Abstract
Rhodococci are common soil heterotrophs that possess diverse functional enzymatic activities with economic and ecological significance. In this study, the correlation between gene expression and biological removal of the water contaminant N-nitrosodimethylamine (NDMA) is explored. NDMA is a hydrophilic, potent carcinogen that has gained recent notoriety due to its environmental persistence and emergence as a widespread micropollutant in the subsurface environment. In this study, we demonstrate that Rhodococcus sp. strain RHA1 can constitutively degrade NDMA and that activity toward this compound is enhanced by approximately 500-fold after growth on propane. Transcriptomic analysis of RHA1 and reverse transcriptase quantitative PCR assays demonstrate that growth on propane elicits the upregulation of gene clusters associated with (i) the oxidation of propane and (ii) the oxidation of substituted benzenes. Deletion mutagenesis of prmA, the gene encoding the large hydroxylase component of propane monooxygenase, abolished both growth on propane and removal of NDMA. These results demonstrate that propane monooxygenase is responsible for NDMA degradation by RHA1 and explain the enhanced cometabolic degradation of NDMA in the presence of propane.
Recently recognized as a drinking water contaminant (19), N-nitrosodimethylamine (NDMA) is now closely monitored by municipal water providers to minimize human exposure (3, 6, 20). Concern has developed due to NDMA's potent mutagenicity and carcinogenicity (11) coupled with increasing awareness of its presence as a groundwater contaminant associated with liquid rocket propellants, certain industrial processes, and chlorine-based water reuse projects (19, 20, 23). The combination of high subsurface mobility coupled with poor attenuation by volatilization, sorption, and abiotic and biological processes (19) has resulted in groundwater plumes that contain measurable quantities of NDMA following decades and miles of subsurface propagation (31). Despite its recalcitrance in groundwater, it has recently been shown that NDMA can be attenuated in wastewater treatment systems (23) and soils (2, 5, 32), presumably through the involvement of microorganisms. This dichotomy between persistence and potential biodegradability necessitates a more detailed understanding of the biochemical mechanisms that contribute to NDMA degradation.
Microorganisms grown on substrates such as propane, methane, and toluene have been shown to rapidly oxidize NDMA in the laboratory (7, 25). In these cases, evidence from inhibition and induction experiments along with observations of requisite oxygen consumption suggests that propane monooxygenases (PrMO), soluble methane monooxygenases (sMMO), and toluene monooxygenases (TMO) are most likely involved in these transformations. In addition, experiments with Escherichia coli clones expressing TMO inserts confirmed the role of toluene 4-monooxygenase (T4MO) in NDMA oxidation, while cupric selection for soluble rather than particulate MMO confirmed the role of sMMO (25). The involvement of PrMO is less understood, as the traditional boundary can blur between enzymes oxidizing gaseous and liquid n-alkanes. Liquid alkanes are typically oxidized by alkane monooxygenases (AlkMO), but AlkMO can be induced by propane in some bacteria but not in others (10, 16). Regardless of the class of monooxygenase involved, NDMA is transformed with little observed benefit to the cells and no evidence of cellular growth, despite the production of oxidized products, including formaldehyde, methylamine, and methanol, that can be incorporated into primary metabolic pathways (7, 24, 33). Limited evidence for metabolism suggests that non-energy-generating transformations, such as cometabolic oxidation reactions, play an important role in the biological attenuation of NDMA.
Rhodococci are soil heterotrophs of the order Actinomycetales with a noted diversity of functional enzymatic activities (9, 14). Collectively, this order is biologically and economically significant for the production of a diverse array of enzymes involved in the production of commercial secondary metabolites, antibiotics, and the metabolism of xenobiotic compounds for environmental and industrial applications (21). Global genomic, transcriptomic, and functional analyses of Rhodococcus sp. strain RHA1 reveal tremendous enzymatic diversity with the potential to grow on a wide variety of aromatic compounds, carbohydrates, nitriles, and steroids as the sole carbon and energy sources (see reference 17 and references therein). Indeed, the genome of Rhodococcus sp. strain RHA1 is predicted to encode over 200 oxygenases, including both PrMO and AlkMO gene clusters. The genetic blueprint provided by the annotated genome and the development of a corresponding global microarray facilitate the identification of genes responsible for physiological traits of RHA1, especially for growth and enzymatic activity. For this reason, strain RHA1 was selected as a model organism to better understand the genetics and biochemistry of NDMA transformation.
To gain insight into NDMA degradation by Rhodococcus sp. strain RHA1, we analyzed the effects of propane on gene expression and NDMA removal. Here, we report the first experimental evidence for NDMA degradation by a PrMO. First, the kinetics associated with NDMA degradation in strain RHA1 are characterized. Then, the candidate genes associated with propane and NDMA oxidation are identified and quantified through differential expression, as assayed by global transcriptional microarray analysis and reverse transcriptase-quantitative PCR (RT-qPCR). Finally, targeted gene disruption is used to confirm the role of PrMO in NDMA degradation and exclude the role of AlkMO in the observed degradation.
MATERIALS AND METHODS
Strains and plasmids.
The bacterial strains and plasmids that were used or made in this study are listed in Table 1.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Characteristic(s) | Source or reference |
|---|---|---|
| Strains | ||
| Rhodococcus sp. | ||
| RHA1 | Wild type | 17 |
| RHA027 | ΔprmA RHA1 with prmA deletion | This study |
| RHA028 | ΔalkB RHA1 with alkB deletion | This study |
| E. coli | ||
| DH5α | Host used for cloning the mutagenic plasmid | Bethesda Research |
| S17-1 | Donor strain for conjugation | 27 |
| TG1/pBS(Kan)PrMO.RHA1 | E. coli host containing gene cluster for PrMO | This study |
| TG1/pBS(Kan)AlkMO.RHA1 | E. coli host containing gene cluster for AlkMO | This study |
| Plasmids | ||
| pK18mobsacB | 5.7-kb mobilizable suicide vector used for triple ligation; sacB alphII | 30 |
| pΔprmA | 2.2-kb fusion PCR fragment flanking ΔprmA cloned into pK18mobsacB; used to make strain RHA027 | This study |
| pΔalkB | 2.2-kb fusion PCR fragment flanking ΔalkB cloned into pK18mobsacB; used to make strain RHA028 | This study |
Cellular growth and harvest conditions.
All strains were grown aerobically in batch flasks at 30°C and 150 rpm to ensure viability, enzyme activity, and adequate mixing unless otherwise noted. For maintaining RHA1 and raising cells for degradation assays, cells were incubated in Luria-Bertani (LB) medium (Becton Dickinson, Sparks, MD) in equilibrium with the atmosphere. Minimal salts medium (25) was amended with 23 mM pyruvate (Fisher Scientific, Fair Lawn, NJ) or a 20% headspace volume addition of 99.5% purity propane (Matheson, Newark, CA) in sealed flasks containing from 12% to 15% (vol/vol) growth medium. The quantity of electron donors was appropriate to prevent oxygen limitation. Deionized water, produced from a Barnstead Nanopure II water-purifying system, was used for preparation of stock solutions, buffer, and medium.
