Virginia. Estuaries 14:113-121. [Google Scholar]
- 43.Li, M., L. J. Zhong, and W. C. Boicourt. 2005. Simulations of Chesapeake Bay estuary: sensitivity to turbulence mixing parameterizations and comparison with observations. J. Geophys. Res. Oceans 110:Article C12004. [Google Scholar]
- 44.Madrid, V. M., G. T. Taylor, M. I. Scranton, and A. Y. Chistoserdov. 2001. Phylogenetic diversity of bacterial and archaeal communities in the anoxic zone of the Cariaco Basin. Appl. Environ. Microbiol. 67:1663-1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Massana, R., C. Pedros-Alio, E. O. Casamayor, and J. M. Gasol. 2001. Changes in marine bacterioplankton phylogenetic composition during incubations designed to measure biogeochemically significant parameters. Limnol. Oceanogr. 46:1181-1188. [Google Scholar]
- 46.McDonough, R. J., R. W. Sanders, K. G. Porter, and D. L. Kirchman. 1986. Depth distribution of bacterial production in a stratified lake with an anoxic hypolimnion. Appl. Environ. Microbiol. 52:992-1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Noble, R. T., and J. A. Fuhrman. 1998. Use of SYBR Green I for rapid epifluorescence counts of marine viruses and bacteria. Aquat. Microb. Ecol. 14:113-118. [Google Scholar]
- 48.Officer, C. B., R. B. Biggs, J. L. Taft, L. E. Cronin, M. A. Tyler, and W. R. Boynton. 1984. Chesapeake Bay anoxia—origin, development, and significance. Science 223:22-27. [DOI] [PubMed] [Google Scholar]
- 49.Paerl, H. W. 1991. Ecophysiological and trophic implications of light-stimulated amino acid utilization in marine picoplankton. Appl. Environ. Microbiol. 57:473-479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Paerl, H. W., J. L. Pinckney, J. M. Fear, and B. L. Peierls. 1998. Ecosystem responses to internal and watershed organic matter loading: consequences for hypoxia in the eutrophying Neuse River estuary, North Carolina, USA. Mar. Ecol. Prog. Ser. 166:17-25. [Google Scholar]
- 51.Parsons, T. R., Y. Maita, and C. M. Lalli. 1984. A manual of chemical and biological methods for seawater analysis. Pergamon Press, Oxford, United Kingdom.
- 52.Pedrós-Alió, C., G.-C. J., and J. I. Calderón. 1993. Bacterial production in anaerobic water columns, p. 519-530. In P. F. Kemp, B. F. Sherr, E. B. Sherr, and J. J. Cole (ed.), Handbook of methods in aquatic microbial ecology. Lewis Publishers, Boca Raton, FL.
- 53.Ramsing, N. B., H. Fossing, T. G. Ferdelman, F. Andersen, and B. Thamdrup. 1996. Distribution of bacterial populations in a stratified fjord (Mariager Fjord, Denmark) quantified by in situ hybridization and related to chemical gradients in the water column. Appl. Environ. Microbiol. 62:1391-1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Roden, E. E., and J. H. Tuttle. 1992. Sulfide release from estuarine sediments underlying anoxic bottom water. Limnol. Oceanogr. 37:725-738. [Google Scholar]
- 55.Shen, G. Z., and D. A. Bryant. 1995. Characterization of a Synechococcus sp. strain PCC-7002 mutant lacking photosystem. 1. Protein assembly and energy-distribution in the absence of the photosystem-I reaction-center core complex. Photosynth. Res. 44:41-53. [DOI] [PubMed] [Google Scholar]
- 56.Simberloff, D. S. 1972. Properties of rarefaction diversity measurement. Am. Nat. 106:414-418. [Google Scholar]
- 57.Singleton, D. R., M. A. Furlong, S. L. Rathbun, and W. B. Whitman. 2001. Quantitative comparisons of 16S rRNA gene sequence libraries from environmental samples. Appl. Environ. Microbiol. 67:4374-4376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Taylor, G. T., M. Iabichella, T. Y. Ho, M. I. Scranton, R. C. Thunell, F. Muller-Karger, and R. Varela. 2001. Chemoautotrophy in the redox transition zone of the Cariaco Basin: a significant midwater source of organic carbon production. Limnol. Oceanogr. 46:148-163. [Google Scholar]
- 59.Ter Braak, C. J. F. 1986. Canonical correspondence analysis—a new eigenvector technique for multivariate direct gradient analysis. Ecology 67:1167-1179. [Google Scholar]
- 60.Thompson, J. R., L. A. Marcelino, and M. F. Polz. 2002. Heteroduplexes in mixed-template amplifications: formation, consequence and elimination by ‘reconditioning PCR.’ Nucleic Acids Res. 30:2083-2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tuominen, L. 1995. Comparison of leucine uptake methods and a thymidine incorporation method for measuring bacterial activity in sediment. J. Microbiol. Methods 24:125-134. [Google Scholar]
- 62.Tuttle, J. H., R. B. Jonas, and T. C. Malone. 1987. Origin, development and significance of Chesapeake Bay anoxia, p. 442-472. In S. K. Majumdar (ed.), Contaminant problems and management of living Chesapeake Bay resources. Pennsylvania Academy of Science, Philadelphia.
- 63.Vetriani, C., H. V. Tran, and L. J. Kerkhof. 2003. Fingerprinting microbial assemblages from the oxic/anoxic chemocline of the Black Sea. Appl. Environ. Microbiol. 69:6481-6488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Viollier, E., P. W. Inglett, K. Hunter, A. N. Roychoudhury, and P. Van Cappellen. 2000. The ferrozine method revisited: Fe(II)/Fe(III) determination in natural waters. Appl. Geochem. 15:785-790. [Google Scholar]