Abstract
Selection of suitable strains for biotechnological purposes is frequently a random process supported by high-throughput methods. Using chitinase production by Hypocrea lixii/Trichoderma harzianum as a model, we tested whether fungal strains with superior enzyme formation may be diagnosed by DNA bar codes. We analyzed sequences of two phylogenetic marker loci, internal transcribed spacer 1 (ITS1) and ITS2 of the rRNA-encoding gene cluster and the large intron of the elongation factor 1-alpha gene, tef1, from 50 isolates of H. lixii/T. harzianum, which were also tested to determine their ability to produce chitinases in solid-state fermentation (SSF). Statistically supported superior chitinase production was obtained for strains carrying one of the observed ITS1 and ITS2 and tef1 alleles corresponding to an allele of T. harzianum type strain CBS 226.95. A tef1-based DNA bar code tool, TrichoCHIT, for rapid identification of these strains was developed. The geographic origin of the strains was irrelevant for chitinase production. The improved chitinase production by strains containing this haplotype was not due to better growth on N-acetyl-β-d-glucosamine or glucosamine. Isoenzyme electrophoresis showed that neither the isoenzyme profile of N-acetyl-β-glucosaminidases or the endochitinases nor the intensity of staining of individual chitinase bands correlated with total chitinase in the culture filtrate. The superior chitinase producers did not exhibit similarly increased cellulase formation. Biolog Phenotype MicroArray analysis identified lack of N-acetyl-β-d-mannosamine utilization as a specific trait of strains with the chitinase-overproducing haplotype. This observation was used to develop a plate screening assay for rapid microbiological identification of the strains. The data illustrate that desired industrial properties may be an attribute of certain populations within a species, and screening procedures should thus include a balanced mixture of all genotypes of a given species.
After cellulose, chitin, a polymer composed of β-(1,4)-linked units of the amino sugar N-acetylglucosamine, is the most abundant organic source of carbon in nature (4). This compound can be industrially renewed by extraction from shellfish waste and can then be processed into many derivatives, which are used for a number of commercial applications, such as medical products (e.g., surgical thread), cosmetics, dietary supplements, agriculture, and water treatment (12, 45, 46). In modern biotechnology, chitin processing is based on microbially secreted chitinases, which account for up to 12% of the total production cost (8). At present, there is no economically profitable technology for chitinase production.
For successful biotechnology design, the choice of the microbial strain is of prime importance. Detection of suitable producer organisms is still based on screening, which, although supported by high-throughput methods (5, 58), is largely random. On the other hand, nearly all wild strains isolated for such surveys are now identified on the basis of certain molecular phylogenetic markers. The quickly growing pool of various core nucleotide gene sequences in public databases and an increasing number of evolutionary studies in microbiology provide clear evidence that the phenotypic (metabolic) diversity in many cases is biased by speciation processes and thus may be correlated with certain phylogenetic markers (36, 37, 52, 57). This means that it should be possible to design diagnostic DNA sequences (DNA bar codes) to identify microorganisms with unique metabolic properties and thus to direct screening and further exploration of selected strains. Interestingly, this idea has not been pursued in biotechnology yet.
Various organisms form chitinolytic enzymes (EC 3.2.1.14), which hydrolyze the β-1,4-glycosidic linkage (21). Several chitinase genes have recently been cloned and characterized from mycoparasitic species of the imperfect soil fungal genus Trichoderma, including Trichoderma harzianum (teleomorph, Hypocrea lixii), Trichoderma virens (teleomorph, Hypocrea virens), Trichoderma asperellum, and Trichoderma atroviride (teleomorph, Hypocrea atroviridis) (7, 14, 22, 24, 31, 47, 54, 56, 59). The most comprehensive biomining of the complete genome of Hypocrea jecorina for chitinase genes and a subsequent survey of detected chitinases of H. atroviridis were performed by Seidl et al. (55). Thus, it has frequently been suggested that members of the genus Hypocrea/Trichoderma are good sources for the production of chitinolytic enzymes (13, 59).
The initial hypothesis of this study was that of all of the mycoparasitic Hypocrea/Trichoderma species studied, the large holomorphic species H. lixii/T. harzianum likely contains superior chitinase-producing strains. We reached this conclusion based on the following ecological and physiological properties. First, under the current environmental conditions this species is obviously an evolutionary “winner” since it dominates the majority of Trichoderma soil communities studied and accounts for up to 20% of the total number of isolates of the genus (I. S. Druzhinina and C. P. Kubicek, unpublished data). Also, T. harzianum strains have been obtained from all forest and agricultural soils studied, as well as from such extreme (for Trichoderma) environments as littoral zones (52) or waste piles in oil production and other heavily polluted areas (V. A. Terekhova, personal communication). This provides the possibility of studying a set of H. lixii/T. harzianum strains from various geographic areas representing different populations. Second, H. lixii/T. harzianum (T. harzianum sensu lato) in a broad taxonomic sense has the highest level of intraspecific genetic diversity in Hypocrea/Trichoderma (37, 38), which, however, is difficult to differentiate even on the basis of detailed multilocus phylogeny (10). Moreover, in an analysis of the growth profiles on 95 carbon sources (Biolog Phenotype MicroArray) performed for about 100 Trichoderma strains from Southeast Asia (35), T. harzianum accounted for 40% of the sample and was the only species with no cumulative species-specific phenotype. The third advantage arises from facts mentioned above and is the large number of phylogenetic marker sequences accumulated for this complex species, together with the availability of reliable methods for molecular identification (15, 18, 32).
We show below (i) that T. harzianum strains having one specific haplotype exhibit statistically supported superior chitinase production compared with strains having the other haplotypes and (ii) that this haplotype can be differentiated by DNA bar codes for at least two phylogenetic markers and/or by a physiological test that is easy to perform.
MATERIALS AND METHODS
T. harzianum strains.
Strains investigated in this study and the GenBank accession numbers of DNA loci sequenced are listed in Table 1. All strains are maintained in the culture collections of the Department of Agricultural Chemical Technology, Technical University of Budapest, Budapest, Hungary, and the Research Area of Gene Technology and Applied Biochemistry, Vienna University of Technology, Vienna, Austria, and can be obtained from either of them upon request. They were grown and maintained on potato dextrose agar slants.
TABLE 1.