Culture growth phase was determined by monitoring optical density at 600 nm (OD600). Cells were harvested from culture medium in the late exponential phase of growth at OD600 of ∼0.7 and 1.5 for cells grown, respectively, on propane or liquid organics. Cells were harvested with a Beckman Avanti J-301 centrifuge (Palo Alto, CA) at 15,000 × g for 5 min. Cells exposed to propane were centrifuged and transferred to a new container to remove residual propane. The resulting pellet was then suspended in 0.1 M phosphate buffer (pH 7) solution. For cells grown with liquid substrates, this washing process was repeated two more times. Washed cells were suspended with buffer to a target density to optimize measurement of transformation rates (adjusted OD600 of between 0.1 and 7.0), and a fraction of the cells were frozen for future protein analysis.
Quantification of NMDA removal.
Cell suspensions to evaluate and quantify the biodegradation of NDMA were incubated in 125-ml bottles sealed with Teflon-lined Miniert valves (Altech, Deerfield, IL). These flasks contained washed cells suspended in phosphate buffer as described above. NMDA (99+%) was purchased from Acros Organics (Geel, Belgium), and additions to experimental cultures, controls, and standards have been described previously (25). NDMA extractions were performed by removing 2-ml samples at each time point followed by equilibration with an equal volume of high-purity methylene chloride (EM Science, Darmstadt, Germany). Methylene chloride extracts containing NDMA were analyzed by previously described methods (18, 25) involving tandem mass spectrometry. The detection limit for the liquid-liquid extraction as determined by standard curve was approximately 5 μg NDMA liter−1.
NDMA degradation rates as a function of protein density were obtained by monitoring initial loss during cellular incubations in phosphate buffer (24). Each rate consisted of an average of four linearly spaced time points run over 2 h. Biomass was quantified as mass of cellular protein, and there was no significant change in cell density during the course of these incubations. Cellular pellets from frozen 1.5-ml samples were suspended in 210 μl of 48 mM NaOH. The mixture was sheared by bead beating for 5 min and boiled at 100°C for 20 min. The digest was then centrifuged in an Eppendorf 5417C (Hamburg, Germany) for 10 min at 10,000 × g to remove cellular debris from the supernatant. Protein mass per volume was quantified from 50 μl or appropriate dilutions of the supernatant using the Coomassie Plus protein assay reagent kit with bovine serum albumin as the standard (Pierce Biotechnology, Rockford, IL). Degradation rates were graphed as a function of substrate concentration to determine the interdependence of these variables. An iterative best fit for nonlinear regression with 95% confidence intervals was applied to the Monod kinetic model for a constant cell density (equation 1):
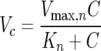 |
(1) |
Components of this equation were defined as follows. Vc is the reaction velocity (μg NDMA mg protein−1 h−1) at NDMA concentration C (μg NDMA liter−1), Vmax,n represents the maximum reaction velocity (μg NDMA mg protein−1 h−1), and Kn is the half-saturation constant (μg NDMA liter−1).
Analysis of global gene expression using spotted microarrays.
For transcriptomic analysis, 65-ml or 80-ml liquid cultures of strain RHA1 were grown in sealed 1-liter flasks containing minimal medium amended, respectively, with 23 mM pyruvate or atmospheric air containing 20% gaseous bulk propane (+99.5% purity) as the sole electron donor and carbon source. Triplicate cultures for each condition were harvested in late-exponential growth. OD600 of approximately 0.7 and 1.3, respectively, were selected to correspond to 70% of the maximal OD600 reached in these incubations as determined by prior growth curves. Upon achieving the target density, 1/10 volumes of “stop solution” (5% phenol [pH 5] in ethanol) were added (1). Cells were collected by centrifugation at 4,900 × g for 10 min at 4°C, suspended in 1.0 ml of the supernatant plus 2.0 ml RNAprotect (QIAGEN), and incubated for 5 min at room temperature. Cells were then centrifuged at 13,000 × g for 2 min at room temperature. Pellets representing 40 ml of culture were each frozen on dry ice and stored at −80°C. RNA extraction was performed on the harvested pellets by adapting previous methods (8). Total RNA isolation involved vortexing with glass beads, hot phenol, and sodium dodecyl sulfate at final concentrations of 14.3% and 0.9% (vol/vol), respectively. Debris was precipitated with acetate followed by the addition of 4.0 ml phenol-chloroform (1:1 [vol/vol]). Nucleic acids were precipitated with acetate plus isopropanol, treated with DNase, and purified with an RNeasy mini column (QIAGEN).
Synthesis of cDNA from the extracted RNA, indirect Cy labeling, and microarray hybridizations were performed as described previously (8), with the following modifications. The cDNA synthesis mixture included 1.5 μg random hexamer primers (Invitrogen) per 6.0 μg RNA, which was brought to 15.3 μl with diethyl pyrocarbonate-treated water. After RNA denaturation for 10 min at 70°C followed by cooling for 5 min on ice, cDNA synthesis components were added to final concentrations of 0.46 mM each dATP, dCTP, and dGTP; 0.19 mM dTTP; 0.28 mM aminoallyl-dUTP (Ambion); 0.01 M dithiothreitol; 10 U RNaseOUT (Invitrogen); and other ingredients as described previously (8). Equal amounts of differentially labeled cDNA, consisting of 50 million pixels measured by ImageQuant 5.2 (Molecular Dynamics), from propane- and pyruvate-grown cells were hybridized at 42°C for 17 h. After the automated washes, the slides were dipped in 0.2× SSC (1×SSC is 0.15 M NaCl plus 0.015 M sodium citrate) and dried by centrifugation at 225 × g for 5 min at room temperature. For one of the three hybridizations, the Cy3 and Cy5 dyes were reversed (i.e., cDNA from the propane treatment was labeled with Cy5 rather than Cy3) to control for dye bias (29). The microarray contained duplicate 70-mer oligonucleotide probes for 8,213 RHA1 genes, representing 89% of the predicted genes (17). The probes were designed and synthesized by Operon Biotechnologies, Inc. (Huntsville, AL).
Microarray spot intensities were quantified using Imagene 6.0 (BioDiscovery, Inc.). Expression ratios were normalized using GeneSpring version 7.2 (Silicon Genetics) by the intensity-dependent Lowess method, with 20% of the data used for smoothing. Average normalized expression ratios were calculated for each gene. Significant differential expression on propane versus pyruvate was defined as absolute ratios of ≥4.0 and Student's t test P < 0.05.
Details of the microarray design, transcriptomic experimental design, and transcriptomic data have been deposited in the NCBI Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/) and are accessible through GEO Series accession no. GSE8480.
Quantification of gene expression by RT-qPCR.
