Abstract
Glycoside linkage (cellobiose versus maltose) dramatically influenced bioenergetics to different extents and by different mechanisms in the hyperthermophilic archaeon Pyrococcus furiosus when it was grown in continuous culture at a dilution rate of 0.45 h−1 at 90°C. In the absence of S0, cellobiose-grown cells generated twice as much protein and had 50%-higher specific H2 generation rates than maltose-grown cultures. Addition of S0 to maltose-grown cultures boosted cell protein production fourfold and shifted gas production completely from H2 to H2S. In contrast, the presence of S0 in cellobiose-grown cells caused only a 1.3-fold increase in protein production and an incomplete shift from H2 to H2S production, with 2.5 times more H2 than H2S formed. Transcriptional response analysis revealed that many genes and operons known to be involved in α- or β-glucan uptake and processing were up-regulated in an S0-independent manner. Most differentially transcribed open reading frames (ORFs) responding to S0 in cellobiose-grown cells also responded to S0 in maltose-grown cells; these ORFs included ORFs encoding a membrane-bound oxidoreductase complex (MBX) and two hypothetical proteins (PF2025 and PF2026). However, additional genes (242 genes; 108 genes were up-regulated and 134 genes were down-regulated) were differentially transcribed when S0 was present in the medium of maltose-grown cells, indicating that there were different cellular responses to the two sugars. These results indicate that carbohydrate characteristics (e.g., glycoside linkage) have a major impact on S0 metabolism and hydrogen production in P. furiosus. Furthermore, such issues need to be considered in designing and implementing metabolic strategies for production of biofuel by fermentative anaerobes.
Because of problems with sufficient access to petroleum and natural gas resources and the emerging threat of global warming, there is increasing interest in alternative energy options to supplement or replace fossil fuels (43). One prospect that has received considerable attention is the conversion of renewable resources (i.e., biomass) to ethanol using biological routes (20). While availability of bioprocess-based ethanol could offset current demands to some extent, problems with significant CO2 emissions upon energy conversion would remain (37). A longer-term option being considered is the production of molecular hydrogen from biomass using fermentative, anaerobic microorganisms (38). For example, many mesophilic Clostridium and Enterobacter species can grow on fermentable sugars and produce hydrogen as a by-product of energy metabolism (8, 11, 16, 28, 38).
Studies suggest that biohydrogen production rates may be enhanced at higher temperatures (9, 23). In fact, the production and consumption of molecular hydrogen drive the microbial physiology and bioenergetics of many hyperthermophilic bacteria and archaea inhabiting hydrothermal environments (1). The potential of these microorganisms for biofuel processes has not gone unnoticed (25). Elevated processing temperatures could facilitate the breakdown of complex carbohydrates to fermentable sugars and, at the same time, minimize the impact of H2-consuming mesophilic acetogens and methanogens associated with heterogeneous biomass feedstocks. Studies of H2 production by a range of high-temperature bacteria and archaea, including Thermotoga neapolitana (58), Caldicellulosiruptor saccharolyticus (24, 57), Pyrococcus furiosus (46), and Thermococcus kodakaraensis KOD1 (25), have been reported. Notably, P. furiosus was found to have high specific hydrogen production rates when it was grown on maltose in a continuous bioreactor configuration (25, 46).
P. furiosus, a facultative sulfur-reducing, fermentative anaerobe, grows optimally at 98 to 100°C on sugars or peptides as the primary carbon and energy source, producing hydrogen, hydrogen sulfide (in the presence of elemental sulfur [S0]), organic acids (primarily acetate when it is grown on sugars), and small amounts of alanine and ethanol (18, 46). The genesis of these products in the presence and absence of reduced inorganic sulfur species is key to understanding the bioenergetics of this microorganism, especially as it relates to sinks and sources of reductants. Three different hydrogenases from P. furiosus have been characterized; two are cytoplasmic (soluble hydrogenase 1 [SH1] and soluble hydrogenase 2 [SH2]) (34, 35), and one is membrane associated (MBH) (45). It has been suggested that the genome encodes a fourth hydrogenase designated MBX (52). However, this is not the case as DNA microarray analyses have shown that the mbx operon is up-regulated in S0-grown cells, which do not produce hydrogen and lack measurable hydrogenase activity (2, 49).