Strains used in this study
| Species | Strain | Origina | GenBank accession no.b
|
|
|---|---|---|---|---|
| ITS1 and ITS2 | tef1 | |||
| T. harzianum sensu stricto | CBS 226.95 (ex-type strain) | England | AY605713 | AY605833 |
| TUB F-684 | Russia, Siberia | EF113574 | AY605828 | |
| TUB F-688 | Russia, Siberia | EF113575 | AY605831 | |
| TUB F-690 | Russia, central region | EF113576 | AY605829 | |
| TUB F-691 | Russia, Siberia | EF593402 | EF593401 | |
| TUB F-699 | Russia, Siberia | AF149855 | AY605826 | |
| TUB F-700 | Russia, central region | EF113577 | EF113553 | |
| TUB F-743 | Russia, Siberia | AF149859 | AY605827 | |
| TUB F-750 | Russia, central region | EF113554 | ||
| TUB F-762 | Russia, central region | AF149866 | EF113555 | |
| TUB F-444 | Turkey | EF113588 | EF113566 | |
| TUB F-464 | Turkey | EF113584 | EF121551 | |
| TUB F-477 | Russia, central region | AF149852 | AY605830 | |
| IMI 359823 | England | EF113587 | AY605832 | |
| TUB F-1020 | Hungary, river sediment (mud), Danube River | EF113589 | EF113569 | |
| DAOM 222343 | Canada, mushroom farm | AY605735 | AY605778 | |
| C.P.K. 1426 | Russia, Central Russian National Park | EF113591 | EF116552 | |
| C.P.K. 1818 | Ethiopia | EF116553 | EF116558 | |
| C.P.K. 2111 | Hungary, mushroom farm | EF116557 | EF116562 | |
| C.P.K. 2218 | Russia, Tatarstan | EF116555 | EF116560 | |
| C.P.K. 2220 | Russia, Tatarstan | EF116556 | EF116561 | |
| H. lixii/T. harzianum sensu lato | TUB F-773 | Nepal | AF149865 | AY605850 |
| TUB F-898 | Malaysia | AF486027 | EF113559 | |
| TUB F-453 | Sri Lanka | EF113583 | EF113567 | |
| TUB F-839 | Mexico | AY857231 | AY857283 | |
| TUB F-1005 | Brazil | AY857234 | AY857285 | |
| TUB F-1006 | Brazil | AY857235 | AY857286 | |
| PPRI 3912 | South Africa | EF113573 | EF113552 | |
| PPRI 3909 | South Africa | EF113572 | EF113551 | |
| C.P.K. 838 | Egypt | EF113570 | ||
| C.P.K. 841 | Egypt | EF113590 | EF113571 | |
| TUB F-873 | Cambodia | EF113579 | EF113557 | |
| TUB F-885 | Cambodia | EF113580 | EF113558 | |
| PPRI 3772 | South Africa | EF113585 | AY605845 | |
| TUB F-768 | Nepal | AF149862 | AY605847 | |
| TUB F-776 | Nepal | AF149868 | AY605848 | |
| TUB F-777 | Nepal | EF113578 | EF113556 | |
| TUB F-947 | Nepal | EF593404 | EF593403 | |
| TUB F-732 | Brazil | AY857214 | EF113568 | |
| TUB F-767 | Nepal | EF113586 | AY605835 | |
| TUB F-771 | Nepal | AF149864 | AY605834 | |
| TUB F-900 | Singapore | AF486024 | EF113560 | |
| TUB F-964 | Burma | AF486023 | EF113561 | |
| TUB F-966 | Taiwan | EF113581 | EF113562 | |
| TUB F-968 | Taiwan | AF486025 | EF113563 | |
| TUB F-972 | Burma | EF113582 | EF113564 | |
| TUB F-1000 | Indonesia | AF486029 | EF113565 | |
| TUB F-832 | Mexico | AY857226 | ||
| TUB F-834 | Mexico | AY857228 | ||
| C.P.K. 2163 | Ivory Coast | EF116554 | EF116559 | |
Unless indicated otherwise, strains were isolated from soil.
GenBank accession numbers in bold type were obtained for this paper; underlined GenBank accession numbers are accession numbers for the ITS1 sequence.
Determination of fungal growth rates.
To determine hyphal growth, plates containing the carbon sources indicated below for the experiments (at a concentration of 0.05% [wt/vol] in modified synthetic nutrient agar containing 2% [wt/vol] agar) were each inoculated with a small piece of agar from the growing front of a stock culture, which was placed 2 cm from the edge of the 11-cm petri dish. The increase in colony extension towards the other side was measured twice daily and plotted against time. The linear part of the resulting graph was used to calculate the growth rate (in mm/h).
SSF.
Chitinase formation was studied in solid-state fermentation (SSF) experiments performed in 750-ml Erlenmeyer flasks containing a mixture of wheat bran (4.5 g; food quality; purchased from a local supermarket) and crude chitin (0.5 g; practical grade; from crab shells; Sigma) wetted with 10 or 15 ml of the following salt solutions to obtain 67% (media 1 and 3) and 75% (media 2 and 4) moisture contents, respectively: 0.5% (wt/vol) NH4NO3, 0.5% (wt/vol) KH2PO4, 0.1% (wt/vol) MgSO4·7H2O, 0.1% (wt/vol) NaCl, and 0.1% (vol/vol) trace element solution (pH 5.0) for media 1 and 2 and 0.5% (wt/vol) (NH4)2HPO4, 0.5% (wt/vol) KNO3, 0.1% (wt/vol) MgSO4·7H2O, 0.1% (wt/vol) NaCl, and 0.1% (vol/vol) trace element solution (pH 5.0) for media 3 and 4. The trace element solution consisted of 0.08% MnSO4, 0.17% ZnSO4·7H2O, and 0.25% FeSO4·7H2O. Viable spores from 6-day-old fully sporulated stock cultures were harvested by washing them with sterile water containing Tween 80 (0.1%, wt/vol). Sterile SSF media were inoculated with 100 spores/g (dry weight) of substrate and incubated at 30°C without shaking (still cultures). All experiments were performed in duplicate.
Determination of fungal biomass dry weight.
Fungal biomass dry weight in SSFs was measured by determining the amount of alkali-extractable protein as described previously (34). To this end, the fermented substrate together with the mycelium (the whole SSF sample) was washed with tap and distilled water, dried at room temperature, and then finally ground in a mortar under liquid nitrogen. The frozen powder (0.25 g) was homogenized in 10 ml of 0.1 N NaOH and centrifuged. The supernatant was used for determination of the soluble protein content by the Bradford assay (6).
Enzyme assays.
To determine extracellular chitinase activity, the contents of each Erlenmeyer flask were suspended in 100 ml (final volume) aqueous Tween 80 (0.1%, wt/vol) and incubated for 2 h at room temperature with occasional shaking. After centrifugation (8,000 rpm, 8 min, 20°C), the supernatant was used as a source of enzyme. As described by Fenice et al. (20), chitinase activity was assayed by measuring the release of reducing sugars from colloidal chitin prepared as described by Roberts and Selitrennikoff (50). One unit of chitinase activity was defined as the amount of enzyme that released 1 μmol N-acetyl-d-glucosamine equivalents per min under assay conditions (50°C, pH 5.5).
Cellulase (carboxymethyl cellulase [CMCase]) activity was determined as described by Mandels et al. (41), using a 2% carboxymethyl cellulose (Sigma) solution as the substrate. One unit of CMCase activity was defined as the amount of enzyme that released 1 μmol d-glucose equivalents per min under the assay conditions (50°C, pH 4.8).
The data obtained were analyzed using factorial analysis of variance (ANOVA) implemented in the STATISTICA 6.1 (StatSoft, Inc., Tulsa, OK) software package. Post hoc comparisons were done using Tukey's honestly significant difference test.
Biolog Phenotype MicroArray technique.
Carbon assimilation profiles were evaluated using a Biolog FF MicroPlate (Biolog Inc., Hayward, CA) as described by Druzhinina et al. (16).
Development of the selective medium for T. harzianum sensu stricto.
In order to develop a visual test for screening T. harzianum sensu stricto, petri plates containing a carbon source (1.5% [wt/vol] glucose, 1.5% [wt/vol] N-acetylglucosamine, or 1.5% [wt/vol] N-acetylmannosamine), 0.1% (wt/vol) KH2PO4, 0.3% (wt/vol) (NH4)2SO4, 0.05% (wt/vol) MgSO4·7 H2O, 0.05% (wt/vol) KCl, 0.03% (wt/vol) CaCl2, and 1% (wt/vol) Phytagel in morpholineethanesulfonic acid (MES) buffer (50 mM, pH 6.1) were used. After autoclaving, iodonitrotetrazolium chloride (150 μM) and menadione bisulfate (20 μM) were added to the sterile medium. Plates were inoculated by point inoculation in the center of the plate and incubated at 30°C.
Electrophoretic analyses.