The above extracted RNA was also used for RT-qPCR analysis. While trace genomic DNA contamination present after DNase/RNeasy mini column cleaning was acceptable for the cDNA-specific Cy labeling used in the microarray study, further removal of contaminating DNA was conducted prior to reverse transcription. This was accomplished through two more rounds of DNase I treatments using the DNA-free kit (Ambion) followed by an additional cleanup step in a RNeasy MinElute Cleanup kit (QIAGEN) to remove any other impurities. All treated RNA was stored at −80°C prior to further use. The transcripts of two genes from the PrMO operon (prmA and prmB) and one gene from the AlkMO operon (alkB) were selected for quantification. TaqMan primer-probe sets labeled with 6-carboxyfluorescein (FAM) using 6-carboxytetramethylrhodamine (TAMRA) as a quencher were purchased from PE Applied Biosystems (Foster City, CA). The primer-probe set for prmA, prmB, alkB, and DNA polymerase IV (DNA pol IV) genes are listed in Table 2. The DNA pol IV gene was used as an internal reference (housekeeping gene) for quantification. Primers and probes were designed for quantitative PCR using ABI Prism Primer Express Software (Applied Biosystems). For each design, sequence specificity was confirmed using the NCBI BLAST algorithm optimized for short nucleotide sequences on the GenBank database (www.ncbi.nlm.nih.gov).
TABLE 2.
RT-qPCR primer and probe set used in this study
| Primer or probea | Sequence |
|---|---|
| prmA | |
| Forward primer | 5′-CGCGGCGAACATCTACCT-3′ |
| Reverse primer | 5′-TGGCTACGAACAGGGTGTTG-3′ |
| Probe | 5′-TGGTCGCCGAGACAGCGTTCA-3′ |
| prmB | |
| Forward primer | 5′-GGACGAGGATTGACGGATTTC-3′ |
| Reverse primer | 5′-CGGCGGGTCCATCGAT-3′ |
| Probe | 5′-CGTTCGTGGCCTGCCTCTCGG-3′ |
| alkB | |
| Forward primer | 5′-TCCCTCACACAGCTGGAACTC-3′ |
| Reverse primer | 5′-TCGCTGTGACGCTGCAA-3′ |
| Probe | 5′-ACCACATCGTGACCAATATCTTCCTGTACCA-3′ |
| DNA pol IV | |
| Forward primer | 5′-GACAACAAGTTACGAGCCAAGATC-3′ |
| Reverse primer | 5′-CCTCCGTCAGCCGGTAGAT-3′ |
| Probe | 5′-CGACGGACTTCGGCAAACCGC-3′ |
All primer-probe sets used 6-carboxyfluorescein (FAM)-labeled TaqMan TAMRA probes.
Differential expression of the target genes prmA, prmB, and alkB was quantified using a two-step RT-qPCR method adapted from a previous study (12). For the first step, cDNA was synthesized using the TaqMan reverse transcription reagents kit (Applied Biosystems). Each 10-μl PCR volume contained 2 μl of RNA (∼0.001 ng total RNA) and 0.5 μM of each reverse primer. For the second step of the RT-qPCR method, the reverse-transcribed samples were amplified on an ABI Prism 7000 sequence detection system (Applied Biosystems). The target and housekeeping genes were quantified in triplicate. Each 25-μl qPCR volume contained 2 μl of the reverse-transcribed RNA samples, 12.5 μl of 2×TaqMan universal PCR master mix (Applied Biosystems), 0.2 μM of probe, and 0.7 μM of each primer (forward and reverse). Thermocycling conditions were as follows: 2 min at 50°C, 10 min at 95°C, and 40 cycles of 15 s at 95°C and 1 min at 60°C. Differential expression was calculated by the Pfaffl method (22), which takes into account the amplification efficiency of qPCR for each target gene. The mass of DNA per volume was quantified using a NanoDrop ND-1000 spectrophotometer (Wilmington, DE), according to the manufacturer's instructions.
Concentrated plasmid DNA standards were synthesized by cloning separately the gene cluster for PrMO and AlkMO into E. coli TG1/pBS(Kan) (4). This resulted in the creation of E. coli TG1/pBS(Kan)PrMO.RHA1 and E. coli TG1/pBS(Kan)AlkMO.RHA1 (Table 1). The 4.3-kb PrMO gene cluster containing prmA was PCR amplified from chromosomal DNA of Rhodococcus sp. strain RHA1 using front primer R.PrMO.f.pBS and rear primer R.PrMO.r.pBS (Table 3). The front primer introduced a restriction site, a new ribosome binding site, a stop codon for the upstream lacZα gene, and an altered start codon for the first gene in the RHA1 PrMO gene cluster; the rear primer introduced an alternate restriction site downstream of the last gene in the cluster. PCR products were gel extracted prior to restriction digestion, and plasmid pBS(Kan) was dephosphorylated using Antarctic phosphatase (New England Biolabs) after digestion. Analogous methods and design were used for constructing pBS(Kan)AlkMO-RHA1, with the 2.9-kb AlkMO cluster containing alkB, except front primer R.AlkMO.f.pBS and rear primer R.AlkMO.r.pBS were used (Table 3). The plasmid inserts were both confirmed by DNA sequencing.
TABLE 3.
Cloning, gene deletion, and knockout confirmation screening PCR primers used in this study
| Primera | Sequenceb | Added restriction site |
|---|---|---|
| Cloning | ||
| PrMO cluster | ||
| R.PrMO.f.pBS | 5′-GCCGGTACCCGATTAAGGAGGCGCACAATGAGTAGGCAAAGCCTG-3′ | KpnI |
| R.PrMO.r.pBS | 5′-GTGTGGCTCTAGACGGCTGCGGTCTACTGCGCTGTGAGG-3′ | XbaI |
| AlkMO cluster | ||
| R.AlkMO.f.pBS | 5′-CGGCGAATTCATAAGGAGGTTCGGATCATGACGACGTCGAATATC-3′ | EcoRI |
| R.AlkMO.r.pBS | 5′-GCGGTCGTCTAGAGACATGACCTCGATGCTAGCGG-3′ | XbaI |
| Gene deletion | ||
| “Up” fragment of ΔprmA | ||
| ΔprmAup-f | 5′-GCTCTAGAATCGCCATCTGGTCCGGTGAGTCG-3′ | XbaI |
| ΔprmAup-r | 5′-CCCAAGCTTCGGATCCCATGACAGTTCGGTGATC-3′ | HindIII |
| “Down” fragment of ΔprmA | ||
| ΔprmAdn-f | 5′-CCCAAGCTTTATGTCCGACGCCGAACGCAAC-3′ | HindIII |
| ΔprmAdn-r | 5′-ATGGATCCCGTGAAATCCGTCAATCCTCGTCC-3′ | BamHI |
| “Up” fragment of ΔalkB | ||
| ΔalkBup-f | 5′-CATTGCATGCTGAAGATCGGCTGGCGACACGACG-3′ | SphI |
| ΔalkBup-r | 5′-ATTAAGCTTCAACCACAGGTAGCGCTTGCGGTC-3′ | HindIII |
| “Down” fragment of ΔalkB | ||
| ΔalkBdn-f | 5′-ATTAAGCTTGTGAACATCCAACCCGGCAAGC-3′ | HindIII |
| ΔalkBdn-r | 5′-GCTCTAGAAATCGGCATCGGCCATCGACC-3′ | XbaI |
| Screening | ||
| prmA gene | ||
| prmA-f | 5′-GTGTGACGTGCTGATGGGCTGTG-3′ | |
| prmA-r | 5′-TTGAGCAGCTCGATGGTGCAGTC-3′ | |
| alkB gene | ||
| alkB-f | 5′-GCACATTGCCGCGATGCTTCA-3′ | |
| alkB-r | 5′-ACAGGAAGTCCTTCGACACCGTCG-3′ |
Cloning primers were used to introduce monooxygenase sequences into the E. coli host. Screening primers were used for knockout confirmation.