Energy conservation in P. furiosus has been linked in part to hydrogen production by the ferredoxin-dependent MBH, which generates a proton gradient across the cellular membrane to drive ATP production (45). Reduced ferredoxin is generated by pyruvate oxidoreductase (POR), which also produces acetyl coenzyme A (acetyl-CoA) (32) that is used to generate acetate and ATP by acetyl-CoA synthetase (36). POR has also been connected to acetaldehyde formation from excess reductant accumulating in the cytoplasm (32). Acetaldehyde can be removed by an aldehyde oxidoreductase to regenerate reduced ferredoxin and acetate, but at the expense of losing the energy conserved as ATP in the acetyl-CoA pathway (5, 27, 42). Acetaldehyde could also be converted into ethanol by alcohol dehydrogenases (31, 54), but at the expense of reducing equivalents in the form of NADPH. This pathway makes ethanol a potential sink when cells are challenged with reductant overflow (33). Another reductant sink is alanine, which is generated from pyruvate by the coordinated action of glutamate dehydrogenase and alanine aminotransferase at the expense of NADPH (60). While other routes to alanine formation are possible, such routes are not apparent from the P. furiosus genome sequence. To reduce the amount of carbon entering the glycolytic pathway, P. furiosus, like other Thermococcales, can produce glycogen (21) and extracellular polysaccharides, which could in turn provide the basis for biofilm formation (40, 41).
Although much has been reported about the role of specific enzymes and proteins in the central metabolic pathways of P. furiosus (17, 44, 45), the mechanism of S0 reduction and the impact of S0 on this archaeon's bioenergetics are still not clear. Sulfur reduction proceeds through polysulfides produced by nucleophilic attack on elemental sulfur (6), but so far the mechanism of S0 reduction has been studied only in maltose-grown cells. S0-grown cells do not produce hydrogen (18), and preliminary transcriptional analysis showed that the three operons encoding the three hydrogenases are dramatically down-regulated in the presence of S0 (49). Conversely, two operons, including the genes encoding MBX and two so-called sulfur-induced proteins (Sips), were up-regulated in S0-grown cells and were proposed to play a key role in H2S evolution (49). Additional transcriptional and biochemical analyses have confirmed these results and have also shown that CoA is involved in S0 reduction (G. J. Schut and M. W. W. Adams, unpublished data).
Given the potential of P. furiosus as an H2 producer, it is important to understand the intricacies of its bioenergetics with different types of biomass. Here, a whole-genome DNA microarray was used in conjunction with high-temperature chemostat experiments to further explore the connection between the utilization of two different types of carbohydrates, product formation, and the role that S0 plays in cellular bioenergetics. Surprisingly, the two carbohydrates examined here (maltose and cellobiose) have very different impacts on the bioenergetics and on H2 production.
MATERIALS AND METHODS
Growth and maintenance of P. furiosus.
P. furiosus DSM3638 was routinely cultured anaerobically at 90°C on a sea salts-based medium supplemented with 1 g/liter yeast extract with or without S0, as described previously (51). In brief, the base medium (950 ml) and yeast extract were autoclaved together, after which 50 ml of a membrane-filtered sugar solution (maltose or cellobiose obtained from Sigma, St. Louis, MO) or tryptone (Fisher Scientific, Pittsburgh, PA) was added at a final concentration of 3.3 g/liter. To achieve anaerobic conditions, the medium was first sparged with N2. Then 0.6% (by volume) of a 10% sodium sulfide solution was added to the culture, which was sparged with N2 again prior to inoculation.