Extracellular proteins were obtained as described above. For electrophoresis, they were centrifuged again (15,000 × g, 15 min, 4°C), and the supernatant was used. Standard protocols (3) were used for sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) and native PAGE. Briefly, protein samples (20 μl) were mixed with 5 μl of SDS-PAGE sample buffer (without 2-mercaptoethanol) and without boiling were subjected to electrophoresis on 12% polyacrylamide gels. In the case of native PAGE, SDS was omitted from the sample buffer, the gels, and the running buffer. After electrophoresis, proteins were stained with colloidal Coomassie blue (42). Enzymatic activities in gels were identified as described by Haran et al. (23); using SDS removal from the gels by washing with a solution containing casein and EDTA, the gels were then equilibrated in sodium acetate (pH 4.8), and the enzymatic activities were subsequently detected by overlaying the gels with a solution containing 300 μg/ml of 4-methylumbelliferyl-N-acetyl-β-d-glucosaminide and with a solution containing 300 μg/ml of 4-methylumbelliferyl-β-d-N,N′,N‴-triacetylchitotriose for detection of N-acetylglucosaminidases and chitinases, respectively, in agarose. The hydrolyzed substrates gave a fluorescent signal which was visualized with a UV transilluminator.
Western blotting was performed as described by Ausubel et al. (3), and CBHI/Cel7A was detected by using monoclonal antibodies (44).
PCR amplification of the ITS1-ITS2 locus and of tef1, sequencing, and allele identification.
Mycelium for DNA extraction was grown on potato dextrose agar covered by sterile cellophane. Genomic DNA was extracted with a plant DNeasy mini kit (QIAGEN GmbH, Hilden, Germany) according to the manufacturer's instructions, using approximately 150 ± 50 mg fresh mycelium. Amplification of the nuclear rRNA gene cluster containing internal transcribed spacer 1 (ITS1), ITS2, and the 5.8S rRNA gene was performed using primer combinations SR6R and LR1 (58, 61) and the protocol of Kullnig-Gradinger et al. (38). A 1.3-kb fragment of the tef1 gene encoding translation elongation factor 1-alpha was amplified using primers EF1728F (9) and TEF1LLErev (5′-AAC TTG CAG GCA ATG TGG-3′). This fragment included the fourth and fifth introns and a significant portion of the last large exon (9, 17). Template DNA (100 μl) was directly prepared from PCR products by purification with a QIAquick PCR purification kit (QIAGEN) and was sequenced with a capillary sequencer (ABI 3730 XL; Applied Biosystems, Foster City, CA).
To identify alleles, DNA sequences were visually aligned using Genedoc 2.6. ITS1, ITS2, and tef1 sequences were used to develop the T. harzianum sensu stricto specific DNA bar code as described by Druzhinina et al. (18). Integration of this bar code into the oligonucleotide bar code program led to release of the TrichoCHIT tool for identification of potential superior chitinase producers among H. lixii/T. harzianum strains (http://www.isth.info/tools/trichochit). The library of oligonucleotide hallmarks is stored in the MySQL database. The TrichoCHIT program was written in PHP scripting language (PHP5) and embedded into HTML (16, 19).
To determine the intraspecific structure, parsimony phylogenetic analyses of tef1 sequences were performed in PAUP* 4.0b10 using a heuristic search, with the starting tree obtained via stepwise addition, with random addition of sequences with 1,000 replicates, and with tree bisection-reconnection as the branch-swapping algorithm. The stability of clades was assessed with 500 bootstrap replications. Bootstrap values less than 60 were not considered. The Bayesian approach to phylogenetic reconstruction (49, 63) was implemented using MrBayes 3.0B4 (25). The interleaved NEXUS file was formatted using PAUP*4.0b10 and was manually formatted for the MrBayes v3.0B4 program. The model of evolution and prior settings for individual loci were used as estimated by Druzhinina and Kopchinskiy (17) for different taxa of Hypocrea/Trichoderma. Metropolis-coupled Markov chain Monte Carlo sampling was performed with four incrementally heated chains that were simultaneously run for 1 × 106 and 3 × 106 generations. Bayesian posterior probabilities (PP) were obtained from the 50% majority rule consensus for trees sampled every 100 generations after removal of the 300 first trees using the “burn-in” command. Based on the protocol of Leache and Reeder (40), PP values less than then 0.95 were not considered significant.
RESULTS
Phylogenetic relationships between strains of H. lixii/T. harzianum sensu lato.
Previous investigations showed that strains of H. lixii/T. harzianum display a complex intraspecific structure and high degree of sequence variability, which nevertheless was not considered to be sufficient to recognize cryptic species (10). Studies by our research group and other research groups (10, 35, 37) showed that there is significant ITS1-ITS2 polymorphism of H. lixii/T. harzianum. Druzhinina et al. (18) reported 24 alleles of this locus and nine sets of diagnostic oligonucleotide hallmarks for a species identification bar code. Therefore, to obtain a representative sample, strains were selected based on their content of different ITS1-ITS2 alleles at appropriate ratios and to obtain a balanced biogeography. To this end, 50 strains were chosen. Fourteen of these strains were obtained from Europe and the Middle East; ten strains were obtained from Southeast Asia; five strains were obtained from Siberia; six strains were obtained from the Himalayas; seven strains were obtained from Africa; three, three, and one strains were obtained from South, Central, and North America, respectively; and one strain was obtained from Sri Lanka (Table 1).
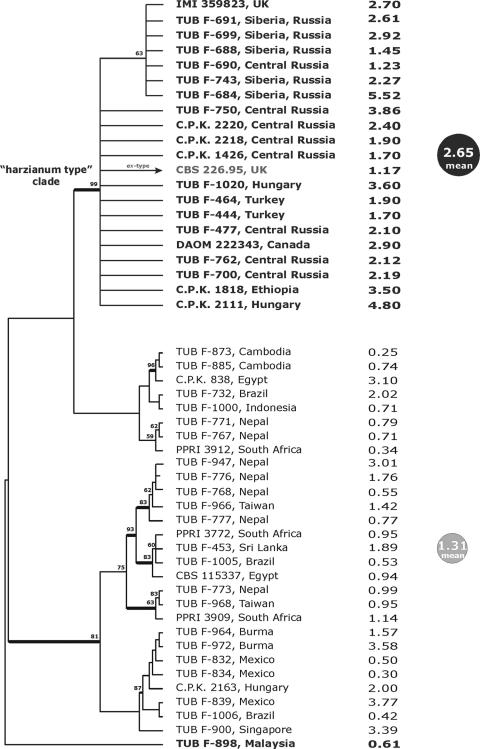
Visual analysis of aligned ITS1-ITS2 sequences revealed the existence of a large set (22) of identical entries, including the ex-type culture of T. harzianum, CBS 226.95, while for the remaining group there were several polymorphic positions in both the ITS1 and ITS2 loci which correspond to “ITS haplotypes” described previously by Kullnig et al. (37). This variability was appropriate for designing a potential bar code, but it was not sufficient for phylogenetic reconstruction. Since development and verification of the bar code require an established phylogeny, another marker locus was needed. The large intron of the tef1 gene coding for the translation elongation 1-alpha protein is usually more variable than ITS1-ITS2. Recently, this intron has been used as a reliable phylogenetic marker for Hypocrea/Trichoderma (26-28, 53). Therefore, we amplified and sequenced the large intron of tef1 for all strains under investigation. Visual analysis of the alignment showed that the strains which have the same ITS1-ITS2 alleles also have identical tef1 sequences. Both parsimony and Bayesian phylogenetic analyses clearly showed the existence of a highly supported homogeneous clade composed of the T. harzianum ex-type strain and 19 other isolates but did not provide clear resolution of the phylogenetic structure for the remaining strains (Fig. 1). Since this so-called “harzianum type” clade was formed by strains from independent isolations from different locations in Russia, Hungary, the United Kingdom, Ethiopia, and Canada, we assumed that it represented a single, cosmopolitan clonal population. The only incongruence between the ITS1-ITS2 alleles and the tef1 clades was noticed for Malaysian strain TUB F-898, which has an allele identical to the ITS1-ITS2 type allele but does not belong to “harzianum type” clade based on tef1.