For cloning primers, the added restriction and start codon sites are underlined. For gene deletion primers, the added restriction sites are underlined.
Construction of knockout mutants.
The prmA and alkB genes were separately deleted in frame (Table 1) using the sacB counterselection system essentially as described previously (30). GeneRunner software was used to design oligonucleotides with appropriate restriction sites that amplified flanking regions of each gene (Table 3). The mutagenic plasmids were transformed into E. coli DH5α by electroporation, verified by PCR, and then transformed into S17 E. coli competent donor cells (27) maintained in 25 μg ml−1 kanamycin (Table 1). Conjunctive plasmid transfer was achieved by coculturing the donor and Rhodococcus sp. strain RHA1 on selective LB peptone plates amended with 30 μg ml−1 nalidixic acid and 50 μg ml−1 kanamycin followed by sacB counterselection. Final confirmation of the removal of the target gene in kanamycin-sensitive, sucrose-resistant colonies was verified by colony PCR using a pair of primers that matched sequences flanking the target gene (Table 3).
RESULTS
NDMA removal by resting wild-type cells.
To assess the constitutive NDMA removal activity of strain RHA1, the cells were grown independently in (i) liquid LB medium, (ii) liquid soy broth, or (iii) liquid minimal salts medium amended with pyruvate as the sole organic substrate. While growth proved most rapid and robust in LB media, cells from each condition that were washed and suspended in phosphate buffer yielded similar NDMA removal rates. The average removal rate for cells grown under these three conditions and exposed to 200 μg NDMA liter−1 was 0.04 ± 0.01 μg NDMA mg protein−1 h−1. In contrast, propane-grown RHA1 cells that were similarly harvested exhibited removal rates that were approximately 500-fold higher (Fig. 1).
FIG. 1.
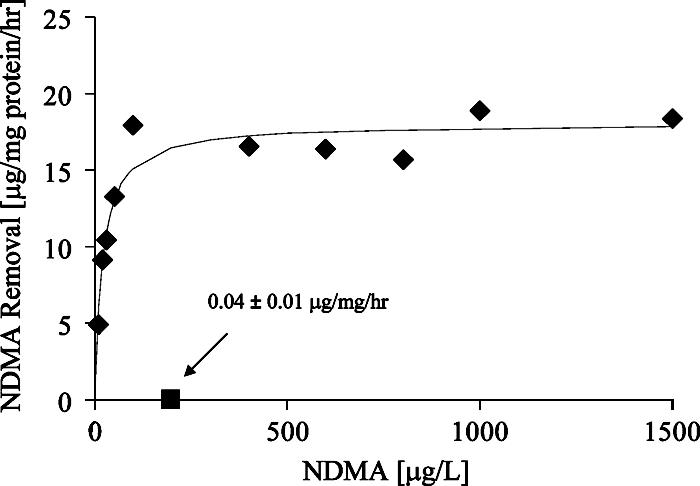
Constitutive removal of NDMA occurs at a fraction of the propane-induced rate. A Monod kinetic model (line) is fit to NDMA removal rates measured after RHA1 was grown on propane (⧫). The constitutive NDMA degradation rate at 200 μg/liter (▪) represents average removal after independent growth on three noninducing substrates (pyruvate, LB medium, or soy broth).
Monod parameters for NDMA degradation by propane-grown RHA1 were calculated by applying a nonlinear fit (R2 = 0.91) of initial disappearance rates to the Monod equation (equation 1) measured at a fixed cellular density. Propane-grown RHA1 cells that were washed and exposed to NDMA in phosphate buffer exhibited a maximum NDMA removal rate (Vmax,n) of 18 ± 3 μg NDMA mg protein−1 h−1 and half-saturation constant (Kn) of 20 ± 17 μg NDMA liter−1.
Effect of propane on gene expression.
The increased NDMA degradation rates observed after growth on propane were explored by investigating the effect of propane on gene expression. Specifically, we employed a microarray with probes for 8,213 of 9,225 predicted genes of Rhodococcus sp. strain RHA1 to identify global transcriptional differences between triplicate batches grown on propane versus those grown on pyruvate. Table 4 lists genes with significant differential expression defined as absolute expression ratios no less than 4.0 and with 95% significance (P < 0.05) according to Student's t test.
TABLE 4.