Determination of product gas composition.
The content of the exhaust gas was determined periodically by using a Gow-Mac G400C online gas chromatograph with a thermoconductivity detector (Gow-Mac, Bethlehem, PA); the H2 content was determined with N2 as the carrier gas, and the CO2 and H2S contents were determined with He as the carrier gas. The exhaust manifold was connected to a cooling tower to minimize water carryover from the heated bioreactor. Before entering the autosampler, the exhaust gas was dried by passage through an adsorber filled with Drierite (Fisher Scientific, Pittsburgh, PA). Results were recorded by using the Chromperfect 5.0 software (Justice Laboratory Software, Denville NJ), and the gas composition was calculated by comparing the peak area to calibration curves.
Continuous culture.
The continuous culture bioreactor configuration used was similar to one described previously (39). The temperature of the reactor was measured with a type K thermocouple and controlled within ±1°C of the desired temperature by a Digi-Sense controller (Cole Palmer Instrument, Vernon Hills, IL) coupled to a heating mantle (Fisher Scientific, Pittsburgh, PA). The culture pH was monitored with an autoclavable pH probe and a Chemcadet pH controller (Cole Palmer Instrument, Vernon Hills, IL). The reactor was placed on a stir plate so that a Teflon stirring bar could be used for mixing. The inlet gas flow rate was controlled with a rotameter at 21 ml/min. N2 or He (NWSCO, Charlotte, NC) was used to sparge the reactor. Product gases (H2, H2S, and CO2) were determined by gas chromatography.
Prior to each set of experiments, P. furiosus was subcultured (four passages) from a stock in 125-ml serum bottles. Starter cultures (30 ml) were prepared and cooled to room temperature prior to inoculation of a 2-liter round-bottom flask with 1 liter (working volume) at an initial density of 6 × 106 cells/ml. After inoculation, the reactor was operated in batch mode for 6 h before continuous operation was initiated at a dilution rate of 0.15 h−1. Once stabilized at 0.15 h−1 for 12 h, the dilution rate was increased to 0.45 h−1. Samples for cell counting and product analysis were taken from the reactor every 2 h. The culture usually achieved steady state after three volume changes, so the final sample was usually harvested roughly 12 h after operation was initiated at a dilution rate of 0.45 h−1. For sulfur-grown cultures, 5 g of elemental sulfur was added to the 1-liter culture at the start. To avoid S0 limitation, 10 g of S0 was added directly to the reactor through a port on the top after 4 h of operation at a dilution rate of 0.45 h−1. In all cases, gas production profiles and exhaust gas concentrations were monitored to ensure that samples used for subsequent analysis were obtained under mechanical and biological steady states.
Determination of cell density and metabolites in spent medium.
Cell densities were determined by epifluorescence microscopy (51). Samples (1 ml) were removed from the reactor and added to 100 μl of a 2.5% glutaraldehyde solution (Sigma, St. Louis, MO). Each mixture was vortexed briefly and let stand for 5 min. From this mixture, 5 to 20 μl was added to 4.9 ml of a 0.004% acridine orange solution, following which the solution containing stained cells was passed through a 25-mm black polycarbonate filter with a pore size of 0.22 μm (GE Water Process Technology, Minnetonka, MN). The filter was placed on a glass slide and examined using an epifluorescence microscope (Nikon, Melville, NY). Ten fields were counted for each sample and cell density. Residual sugar concentrations were determined by first hydrolyzing maltose with α-glucosidase (Roche Diagnostics, Basel, Switzerland) and cellobiose with β-glucosidase from P. furiosus (4). The glucose was then measured with a glucose assay kit (Sigma, St. Louis, MO). Acetate and ethanol concentrations were also determined with assay kits (R-Biopharm, Inc., Marshall, MI). Pyruvate was assayed by examining the oxidation of NADH (Sigma, St. Louis, MO) to NAD+ in the presence of lactate dehydrogenase (Sigma, St. Louis, MO). The subsequent changes in the optical density at 340 nm were determined spectrophotometrically (Perkin-Elmer, Boston, MA).