FIG. 1.
Correlation between the haplotype of T. harzianum sensu stricto and superior production of chitinases. The phylogram on the left corresponds to 1 of 100 saved equally parsimonious trees obtained after 1,000 replicates of a heuristic search applied to the tef1 last large intron locus were examined. Bold branches lead to nodes which were significantly supported in Bayesian analysis (1,000,000 generations; PP > 0.95). The values above branches indicate bootstrap coefficients after 500 replicates. Bold type indicates strains which share the same allele for ITS1-ITS2 sequences with T. harzianum ex-type culture CBS 226.95. The numbers on the right are the levels of extracellular chitinase production after 3 days of SSF (expressed in IU/g [dry weight]) on medium 1.
Chitinase formation by a worldwide collection of H. lixii/T. harzianum strains.
To examine chitinase production by the strains, they were grown on chitin-wheat bran as a carbon source. SSF was used because Trichoderma has been shown to produce only a little chitinase in submerged cultures. Also, SSF may to some extent resemble the lifestyle of a filamentous fungus in nature.
The chitinase activities are shown in Fig. 1. The majority of efficient chitinase producers were found among the strains in the “harzianum type” clade, all of which had the same allele of the tef1 large intron, which was identical to that of T. harzianum ex-type culture CBS 226.95. Statistical analyses confirmed that these strains were significantly more efficient chitinase producers on all media tested [as determined by ANOVA, for medium 1, F(1,47) = 18.23 and P = 0.0001; for medium 2, F(1,47) = 24.055 and P = 0.0000; for medium 3, F(1,47) = 12.695 and P = 0.0009; and for medium 4, F(1,47) = 31.463 and P = 0.0000]. This was most apparent during the linear phase of growth after 3 days of incubation, but it was still evident after 5 days. A representative time course of growth and chitinase formation for two strains belonging to the “harzianum type” clade is shown in Fig. 2. Maximal extracellular chitinase production occurred in 4-, 5-, and 6-day SSFs, and the peak activities were 5.5 and 4.8 IU/g (dry weight) with isolates TUB F-684 and C.P.K. 2111, respectively. The mycelia developing in SSF exhibited maximal growth after 2 to 3 days of incubation at 30°C. A comparison of chitinase production in the four media did not reveal any significant difference [as determined by ANOVA, F(3,192) = 2.0456 and P = 0.109]. Therefore, the data show that improved chitinase production correlates with a unique haplotype of T. harzianum sensu stricto.
FIG. 2.
Time course of chitinase production by two strains belonging to the “harzianum type” clade. The solid line indicates the results for T. harzianum ex-type strain CBS 226.95, and the dashed line indicates the results for strain TUB F-750. Standard deviations for three independent cultivations are indicated by the error bars. DM, dry weight.
Physiological properties of the superior chitinase producers.
Having identified a genetically distinct clade of T. harzianum with increased chitinase production, we were interested in identifying some physiological properties which may explain this trait. One of the reasons for the improved chitinase production by strains in the “harzianum type” clade could be that they have a different rate of assimilation of the chitin monomers N-acetyl-β-d-glucosamine and glucosamine, which (assuming that all the chitinases could be induced by chitin degradation products) could give rise improved intracellular pools of chitinase inducers. To test this hypothesis, we compared the utilization of these two chitin monomers with that of glucose by strains with different ITS and tef1 alleles. As shown in Table 2, there was no allele-specific variation in the rate of growth on any of the carbon sources. However, when individual growth rates on either glucosamine or N-acetyl-β-d-glucosamine were plotted against the chitinase activities shown in Fig. 1, no correlation was obtained (data not shown). We therefore concluded that the superior chitinase-producing strains do not have a different rate of assimilation of chitin monomers.
TABLE 2.
Growth of H. lixii/T. harzianum on glucose, glucosamine, and N-acetylglucosamine
| Taxon | No. of strains | Growth (mm/24 h) on:
|
||
|---|---|---|---|---|
| Glc | GlcN | GlcNAc | ||
| “harzianum type” clade | 11 | 18.1 ± 1.9 | 17.3 ± 1.67 | 18.1 ± 1.56 |
| Other H. lixii/T. harzianum | 18 | 19.6 ± 2.0 | 18.7 ± 2.0 | 18.5 ± 1.96 |
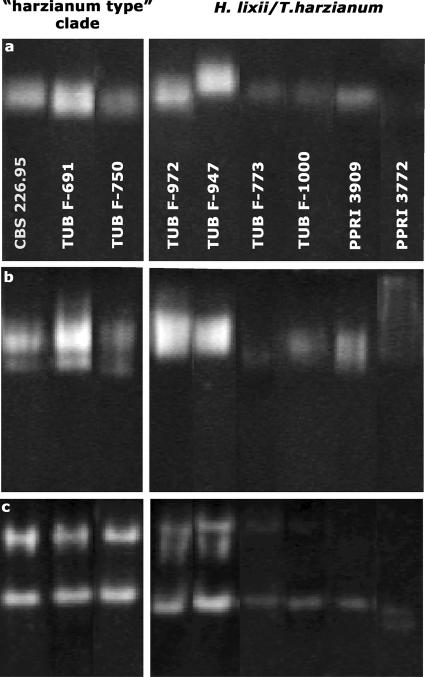
The total chitinase activity of Trichoderma is the result of the activity of several isoenzymes (54). Overproduction of selected components of this chitinase system could therefore also result in increased chitinase activity. In order to find out whether the superiority of the strains bearing the allele of the “harzianum type” clade is due to increased formation of one or more of the chitinases, we performed electrophoresis with three and six representative isolates of the “harzianum type” clade and other H. lixii/T. harzianum strains, respectively, and stained the gels for N-acetylglucosaminidase and endochitinase. Both SDS-PAGE (without 2-mercaptoethanol and without the boiling step but with subsequent renaturation) and nondenaturing PAGE were used. The results are shown in Fig. 3. Native PAGE revealed a diffuse band of activity which migrated only poorly and which sometimes split into two bands representing the two N-acetylglucosaminidases, Nag1 and Nag2 (56). In contrast, SDS-PAGE and renaturation revealed only a single band. This was due to the fact that Nag2 is not renaturable (V. Seidl, unpublished data) and thus only Nag1 was measured. However, the different staining intensities were comparable for the two gels. The intensity of staining of Nag1 and Nag2 did not correlate with the total chitinase activities measured.
FIG. 3.
Chitinase isoenzyme pattern. (a) SDS-PAGE with staining for N-acetyl-β-d-glucosaminidase; (b) PAGE with staining for N-acetyl-β-d-glucosaminidase; (c) PAGE with staining for chitinase.
Most of the endochitinase isoenzymes were not renaturable, and therefore only the results of native PAGE are shown; different strains produced only one or two bands upon zymostaining with 4-methylumbelliferyl-N-acetyl-β-d-glucosaminide. Interestingly, despite the different isoenzyme patterns which were observed, these patterns were similar for the “harzianum type” strains. However, a comparison with Fig. 1 shows that none of the patterns correlated with chitinase activity.