Genes up-regulated or down-regulated for growth on propane relative to pyruvate
| Gene identification no.a | Fold change | Gene/annotation |
|---|---|---|
| Up-regulated | ||
| ro00441 | 125 | prmA/PrMO hydroxylase large subunit |
| ro00442 | No probe | prmB/PrMO reductase |
| ro00443 | 13 | prmC/PrMO hydroxylase small subunit |
| ro00444 | 21 | prmD/PrMO coupling protein |
| ro00445 | 50 | Conserved hypothetical protein |
| ro00446 | 14 | Conserved hypothetical protein |
| ro00447 | 29 | prmE/alcohol dehydrogenase |
| ro00448 | 13 | groL1/60-kDa chaperonin GroEL |
| ro00449 | 4 | Conserved hypothetical protein |
| ro00450 | 8 | Probable glycolate oxidase flavin adenine dinucleotide-linked subunit |
| ro00455 | 4 | Conserved hypothetical protein |
| ro00521 | 5 | Metabolite transporter, major facilitator superfamily |
| ro00636 ro00553 | 4 | Conserved hypothetical protein |
| ro01183 | 4 | Conserved hypothetical protein |
| ro02242 | 10 | Probable 2-pyrone-4,6-dicarboxylic acid hydrolase |
| ro02764 | 4 | Hypothetical protein |
| ro02795 | 4 | mdlC/benzoylformate decarboxylase |
| ro03490 | 4 | Probable carbon monoxide dehydrogenase small subunit (ferredoxin) |
| ro03894 | 7 | pcal2/3-oxoacid coenzyme A-transferase alpha subunit |
| ro04027 ro06995 ro08131 ro08421 ro09095 | 5 | Probable triacylglycerol or secretory lipase or conserved hypothetical protein |
| ro04062 | 7 | Conserved hypothetical protein |
| ro04527 | 6 | Possible magnesium chelatase |
| ro04843 | 9 | Conserved hypothetical protein |
| ro04898 | 5 | Probable organic hydroperoxide resistance protein |
| ro05000 | 8 | Sensor kinase, two-component system |
| ro06099 | 6 | Citrate (pro-3S)-lyase |
| ro06305 | 8 | rfbD/dTDP-4-dehydrorhamnose reductase |
| ro06664 | 4 | Nonribosomal peptide synthetase |
| ro08036 | 8 | Hypothetical protein |
| ro08409 ro09134 ro08121 | 4 | Possible glycosyl hydrolase or metallopeptidase/glycoside hydrolase |
| ro09108 | 7 | Hypothetical protein |
| ro10116 | 8 | bphG4/acetaldehyde dehydrogenase |
| ro10126 | 11 | bphB2/cis-3-phenylcyclohexa-3,5-diene-1,2-diol dehydrogenase |
| ro10127 | 6 | chnE/6-oxohexanoate dehydrogenase |
| ro10135 | 31 | etbC/2,3-dihydroxybiphenyl 1,2-dioxygenase |
| ro10136 | 18 | bphD1/2-hydroxy-6-oxo-6-phenylhexa-2,4-dienoate hydrolase |
| ro10137 | 7 | bphE2/2-oxopent-4-enoate hydratase |
| ro10138 | 10 | bphF2/4-hydroxy-2-oxovalerate aldolase |
| ro10139 | 14 | Possible ketosteroid isomerase-related protein |
| ro10140 | 14 | Oxidoreductase |
| ro10143 ro10133 | 29 | etbAa1 or etbAa2/ethylbenzene dioxygenase alpha subunit |
| ro10144 ro10134 | 20 | etbAa1 or etbAa2/ethylbenzene dioxygenase alpha subunit |
| ro10145 | 12 | etbAc/ethylbenzene dioxygenase, ferredoxin component |
| ro10147 | 5 | Transporter, major facilitator superfamily |
| ro10422 ro10416 | 5 | Hypothetical protein |
| ro11069 | 4 | Cytochrome P450 CYP257 |
| Down-regulated | ||
| ro00995 | −7 | Probable branched-chain amino acid ABC transporter binding protein |
| ro01066 | −7 | Pyruvate dehydrogenase (cytochrome) |
| ro01361 | −5 | Sugar transporter, major facilitator superfamily |
| ro02000 | −6 | Conserved hypothetical protein |
| ro02071 | −5 | Long-chain-fatty acid-coenzyme A ligase |
| ro02146 | −6 | groL2/60-kDa chaperonin GroEL |
| ro02448 | −5 | Probable tellerium resistance protein |
| ro03152 ro08804 | −5 | Conserved hypothetical protein |
| ro03258 ro03083 ro08112 | −5 | Possible heat shock protein or MerR transcriptional regulator |
| ro04165 | −7 | Possible vanillate monooxygenase oxygenase subunit |
| ro04524 | −5 | trpB2/tryptophan synthase beta subunit |
| ro04557 | −4 | hisD2/histidinol dehydrogenase |
| ro04739 ro04740 | −4 | Hypothetical protein |
| ro05011 | −4 | Acetyl coenzyme A carboxylase carboxyl transferase alpha and beta subunits |
| ro05146 | −4 | Hydrolase |
| ro05181 | −5 | Hypothetical protein |
| ro05912 | −5 | Probable NADH dehydrogenase subunit D |
| ro06034 | −5 | Long-chain fatty acid-coenzyme A ligase |
| ro06190 | −5 | Chaperone protein |
| ro08019 | −6 | Aldehyde dehydrogenase |
| ro08148 | −8 | Possible transposase |
| ro08345 | −4 | dnaJ4/chaperone protein |
| ro08348 ro03566 | −12 | Heat shock protein |
| ro08449 | −7 | Short-chain dehydrogenase |
| ro08610 | −9 | Probable integration host factor |
| ro08821 | −6 | Hypothetical protein |
| ro08824 | −5 | Fumarate hydratase, class I |
| ro08825 | −5 | Probable succinate dehydrogenase hydrophobic membrane anchor protein |
| ro08826 | −8 | sdhC/succinate dehydrogenase cytochrome b subunit |
| ro08827 | −6 | Succinate dehydrogenase flavoprotein subunit |
| ro08828 | −7 | sdhB3/succinate dehydrogenase Fe-S protein subunit |
| ro08830 | −10 | Metabolite transporter, major facilitator superfamily |
| ro08833 | −5 | Epoxide hydrolase |
| ro10046 | −4 | Conserved hypothetical protein |
| ro10375 | −6 | Conserved hypothetical protein |
| ro11166 | −5 | Probable glucose-6-phosphate 1-dehydrogenase |
Up-regulated, ≥4-fold change (P < 0.05); down-regulated, ≤−4-fold chance (P < 0.05). Under circumstances in which there were multiple probes for a given gene, only the last one was selected for display.
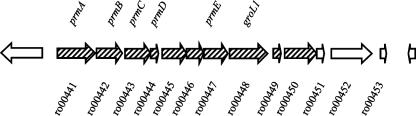
A number of features of this data set are striking. First, growth on propane affects expression of a limited number of genes, many of which cluster in both proximity and function. More specifically, 45 genes were up-regulated in response to propane (Table 4), nine of which occur in a chromosomal gene cluster associated with a putative PrMO (Fig. 2). Another nine genes up-regulated on propane (ro10135 to 10140 and ro10143 to 10145) encode components of an ethylbenzene dioxygenase. These genes are found on the linear plasmid pRHL2 and belong to two operons that were previously found to be coregulated (8). Thirty-six genes were down-regulated in response to propane (Table 4), of which 5 are in a putative operon associated with the tricarboxylic acid cycle (ro08824 to ro08828). Several additional down-regulated genes are involved in the metabolism of simple sugars, including glucose and pyruvate.
FIG. 2.
Putative operon containing prm genes. Gene annotations are available in Table 4. Hatched arrows represent genes that had significant expression ratios, as determined by microarray and/or RT-qPCR, indicating up-regulation on propane.
A second notable feature of the data set is that the most highly up-regulated gene in response to propane, with an expression ration of 125, was prmA, encoding the large hydroxylase subunit of PrMO. The prmA gene is part of a 13-gene cluster (Fig. 2), of which 7 additional genes had expression ratios greater than 10 (Table 4). One of the genes in the cluster, prmB, lacks a probe on the microarray.
Finally, significant differential expression was not demonstrated for a cluster of genes encoding a putative AlkMO (ro02534 to ro02538), despite its predicted functional similarity to PrMO (16).
RT-qPCR analysis of the expression of PrMO and AlkMO genes.