The increased production of chitinases may also be due to an improved secretion capability of strains of the clade identified. If this was the case, there should also be superior production of other extracellular enzymes. We examined the concomitant production of cellulases by selected strains used in this study. A comparison showed that for 10 strains of the “harzianum type” clade there was decreased activity of the cellulases; the CMCase activity of these strains was 4.82 ± 1.92 IU/g (dry weight), compared with 6.99 ± 2.09 IU/g (dry weight) for 18 other H. lixii/T. harzianum strains [as determined by ANOVA, F(1,12) = 5.0580 and P = 0.0441]. Similarly, SDS-PAGE and Western blotting of the major cellulase isoenzyme Cel7A showed that strains of the “harzianum type” clade sometimes had a significantly lower concentration of this enzyme in the culture fluid than strains with other haplotypes (data not shown). We concluded that strains of the “harzianum type” clade are probably not characterized by higher protein secretion.
Mycoparasitic activity of T. harzianum sensu stricto strains.
Since chitinases have been shown to be involved in mycoparasitism and consequently biological control of plant-pathogenic fungi by Trichoderma spp., we reasoned that the isolates of the “harzianum type” clade would also display enhanced mycoparasitic abilities. To test this hypothesis, we used plate confrontation assays with Rhizoctonia solani, Botrytis cinerea, and Sclerotinia sclerotiorum as plant-pathogenic hosts. The data (Table 3) demonstrated that there was quite some variability between individual isolates and also with respect to individual hosts. However, on average, strains producing more chitinase in SSF were neither better nor worse than strains producing less chitinase.
TABLE 3.
Mycoparasitism of H. lixii/T. harzianum strains used in this studya
| Overgrowth | No. of strains with different hosts
|
|||||
|---|---|---|---|---|---|---|
| “harzianum type” clade
|
Other H. lixii/T. harzianum
|
|||||
| R. solani | B. cinerea | S. sclerotiorum | R. solani | B. cinerea | S. sclerotiorum | |
| Strong | 6 | 8 | 6 | 7 | 8 | 8 |
| Moderate | 9 | 6 | 7 | 7 | 4 | 6 |
| None | 6 | 7 | 8 | 5 | 7 | 5 |
The data are the results of plate confrontation assays. Strong overgrowth indicates overgrowth and sporulation within 4 days; moderate overgrowth indicates overgrowth within 7 days and occasional sporulation; no overgrowth indicates that growth stopped before or at contact with the host. Experiments were repeated at least twice, and only consistent data were recorded.
Development of a screening method for T. harzianum sensu stricto strains.
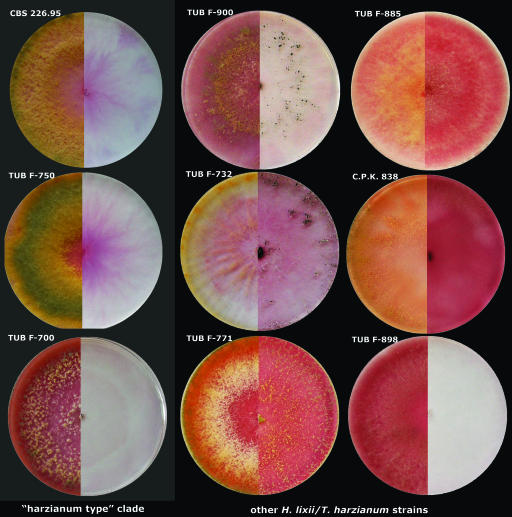
The correlation of two unlinked gene markers with improved chitinase production suggested that there are also phenotypic traits which could be used for rapid screening of strains of the “harzianum type” clade. To identify such a trait, we used Biolog Phenotype MicroArrays to find specific carbon utilization properties which could be used for diagnostic purposes. The results of this analysis (data not shown) revealed only 1 of 95 carbon sources which is utilized qualitatively differently by “harzianum type” strains and all other strains of the species: strains of T. harzianum sensu stricto are unable to grow on N-acetyl-β-d-mannosamine [as determined by ANOVA, F(1,47) = 9.011 and P = 0.0000]. Consequently, this finding was used to establish a plate test for identification of strains of the “harzianum type” clade. This test consists of parallel plating on α-d-glucose and N-acetyl-β-d-mannosamine. We tested 15 strains, including five strains of the “harzianum type” clade. The data showed that a lack of growth on N-acetyl-β-d-mannosamine correlated with the “harzianum type” clade and therefore with superior chitinase production (Fig. 4). The only exception was strain TUB F-898, which has the above-mentioned incongruence between the ITS1-ITS2 allele and the tef1 allele and low levels of extracellular chitinases. Similar to the “harzianum type” clade, this strain did not grow on N-acetyl-β-d-mannosamine. Thus, the proposed selective conditions may be used to prescreen for superior chitinase producers of H. lixii/T. harzianum.
FIG. 4.
Nine selected examples of plate assays for screening T. harzianum strains for superior chitinase producers based on nonutilization of N-acetyl-β-d-mannosamine. Strains in the first column belong to the “harzianum type” clade, while the two other columns contained other strains of the species. The left side of each plate shows the colony morphology on α-d-glucose, while the right side shows the growth on N-acetyl-β-d-mannosamine. The red color was formed due to enzymatic reduction of the iodonitrotetrazolium chloride indicator of microbial metabolic activity (in a reaction catalyzed by succinate dehydrogenase from the citric acid cycle) to the insoluble dye formazan red. Note that the negative reaction for isolate TUB F-898 may be explained by the fact that this strain has the type allele of the ITS1-ITS2 sequences but does not share identity to the “harzianum type” clade based on tef1.
Bar code for identification of superior T. harzianum sensu stricto chitinase producers.
The unique ITS1-ITS2 sequences and particularly the tef1 sequences of strains of the “harzianum type” clade were used to develop the first online screening system based on the oligonucleotide bar code. Since the correlation between tef1 alleles and superior levels of extracellular chitinases was absolute, while the results for the ITS1-ITS2 alleles were incongruent in one case, we recommend that the large fourth intron of tef1 should be used as the first locus for molecular screening of H. lixii/T. harzianum strains. However, when a sample is composed of various species, including T. harzianum, we recommend that ITS1-ITS2 and TrichOKEY 2 (18) should be used since besides superior chitinase producers of the “harzianum type” clade the majority of other sequences are identified on the species level. We developed the TrichoCHIT web tool (www.isth.info/tools/trichochit), which recognizes the multiple sets of either ITS1-ITS2 sequences or tef1 sequences and selects the records which may indicate superior chitinase production. In the case of the ITS1-ITS2 locus, each positive record can be directly checked with the TrichOKEY program (18; www.isth.info/molkey), while both ITS1-ITS2 sequences and tef1 sequences may be analyzed by the similarity search integrated in TrichoBLAST (32; www.isth.info). Similar options are suggested for negative results as well.
DISCUSSION
In this study, we used chitinase production to test whether the principle that the DNA bar code could be employed for screening for improved product formation by environmental isolates of industrially important fungal species. Our data proved the hypothesis that physiological traits are not randomly distributed within a given species; a single subpopulation of H. lixii/T. harzianum indeed exhibits statistically supported enhanced chitinase formation. This subpopulation also contains the ex-type strain of T. harzianum, and therefore we call it “T. harzianum sensu stricto.” The global distribution and the highly similar or identical nucleotide sequences of the two molecular markers used (and also other molecular markers [I. S. Druzhinina and C. P. Kubicek, unpublished data]) indicate that this subpopulation is essentially clonal. Data to be presented elsewhere show that populations composed of strains outside this clade (which include the ex-type strain of the teleomorph and therefore should be called H. lixii) have a strongly sexually recombining structure (I. S. Druzhinina and C. P. Kubicek, unpublished data). This finding may also explain why T. harzianum sensu stricto shows enhanced chitinase production, because specific traits, once obtained, become rapidly fixed in clonal populations. It is therefore likely that similar gradients for the production of enzymes and metabolites may be found in other filamentous fungi which are known to consist of cryptic species with varying degrees of sexual recombination (11, 33, 43, 48).
The above explanation, if correct, implies that chitinase production must be (or least has been) an important trait for evolution of T. harzianum sensu stricto. One could speculate that this is due to the fact that an essential part of the life cycle of this fungus is spent in mycoparasitism, which would be favored by increased chitinase formation. However, our data did not prove the assumption that T. harzianum sensu stricto strains are superior antagonists (see below) in vitro, and therefore, they may also not be superior antagonists in their natural habitat. In order to understand the evolution of this trait, the biogeographic distribution of T. harzianum sensu stricto and the biochronology of its emergence need to be considered. For the first of these characterisitics, global screening of more than 300 isolates of T. harzianum revealed that T. harzianum sensu stricto is abundant in Canada, Europe, and Siberia and that only a few isolates were obtained from other areas. For the second, T. harzianum sensu stricto differs in only one to three nucleotides from the most closely related isolates of H. lixii sensu lato in ITS1-ITS2. Using the estimates of Kasuga et al. (29, 30) as a basis, the divergence time of the “harzianum type” clade from the rest of the strains must therefore have taken place some 0.6 to 4 million years ago (100% estimation error included). Consequently, the speciation of T. harzianum sensu stricto was a late Pleistocene event and correlated with the occurrence of up to 10 glaciation periods in its habitat, the earliest one starting 2.4 million years ago with the emergence of the isthmus of Panama. In fact, such severe cold periods have been shown to cause restriction of various organisms to biotic refuges, loss of genetic variability, and fixation of specific traits (62), which fits well with the present data. We speculate that during this time T. harzianum sensu stricto specialized in habitats enriched in chitinous debris, which favored natural selection towards the enhanced chitinase formation observed in this study.
Various physiological tests applied to strains of the “harzianum type” clade did not reveal other properties that could explain the enhanced chitinase formation. Our results indicate that the metabolism of the monomers, the rough overall compositions of the chitinase mixture, and the general secreting capacities of T. harzianum sensu stricto and H. lixii sensu lato are similar or at least not different enough to explain the enhanced chitinase production. A characteristic not investigated here is the induction mechanism, which is a topic for further studies.
A remarkable finding was the fact that, despite its increased chitinase activities, T. harzianum sensu stricto was not a better antagonist of several plant pathogens than other strains of the species. This agrees well with recent data that the total amount of chitinases, although undoubtedly important for digestion of the host cell wall during mycoparasitism, is less critical for the process (55). In fact, the major band of chitinase seen in the electropherograms is likely due to chi18-5 (previously ech42 or chit42), because it accounts for the major extracellular chitinase activity in Hypocrea/Trichoderma (22, 24). According to phylogenetic analysis, the chi18-5 gene is a housekeeping gene rather than a tool developed for mycoparasitism (55). It is likely that some of the other chitinase isoenzymes, which are expressed only poorly (such as that encoded by chi18-10 [55]), have more subtle relevance to this process.
To the best of our knowledge, this study is the first study to apply the bar code concept for strain screening in biotechnology. However, some previous work pointed to potential success in this direction; for example, Kubicek et al. (36) selected strains of Trichoderma section Longibrachiatum species for their ability to produce cellulases and found clearly enhanced production by isolates of H. jecorina and Trichoderma longibrachiatum. Similarly, Arisan-Atac et al. (2) showed that the ability to biocontrol Cryphonectria parasitica causing cancer of chestnut trees is restricted to only a few molecular species of Trichoderma. Also, some evidence is available for other fungi. In Rhizopus oryzae, rRNA sequence differences specify higher lactic acid producers and the type of lactate dehydrogenase allele (1, 51). In general, our data also support the concept that there is a correlation between secondary metabolites and taxonomy (39), and we suggest that bar codes may also aid in screening for new secondary metabolites. We describe here the possibility to screen using two molecular phylogenetic markers for superior chitinase-producing strains of the “harzianum type” clade. Sequencing of ITS1-ITS2 should be chosen when the sample contains mixed and unidentified strains of Hypocrea/Trichoderma. In this case we recommend submitting the entire file with multiple ITS1-ITS2 sequences in FASTA format to the TrichOKEY 2 program (18, 19; www.isth.info/molkey) and obtaining identification of all sequences at the species level. Potential superior chitinase producers should be identified as “Trichoderma harzianum sensu stricto” and should not be confused with a general result for “H. lixii/T. harzianum.” For cases of more specific screening of already identified strains of H. lixii/T. harzianum, we recommend sequencing the large intron of the tef1 gene and submitting it to TrichoCHIT (www.isth.info/tools/trichochit), the first version of the tef1-based bar code for superior chitinase producers. The program undoubtedly selects such strains from the pool of submitted sequences. Since a negative result may be due to various reasons, including poor sequence quality in the diagnostic bar code areas, we recommend testing all results by the similarity search tool implemented in TrichoBLAST. This should also reveal whether the sequence belongs to the correct species and includes the diagnostic locus. All known strains of the “harzianum type” clade contain one allele of tef1 sequences. Therefore, positive identification should be concluded either based on an identical sequence match or based on a high-similarity result with no more than two mismatches.
The morphology of T. harzianum has been shown to be polyphyletic (compare, for example, the morphologically indistinguishable species T. aggressivum), and therefore identification of this taxon at the species level based on morphological characteristics is uncertain by definition. Here we provide a set of reliable molecular and physiological methods which either individually or in a combination should lead to successful screening for superior chitinase producers among the widely distributed soil saprotrophs in the genus T. harzianum.
Acknowledgments
We thank Alexey G. Kopchinskiy for programming the TrichoCHIT web tool and for integrating it into the ISTH portal.
This work was supported by grant FWF P-1660 from the Austrian Science Foundation to C.P.K. and by grant FWF P-17895-B06 to I.S.D.
Footnotes
Published ahead of print on 7 September 2007.
REFERENCES
- 1.Abe, A., T. Sone, I. N. Sujaya, K. Saito, Y. Oda, K. Asano, and F. Tomita. 2003. rDNA ITS sequence of Rhizopus oryzae: its application to classification and identification of lactic acid producers. Biosci. Biotechnol. Biochem. 67:1725-1731. [DOI] [PubMed] [Google Scholar]
- 2.Arisan-Atac, I., E. Heidenreich, and C. P. Kubicek. 1995. Randomly amplified polymorphic DNA fingerprinting identifies subgroups of Trichoderma viride and other Trichoderma sp. capable of chestnut blight biocontrol. FEMS Microbiol. Lett. 126:249-256. [DOI] [PubMed] [Google Scholar]
- 3.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Stuhl. 2003. Current protocols in molecular biology. Greene Publishing Associates and Wiley Interscience, New York, NY.
- 4.Ballenweg, S. 2005. Roempp online. Thieme Chemistry, Stuttgart, Germany.