To better quantify the differential expression of genes encoding PrMO and AlkMO, we employed RT-qPCR. Specifically, three genes were targeted: prmA, encoding the large hydroxylase subunit of PrMO; prmB, encoding a reductase component of PrMO not represented by a probe on the microarray; and alkB, encoding the large subunit of AlkMO. RNA extracts used for the prior microarray experiment were also used for RT-qPCR. The relative amount of each gene transcript was normalized to that of a housekeeping gene coding for polymerase IV (DNA pol IV).
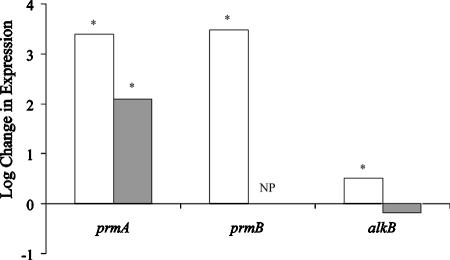
As shown in Fig. 3, the RT-qPCR results are generally consistent with those from the microarray. Using the Pfaffl method (22) of relative quantification, the prmA and prmB genes of the PrMO had propane/pyruvate expression ratios of 2,450 and 3,020, respectively. Conversely, the alkB gene had a much lower expression ratio of 3.2. The levels of expression of each of these genes were significantly different on the two substrates, as determined by Student's t test (α = 0.05 and n = 9). For prmA, the RT-qPCR expression ratio is more than 10-fold greater than the microarray value. This is not unusual for such highly expressed genes and probably indicates that the change in expression exceeded the dynamic range of the microarray analysis (8). The results for prmB confirm that, like the other genes in the putative PrMO operon, it is also up-regulated on propane (Fig. 2).
FIG. 3.
Effect of propane on transcription of three aliphatic monooxygenase components as quantified by RT-qPCR (□; Pfaffl method) and spotted microarray (▪). The microarray did not code for prmB (NP), preventing its quantification by that method. Asterisks denote that the propane-grown values are statistically different from those of the pyruvate-grown controls based upon Student's t test (P < 0.05; n = 9 analytical replicates for RT-qPCR and n = 6 for microarray).
Deletion strains for monooxygenase genes.
To confirm that PrMO is the primary catalyst for NDMA oxidation in RHA1, knockout mutant strains were generated with deletions in prmA and alkB, respectively. As shown in Table 1, this resulted in the mutant Rhodococcus sp. strains RHA027 (prmA mutant) and RHA028 (alkB mutant). Growth of the engineered mutants on both solid- and liquid-phase LB media proved rapid and reproducible, and their morphology mirrored that of the wild-type strain. Both wild-type RHA1 and the alkB mutant grew robustly on liquid minimal medium with propane as the sole organic substrate. In contrast, the prmA mutant did not grow on propane.
The constitutive removal of NDMA (Fig. 1) enabled screening of these knockout mutants. Parallel batch cultures of wild-type RHA1 and the two mutants were grown in liquid LB medium and harvested in the late exponential phase of growth. The cells were washed, suspended in phosphate buffer containing 200 μg NDMA liter−1, and assayed for NDMA removal (Fig. 4). In less than 4 h, both the wild-type strain and the alkB mutant removed NDMA to below detection limits. In contrast, NDMA removal by the prmA mutant was indistinguishable from that of the abiotic control. After 19 h, an additional sample was analyzed and still no significant biological NDMA removal by the prmA mutant was detected (not shown).
FIG. 4.
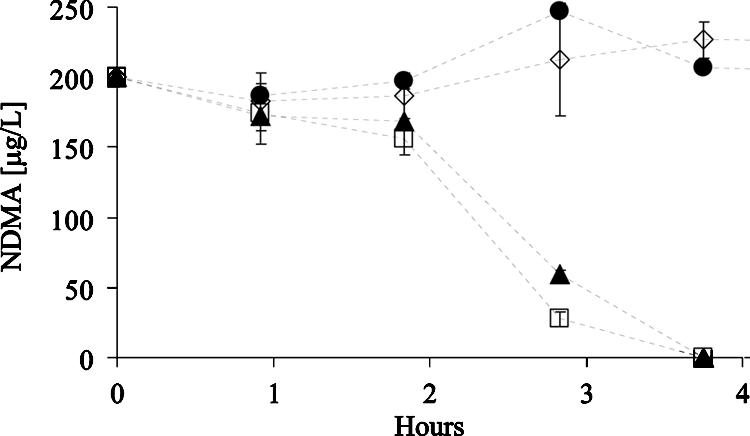
Excision of the prmA component of Rhodococcus sp. strain RHA1 eliminates this strain's ability to transform NDMA. □, wild-type RHA1; ▴, alkB knockout (RHA028); ⋄, prmA knockout (RHA027); and •, no-cell control. LB-grown cells were suspended in phosphate buffer (amended with 200 μg NDMA liter−1) to cellular densities of 510, 530, 730, and 0 mg protein liter−1, respectively. Error bars portray the mean deviation of biological replicates.
DISCUSSION
In this study, we demonstrated that the PrMO gene cluster in Rhodococcus sp. strain RHA1 encodes both for growth on propane and removal of NDMA. RHA1 has the capability to remove NDMA when grown on pyruvate, soy broth, LB medium, or propane. However, transformation rates (Vmax,n) were enhanced by approximately 500-fold after growth on propane relative to these other noninducing substrates (Fig. 1 and 4). This pattern of NDMA removal and propane induction is mirrored in Rhodococcus sp. strain RR1 (GenBank accession no. DQ889725) and Rhodococcus ruber (DSM no. 43338) but not in all rhodococci tested (24). Similarities in induction and NDMA removal kinetics among these three organisms suggest that an analogous PrMO is employed by all three. A comparison of Monod constants for propane-induced versus noninduced cells of strain RR1 (J. O. Sharp, C. M. Sales, and L. Alvarez-Cohen, unpublished data) revealed that Vmax,n was approximately 300-fold higher after growth on propane (44 ± 5 versus 0.14 ± 0.02 μg NDMA mg protein−1 h−1), while Kn was similar for each growth condition (36 ± 10 μg NDMA liter−1 versus 45 ± 10 μg NDMA liter−1). The conservation of Kn values between induced and uninduced conditions in RR1 is consistent with our finding that RHA1's PrMO is responsible for NDMA oxidation under either growth condition.
The prm genes of RHA1 are part of a gene cluster (Fig. 2) which appears to be conserved in other actinobacteria. The first eight genes of this cluster have homologs in Gordonia sp. strain TY-5 (13) and in Mycobacterium smegmatis (GenBank accession no. NC008596). The orders of the genes are identical in the three organisms, and the encoded proteins of RHA1 are 64% to 93% identical to their homologs in the other two organisms. The prmABCD genes in TY-5 are part of a cotranscript induced by propane. Knockout mutagenesis in TY-5 showed that prmB and the seventh gene in the cluster, which we named prmE, are involved in propane catabolism. The prmE gene appears to encode a secondary alcohol dehydrogenase that catalyzes the second step of the catabolic pathway.