- 5.Beydon, M. H., A. Fournier, L. Drugeault, and J. Becquart. 2000. Microbiological high throughput screening: an opportunity for the lead discovery process. J. Biomol. Screen. 5:13-22. [DOI] [PubMed] [Google Scholar]
- 6.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 7.Carsolio, C., A. Gutierrez, B. Jimenez, M. Van Montagu, and A. Herrera-Estrella. 1994. Characterization of ech-42, a Trichoderma harzianum endochitinase gene expressed during mycoparasitism. Proc. Natl. Acad. Sci. USA 91:10903-10907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Casio, I. G., R. A. Fisher, and P. A. Carroad. 1982. Bioconversion of shellfish chitin waste: waste pre-treatment, enzyme production, process design and economic analysis. J. Food. Sci. 47:901-911. [Google Scholar]
- 9.Chaverri, P., L. A. Castlebury, B. E. Overton, and G. J. Samuels. 2003. Hypocrea/Trichoderma: species with conidiophore elongations and green conidia. Mycologia 95:1100-1140. [PubMed] [Google Scholar]
- 10.Chaverri, P., L. A. Castlebury, G. J. Samuels, and D. M. Geiser. 2003. Multilocus phylogenetic structure within the Trichoderma harzianum/Hypocrea lixii complex. Mol. Phylogenet. Evol. 27:302-313. [DOI] [PubMed] [Google Scholar]
- 11.Dettman, J. R., D. J. Jacobson, and J. W. Taylor. 2003. A multilocus genealogical approach to phylogenetic species recognition in the model eukaryote Neurospora. Evolution 57:2703-2720. [DOI] [PubMed] [Google Scholar]
- 12.Di Martino, A., M. Sittinger, and M. V. Risbud. 2005. Chitosan: a versatile biopolymer for orthopaedic tissue-engineering. Biomaterials 26:5983-5990. [DOI] [PubMed] [Google Scholar]
- 13.Donzelli, B. G., G. Ostroff, and G. E. Harman. 2003. Enhanced enzymatic hydrolysis of langostino shell chitin with mixtures of enzymes from bacterial and fungal sources. Carbohydr. Res. 338:1823-1833. [DOI] [PubMed] [Google Scholar]
- 14.Draborg, H., S. Kauppinen, H. Dalboge, and S. Christgau. 1995. Molecular cloning and expression in S. cerevisiae of two exochitinases from Trichoderma harzianum. Biochem. Mol. Biol. Int. 36:781-791. [PubMed] [Google Scholar]
- 15.Druzhinina, I., A. Kopchinskiy, M. Komon, J. Bissett, G. Szakacs, and C. P. Kubicek. 2005. An oligonucleotide barcode for species identification in Trichoderma. Fungal Genet. Biol. 42:813-828. [DOI] [PubMed] [Google Scholar]
- 16.Druzhinina, I., P. Chaverri, P. Fallah, C. P. Kubicek, and G. J. Samuels. 2004. Hypocrea flavoconidia, a new species from Costa Rica with yellow conidia. Stud. Mycol. 50:401-407. [Google Scholar]
- 17.Druzhinina, I. S., and A. G. Kopchinskiy. 2006. TrichOKEY v. 2—a DNA oligonucliotide BarCode program for the identification of multiple sequences of Hypocrea and Trichoderma, p. 53-59. In Proceedings of the 8th International Mycological Congress, Cairns, Australia. Medimond S.r.l., Bologna, Italy.
- 18.Druzhinina, I. S., M. Schmoll, B. Seiboth, and C. P. Kubicek. 2006. Global carbon utilization profiles of wild-type, mutant, and transformant strains of Hypocrea jecorina. Appl. Environ. Microbiol. 72:2126-2132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Druzhinina, I. S., A. Kopchinskiy, and C. P. Kubicek. 2006. The first one hundred Trichoderma species characterized by molecular data. Mycoscience 47:55-64. [Google Scholar]
- 20.Fenice, M., J.-L. Leuba, and F. Federici. 1998. Chitinolytic enzyme activity of Penicillium janthinellum P9 in bench-top bioreactor. J. Ferment. Bioeng. 86:620-623. [Google Scholar]
- 21.Flach, J., P. E. Pilet, and P. Jolles. 1992. What's new in chitinase research? Experientia 48:701-716. [DOI] [PubMed] [Google Scholar]
- 22.Garcia, I., J. M. Lora, J. de la Cruz, T. Benitez, A. Llobell, and J. A. Pintor-Toro. 1994. Cloning and characterization of a chitinase (chit42) cDNA from the mycoparasitic fungus Trichoderma harzianum. Curr. Genet. 27:83-89. [DOI] [PubMed] [Google Scholar]
- 23.Haran, S., H. Schickler, A. Oppenheim, and I. Chet. 1995. New components of the chitinolytic system of Trichoderma harzianum. Mycol. Res. 99:441-446. [Google Scholar]
- 24.Hayes, C. K., S. Klemsdal, M. Lorito, A. Di Pietro, C. K. Peterbauer, J. P. Nakas, A. Tronsmo, and G. E. Harman. 1994. Isolation and sequence of an endochitinase-encoding gene from a cDNA library of Trichoderma harzianum. Gene 138:143-148. [DOI] [PubMed] [Google Scholar]
- 25.Huelsenbeck, J. P., and F. Ronquist. 2001. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17:754-755. [DOI] [PubMed] [Google Scholar]
- 26.Jaklitsch, W. M., G. J. Samuels, S. L. Dodd, B. S. Lu, and I. S. Druzhinina. 2006. Hypocrea rufa/Trichoderma viride: a reassessment, and description of five closely related species with and without warted conidia. Stud. Mycol. 56:135-177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jaklitsch, W. M., M. Komon, C. P. Kubicek, and I. S. Druzhinina. 2005. Hypocrea voglmayrii sp. nov. from the Austrian Alps represents a new phylogenetic clade in Hypocrea/Trichoderma. Mycologia 97:1365-1378. [DOI] [PubMed] [Google Scholar]
- 28.Jaklitsch, W. M., M. Komon, C. P. Kubicek, and I. S. Druzhinina. 2006. Hypocrea crystalligena sp. nov., a common European species with a white-spored Trichoderma anamorph. Mycologia 98:499-513. [DOI] [PubMed] [Google Scholar]
- 29.Kasuga, T., T. J. White, T. J., and J. W. Taylor. 2002. Estimation of nucleotide substitution rates in Eurotiomycete fungi. Mol. Biol. Evol. 19:2318-2324. [DOI] [PubMed] [Google Scholar]
- 30.Kasuga, T., T. J. White, G. Koenig, J. McEwen, A. Restrepo, E. Castaneda, C. Da Silva Lacaz, E. M. Heins-Vaccari, R. S. De Freitas, R. M. Zancope-Oliveira, Z. Qin, R. Negroni, D. A. Carter, Y. Mikami, M. Tamura, M. L. Taylor, G. F. Miller, N. Poonwan, and J. W. Taylor. 2003. Phylogeography of the fungal pathogen Histoplasma capsulatum. Mol. Ecol. 12:3383-3401. [DOI] [PubMed] [Google Scholar]
- 31.Kim, D. J., J. M. Baek, P. Uribe, C. M. Kenerley, and D. R. Cook. 2002. Cloning and characterization of multiple glycosyl hydrolase genes from Trichoderma virens. Curr. Genet. 40:374-384. [DOI] [PubMed] [Google Scholar]
- 32.Kopchinskiy, A., M. Komon, C. P. Kubicek, and I. S. Druzhinina. 2005. TrichoBLAST: a multiloci database of phylogenetic markers for Trichoderma and Hypocrea powered by sequence diagnosis and similarity search tools. Mycol. Res. 109:658-660. [DOI] [PubMed] [Google Scholar]
- 33.Koufopanou, V., A. Burt, T. Szaro, and J. W. Taylor. 2001. Gene genealogies, cryptic species, and molecular evolution in the human pathogen Coccidioides immitis and relatives (Ascomycota, Onygenales). Mol. Biol. Evol. 18:1246-1258. [DOI] [PubMed] [Google Scholar]