Our results provide both transcriptomic and phenotypic evidence for the involvement of the annotated PrMO in NDMA biotransformation. Combined oligonucleotide microarray and RT-qPCR demonstrate that transcripts from the prm gene cluster increased by orders of magnitude following growth on propane (Table 4 and Fig. 3). Next, partial excision of prmA prevented an otherwise genetically identical bacterium from growing on propane and eliminated both its constitutive and induced capability to degrade NDMA (Fig. 4). Despite the presumptive functional similarity between PrMO and AlkMO, deletion of alkB, encoding the large catalytic subunit of the latter enzyme, had no appreciable effect on either growth on propane or removal of NDMA (Fig. 4). Accordingly, the alk operon was not significantly up-regulated during growth (Table 4 and Fig. 3). These findings are consistent with observations of Nocardioides in which degradation of C2-to-C16 n-alkanes was the result of two distinct systems which included one alkane hydroxylase with a homolog to alkB (65% nucleotide identity by pairwise alignment) that was active on alkanes larger than C6 (10). However, a survey of alkB expression in three strains of Mycobacterium austroafricanum has shown that expression of this gene can correlate with the transformation of smaller gaseous alkanes, including propane (16). NDMA removal after growth on propane and the presumed expression of AlkMO was observed in one of these strains, Mycobacterium austroafricanum JOB5 (J. O. Sharp, C. M. Sales, and L. Alvarez-Cohen, unpublished data); however, the strain did not share RHA1's ability to constitutively remove NDMA. Though environmental NDMA degradation through induction on propane appears to extend beyond homologues of prmA, degradation and gene expression in systems not exposed to propane are less well understood.
Since the phenotype of the prmA deletion strain indicates that PrMO is solely responsible for both propane and NDMA oxidation in RHA1, the 20- to 30-fold up-regulation of etb genes on propane was surprising. Interestingly, EtbA has been implicated in the transformation of polychlorinated biphenyls, biphenyl, and ethylbenzene (8). Thus it is possible that propane, a comparatively inexpensive, benign, and mobile carbon source could serve as an effective alternative to ethylbenzene or biphenyl to induce expression of EtbA in environmental settings. It has been suggested (17) that large genomes with multiple broad-specificity catabolic enzymes such as those reported in strain RHA1 could have a competitive advantage in constantly changing soil environments. Such metabolic diversity could result in bacteria that can sustain growth by simultaneously metabolizing an array of compounds present in trace quantities. The coactivation of multiple oxygenase enzymes, while a surprising allocation of biochemical resources, could contribute to such a strategy.
Rhodococci and other members of the Actinomycetales are common soil bacteria. Given the involvement of PrMO in NDMA degradation and the previously discussed identification of similar activity in related strains, quantification of genes such as prmA in uncharacterized communities could provide a proxy for NDMA transformation potential. Furthermore, PrMOs have been reported to degrade a diverse array of organic compounds, including chlorinated C1-to-C6 alkanes, vinyl chloride, chlorinated ethylenes, methyl and ethyl tert-butyl ether, and tert-amyl methyl ether (26, 28), suggesting broader applications.
Interestingly, a correlation between desiccation-induced cell stress and induction of the prm operon in RHA1 was previously observed (15). The reason for up-regulation of prmA under these conditions is not obvious. However, other genes in the operon, such as groEL encoding a chaperone protein, may be part of a general stress response. Due to this response, stressed RHA1 cells, such as those likely to occur in a subsurface vadose zone experiencing alternating wet and dry cycles, varying oxygen content, or periods of growth and starvation, could have increased activity toward low-concentration environmental contaminates such as NDMA. While it is unclear how common this feature of prmA regulation is, it is possible that attenuation strategies involving stressed biomass could hold promise for remediating aquifers containing analogous micropollutants without the introduction of exogenous inducers.
ADDENDUM IN PROOF
Since submission of this article, strain RHA1 has been assigned the species name Rhodococcus jostii RHA1 (A. L. Jones and M. Goodfellow, personal communication).
Acknowledgments
Funding for this project was supplied by Strategic Environmental Research and Development Program grant 1417 (L.A.-C.), National Institutes of Health grant 3 P42 ES004705-19S1 (L.A.-C. and T.K.W.), National Science Foundation grant BES-0529048 (T.K.W.), and a grant from Genome Canada and Genome British Columbia (W.W.M. and L.D.E.). J.C.L. was supported by a postgraduate fellowship from the Natural Sciences and Engineering Research Council of Canada.
Plasmid vector pK18mobsacB was provided by the van der Geize laboratory (Haren, The Netherlands). Aid in the form of microbial and analytical assistance was supplied by Gordon Stewart, Shaily Mahendra, Rebecca Davis, Kristin Robrock, and Wenjin Liu. Two anonymous reviewers provided helpful comments and criticisms during the submission process.
Footnotes
Published ahead of print on 14 September 2007.
REFERENCES
- 1.Bernstein, J. A., A. B. Khodursky, P. H. Lin, S. Lin-Chao, and S. N. Cohen. 2002. Global analysis of mRNA decay and abundance in Escherichia coli at single-gene resolution using two-color fluorescent DNA microarrays. Proc. Natl. Acad. Sci. USA. 99:9697-9702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bradley, P., S. Carr, R. Baird, and F. Chapelle. 2005. Biodegradation of N-nitrosodimethylamine in soil from a water reclamation facility. Bioremediat. J. 9:115-120. [Google Scholar]
- 3.CalEPA. 2006. Public health goals for chemicals in drinking water: N-nitrosodimethylamine. California Environmental Protection Agency, Sacramento, CA.
- 4.Canada, K. A., S. Iwashita, H. Shim, and T. K. Wood. 2002. Directed evolution of toluene ortho-monooxygenase for enhanced 1-naphthol synthesis and chlorinated ethene degradation. J. Bacteriol. 184:344-349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Drewes, J. E., C. Hoppe, and T. Jennings. 2006. Fate and transport of N-nitrosamines under conditions simulating full-scale groundwater recharge operations. Water Environ. Res. 78:2466-2473. [DOI] [PubMed] [Google Scholar]
- 6.EBMUD. 2006. Annual water quality report. East Bay Municipal Utility District, Oakland, CA.