- 34.Kubicek, C. P. 1981. Release of carboxymethyl-cellulase and β-glucosidase from the cell walls of Trichoderma reesei. Eur. J. Appl. Microbiol. Biotechnol. 13:226-231. [Google Scholar]
- 35.Kubicek, C. P., J. Bissett, C. M. Kullnig-Gradinger, I. S. Druzhinina, and G. Szakacs. 2003. Genetic and metabolic diversity of Trichoderma: a case study on South-East Asian isolates. Fungal Genet. Biol. 38:310-317. [DOI] [PubMed] [Google Scholar]
- 36.Kubicek, C. P., U. M. Bölzlbauer, W. Kovacs, R. L. Mach, K. Kuhls, E. Lieckfeldt, T. Börner, and G. J. Samuels. 1996. Cellulase formation by species of Trichoderma sect. Longibrachiatum and of Hypocrea spp. with anamorphs referable to Trichoderma sect. Longibrachiatum. Fungal Genet. Biol. 20:105-114. [DOI] [PubMed] [Google Scholar]
- 37.Kullnig, C. M., G. Szakacs, and C. P. Kubicek. 2000. Molecular identification of Trichoderma species from Russia, Siberia and the Himalaya. Mycol. Res. 104:1117-1125. [Google Scholar]
- 38.Kullnig-Gradinger, C. M., G. Szakacs, and C. P. Kubicek. 2002. Phylogeny and evolution of the fungal genus Trichoderma—a multigene approach. Mycol. Res. 106:757-767. [Google Scholar]
- 39.Larsen, T. O., J. Smedsgaard, K. F. Nielsen, M. E. Hansen, and J. C. Frisvad. 2005. Phenotypic taxonomy and metabolite profiling in microbial drug discovery. Nat. Prod. Rep. 22:672-695. [DOI] [PubMed] [Google Scholar]
- 40.Leache, A. D., and T. W. Reeder. 2002. Molecular systematics of the eastern fence lizard (Sceloporus undulatus): a comparison of parsimony, likelihood and Bayesian approaches. Syst. Biol. 51:44-68. [DOI] [PubMed] [Google Scholar]
- 41.Mandels, M., R. Andreotti, and C. Roche. 1976. Measurement of saccharifying cellulase. Biotechnol. Bioeng. Symp. 6:21-33. [PubMed] [Google Scholar]
- 42.Matsui, N. M., D. M. Smith-Beckerman, and L. B. Epstein. 1999. Staining of preparative 2-D gels. Coomassie blue and imidazole-zinc negative staining. Methods Mol. Biol. 112:307-311. [DOI] [PubMed] [Google Scholar]
- 43.Matute, D. R., J. G. McEwen, R. Puccia, B. A. Montes, G. San-Blas, E. Bagagli, J. T. Rauscher, A. Restrepo, F. Morais, G. Nino-Vega, and J. W. Taylor. 2006. Cryptic speciation and recombination in the fungus Paracoccidioides brasiliensis as revealed by gene genealogies. Mol. Biol. Evol. 23:65-73. [DOI] [PubMed] [Google Scholar]
- 44.Mischak, H., F. Hofer, R. Messner, E. Weissinger, M. Hayn, P. Tomme, H. Esterbauer, E. Küchler, M. Claeyssens, and C. P. Kubicek. 1989. Monoclonal antibodies against different domains of cellobiohydrolase I and II from Trichoderma reesei. Biochim. Biophys. Acta 990:1-7. [DOI] [PubMed] [Google Scholar]
- 45.Muzzarelli, R. A. 1999. Native, industrial and fossil chitins. EXS 87:1-6. [DOI] [PubMed] [Google Scholar]
- 46.Muzzarelli, R. A., M. Guerrieri, G. Goteri, C. Muzzarelli, T. Armeni, R. Ghiselli, and M. Cornelissen. 2005. The biocompatibility of dibutyryl chitin in the context of wound dressings. Biomaterials 26:5844-5854. [DOI] [PubMed] [Google Scholar]
- 47.Peterbauer, C. K., M. Lorito, C. K. Hayes, G. E. Harman, and C. P. Kubicek. 1996. Molecular cloning and expression of the nag1 gene (N-acetyl-β-d-glucosaminidase-encoding gene) from Trichoderma harzianum P1. Curr. Genet. 30:325-331. [DOI] [PubMed] [Google Scholar]
- 48.Pringle, A., D. M. Baker, J. L. Platt, J. P. Wares, J. P. Latge, and J. W. Taylor. 2005. Cryptic speciation in the cosmopolitan and clonal human pathogenic fungus Aspergillus fumigatus. Evolution 59:1886-1899. [PubMed] [Google Scholar]
- 49.Rannala, B., and Z. Yang. 1996. Probability distribution of molecular evolutionary trees: a new method of phylogenetic interference. J. Mol. Evol. 43:304-311. [DOI] [PubMed] [Google Scholar]
- 50.Roberts, W. K., and C. P. Selitrennikoff. 1988. Plant and bacterial chitinases differ in antifungal activity. J. Gen. Microbiol. 134:169-176. [Google Scholar]
- 51.Saito, K., A. Saito, M. Ohnishi, and Y. Oda. 2004. Genetic diversity in Rhizopus oryzae strains as revealed by the sequence of lactate dehydrogenase genes. Arch. Microbiol. 182:30-36. [DOI] [PubMed] [Google Scholar]
- 52.Sallenave-Namont, C., Y. F. Pouchus, T. Robiou du Pont, P. Lassus, and J. F. Verbist. 2000. Toxigenic saprophytic fungi in marine shellfish farming areas. Mycopathologia 149:21-25. [DOI] [PubMed] [Google Scholar]
- 53.Samuels, G. J., S. Dodd, B. Lu, O. Petrini, H.-J. Schroers, and I. S. Druzhinina. 2006. The Trichoderma koningii morphological species. Stud. Mycol. 56:67-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sanz, L., M. Montero, I. Grondona, J. A. Vizcaino, A. Llobell, R. Hermosa, and E. Monte. 2004. Cell wall-degrading isoenzyme profiles of Trichoderma biocontrol strains show correlation with rDNA taxonomic species. Curr. Genet. 46:277-826. [DOI] [PubMed] [Google Scholar]
- 55.Seidl, V., B. Huemer, B. Seiboth, and C. P. Kubicek. 2005. A complete survey of Trichoderma chitinases reveals a new family 18 subgroup. FEBS J. 272:5923-5939. [DOI] [PubMed] [Google Scholar]
- 56.Seidl, V., I. S. Druzhinina, and C. P. Kubicek. 2006. A screening system for carbon sources enhancing β-N-acetylglucosaminidase formation in Hypocrea atroviridis (Trichoderma atroviride). Microbiology 152:2003-2012. [DOI] [PubMed] [Google Scholar]
- 57.Talbot, N. J., P. Vincent, and H. G. Wildman. 1996. The influence of genotype and environment on the physiological and metabolic diversity of Fusarium compactum. Fungal Genet. Biol. 20:254-267. [DOI] [PubMed] [Google Scholar]
- 58.Thiericke, R. 2003. High-throughput screening technologies. EXS 93:71-85. [DOI] [PubMed] [Google Scholar]
- 59.Viterbo, A., M. Montero, O. Ramot, D. Friesem, E. Monte, A. Llobell, and I. Chet. 2002. Expression regulation of the endochitinase chit36 from Trichoderma asperellum (T. harzianum T-203). Curr. Genet. 42:114-122. [DOI] [PubMed] [Google Scholar]
- 60.Reference deleted.
- 61.White, T. J., T. Bruns, S. Lee, and J. Taylor. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics, p. 315-322. In M. A. Innis, D. H. Gelfand, J. J. Sninsky, and T. J. White (ed.), PCR protocols. A guide to methods and applications. Academic Press, San Diego, CA.
- 62.Willis, K. J., and R. J. Whitacker. 2000. The refugial debate. Science 287:1406-1407. [DOI] [PubMed] [Google Scholar]
- 63.Yang, Z., and B. Rannala. 1997. Bayesian phylogenetic interference using DNA sequences: a Markov chain Monte Carlo method. Mol. Biol. Evol. 14:717-724. [DOI] [PubMed] [Google Scholar]