- 7.Fournier, D., J. Hawari, S. H. Streger, K. McClay, and P. B. Hatzinger. 2006. Biotransformation of N-nitrosodimethylamine by Pseudomonas mendocina KR1. Appl. Environ. Microbiol. 72:6693-6698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gonçalves, E. R., H. Hara, D. Miyazawa, J. E. Davies, L. D. Eltis, and W. W. Mohn. 2006. Transcriptomic assessment of isozymes in the biphenyl pathway of Rhodococcus sp. strain RHA1. Appl. Environ. Microbiol. 72:6183-6193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gurtler, V., B. C. Mayall, and R. Seviour. 2004. Can whole genome analysis refine the taxonomy of the genus Rhodococcus? FEMS Microbiol. Rev. 28:377-403. [DOI] [PubMed] [Google Scholar]
- 10.Hamamura, N., C. M. Yeager, and D. J. Arp. 2001. Two distinct monooxygenases for alkane oxidation in Nocardioides sp. strain CF8. Appl. Environ. Microbiol. 67:4992-4998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.IARC. 1987. Relevance of N-nitroso compounds to human cancer: exposures and mechanisms. In H. Bartsch, I. K. O'Neill, and R. Schulte-Hermann (ed.), International Agency for Research on Cancer Proceedings of the IXth International Symposium on N-Nitroso Compounds, Baden, Austria. Oxford University Press, New York, NY.
- 12.Johnson, D. R., P. K. H. Lee, V. F. Holmes, and L. Alvarez-Cohen. 2005. An internal reference technique for accurately quantifying specific mRNAs by real-time PCR with application to the tceA reductive dehalogenase gene. Appl. Environ. Microbiol. 71:3866-3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kotani, T., T. Yamamoto, H. Yurimoto, Y. Sakai, and N. Kato. 2003. Propane monooxygenase and NAD+-dependent secondary alcohol dehydrogenase in propane metabolism by Gordonia sp. strain TY-5. J. Bacteriol. 185:7120-7128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Larkin, M. J., R. De Mot, L. A. Kulakov, and I. Nagy. 1998. Applied aspects of Rhodococcus genetics. Antonie Leeuwenhoek 74:133-153. [DOI] [PubMed] [Google Scholar]
- 15.LeBlanc, J. C., E. R. Gonçalves, and W. W. Mohn. 2006. Transcriptomic response of Rhodococcus sp. RHA1 to dessication stress, abstr. 52, p. 57. Res. Exchange 2006. Genome BC, Vancouver, British Columbia, Canada.
- 16.Lopes Ferreira, N., H. Mathis, D. Labbe, F. Monot, C. W. Greer, and F. Fayolle-Guichard. 2007. N-Alkane assimilation and tert-butyl alcohol (TBA) oxidation capacity in Mycobacterium austroafricanum strains. Appl. Microbiol. Biotechnol. 75:909-919. [DOI] [PubMed] [Google Scholar]
- 17.McLeod, M. P., R. L. Warren, W. W. Hsiao, N. Araki, M. Myhre, C. Fernandes, D. Miyazawa, W. Wong, A. L. Lillquist, D. Wang, M. Dosanjh, H. Hara, A. Petrescu, R. D. Morin, G. Yang, J. M. Stott, J. E. Schein, H. Shin, D. Smailus, A. S. Siddiqui, M. A. Marra, S. J. Jones, R. Holt, F. S. Brinkman, K. Miyauchi, M. Fukuda, J. E. Davies, W. W. Mohn, and L. D. Eltis. 2006. The complete genome of Rhodococcus sp. RHA1 provides insights into a catabolic powerhouse. Proc. Natl. Acad. Sci. USA 103:15582-15587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mitch, W. A., and D. L. Sedlak. 2002. Formation of N-nitrosodimethylamine (NDMA) from dimethylamine during chlorination. Environ. Sci. Technol. 36:588-595. [DOI] [PubMed] [Google Scholar]
- 19.Mitch, W. A., J. O. Sharp, R. R. Trussell, R. L. Valentine, L. Alvarez-Cohen, and D. L. Sedlak. 2003. N-Nitrosodimethylamine (NDMA) as a drinking water contaminant: a review. Environ. Eng. Sci. 20:389-404. [Google Scholar]
- 20.MOE. 2006. Technical support document for Ontario drinking water standards, objectives, and guidelines. Ontario Ministry of the Environment report PIBS 4449e01. Ontario Ministry of the Environment, Ontario, Canada.
- 21.O'Keefe, D. P., and P. A. Harder. 1991. Occurrence and biological function of cytochrome P450 monooxygenases in the actinomycetes. Mol. Microbiol. 5:2099-2105. [DOI] [PubMed] [Google Scholar]
- 22.Pfaffl, M. W. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29:e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sedlak, D. L., R. A. Deeb, E. L. Hawley, W. A. Mitch, T. D. Durbin, S. Mowbray, and S. Carr. 2005. Sources and fate of nitrosodimethylamine and its precursors in municipal wastewater treatment plants. Water Environ. Res. 77:32-39. [DOI] [PubMed] [Google Scholar]
- 24.Sharp, J. O. 2006. Ph.D. dissertation. University of California, Berkeley.
- 25.Sharp, J. O., T. K. Wood, and L. Alvarez-Cohen. 2005. Aerobic biodegradation of N-nitrosodimethylamine (NDMA) by axenic bacterial strains. Biotechnol. Bioeng. 89:608-618. [DOI] [PubMed] [Google Scholar]
- 26.Shennan, J. L. 2006. Utilization of C2-C4 gaseous hydrocarbons and isoprene by microorganisms. J. Chem. Technol. Biotechnol. 81:237-256. [Google Scholar]
- 27.Simon, R., V. Priefer, and A. Puhler. 1983. A broad-host-range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Biotechnology 1:784-791. [Google Scholar]
- 28.Steffan, R. J., K. McClay, S. Vainberg, C. W. Condee, and D. L. Zhang. 1997. Biodegradation of the gasoline oxygenates methyl tert-butyl ether, ethyl tert-butyl ether, and tert-amyl methyl ether by propane-oxidizing bacteria. Appl. Environ. Microbiol. 63:4216-4222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tseng, G. C., M. K. Oh, L. Rohlin, J. C. Liao, and W. H. Wong. 2001. Issues in cDNA microarray analysis: quality filtering, channel normalization, models of variations and assessment of gene effects. Nucleic Acids Res. 29:2549-2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van der Geize, R., G. I. Hessels, R. van Gerwen, P. van der Meijden, and L. Dijkhuizen. 2001. Unmarked gene deletion mutagenesis of kstD, encoding 3-ketosteroid Delta1-dehydrogenase, in Rhodococcus erythropolis SQ1 using sacB as counter-selectable marker. FEMS Microbiol. Lett. 205:197-202. [DOI] [PubMed] [Google Scholar]
- 31.WateReuse. 2006. Investigation of N-nitrosodimethylamine (NDMA) fate and transport. WateReuse Foundation report 02-002-01. WateReuse Foundation, Alexandria, VA.
- 32.Yang, W. C., J. Gan, W. P. Liu, and R. Green. 2005. Degradation of N-nitrosodimethylamine (NDMA) in landscape soils. J. Environ. Qual. 34:336-341. [DOI] [PubMed] [Google Scholar]
- 33.Yoshinari, T., and D. Shafer. 1990. Degradation of dimethyl nitrosamine by Methylosinus trichosporium OB3b. Can. J. Microbiol. 36:834-838. [DOI] [PubMed] [Google Scholar]