Analysis of liquid phase fermentation products.
An HPX-87H cation-exchange column and guard column (Bio-Rad Laboratories, Hercules, CA) were used to quantify the residual sugar and fermentative products by high-performance liquid chromatography (Waters Corp., Milford, MA). The mobile phase was a 0.008 N sulfuric acid solution, and the column temperature was maintained at 35°C. Standards composed of freshly prepared sea salts-based medium containing known amounts of maltose or cellobiose were injected and used to generate standard curves. Alanine concentrations were determined in culture supernatant samples separated on a Symmetry C18 reverse-phase column, using phenyl isothiocyanate derivatization (Pierce Biotechnology, Rockford, IL).
RNA isolation and purification.
Cells from the reactor were harvested into a glass vessel in a dry ice-ethanol bath. Cells were separated from the spent medium by centrifugation at 10,000 × g. RNA extraction and purification were carried out as described previously (29, 50).
Microarray experiment and data analysis.
RNA samples were converted to fluorescence-labeled cDNA and hybridized to a whole-genome P. furiosus microarray according to a modified protocol from TIGR (10). The amount of cDNA hybridized on a slide was normalized to minimize operational variations. Each slide was hybridized for 18 h, washed, and read (ScanExpress scanner; Perkin-Elmer, Boston, MA). Spot intensities were quantified by the vendor-supplied software, and quality control was confirmed by using MA plots (13) of raw intensity values. Raw intensities were normalized by using a mixed-effects analysis of variance model described previously (10). Data for Venn diagrams were generated by JMP software (SAS Institute, Cary, NC).
RESULTS
Influence of growth substrate and S0 on the bioenergetics of chemostat-grown P. furiosus.
Chemostat cultures grown at 90°C (dilution rate, 0.45 h−1) were used to assess the influence of the carbon source and S0 on the bioenergetics of P. furiosus. Tables 1 and 2 summarize the growth characteristics and stoichiometry for cultures grown on maltose or cellobiose in the presence and absence of S0. Data for growth on tryptone (peptides) in the presence of S0 are also included; no growth was observed on tryptone in the absence of S0. The impact of glycoside linkage on P. furiosus growth and bioenergetics was profound. In the absence of S0, the cell densities for growth on cellobiose or maltose were comparable, although cellobiose generated 50% more protein. Equivalent increases in acetate, CO2, and H2 production were also observed. While no ethanol was detected for growth on maltose, small amounts were measured for growth on cellobiose. As expected, the presence of S0 in the medium of maltose-grown cells led to complete cessation of H2 production; the H2S levels were comparable to the H2 levels in the absence of S0. However, the protein production on a medium containing maltose and S0 increased nearly fourfold, even in the face of reduced specific sugar consumption, compared to the protein production by S0-free cells. Specific rates of production of acetate and CO2 for growth on maltose were not affected by S0. For cells grown on cellobiose in the presence of S0, the cell density and protein yields were greater than the cell density and protein yields of cells grown in the absence of S0, while the specific rates of CO2 and acetate production decreased by one-third. Surprisingly, while H2 production decreased in the presence of S0, it decreased only by about 50% (from 94.6 to 43.6 mmol/g cell protein/h) and did not decrease to zero, as observed with cells grown on maltose plus S0. The effect of S0 on maltose-grown cells was more profound than the effect of S0 on cellobiose-grown cells. Compared to a culture grown with maltose plus S0 or with cellobiose plus S0, the culture grown with tryptone plus S0 had a much lower cell density but comparable protein production levels. This comparison also showed that much less acetate and slightly less CO2 were produced in the culture grown with tryptone plus S0. No H2 could be detected in cultures grown on tryptone plus S0.
TABLE 1.
Bioenergetic parameters for P. furiosus growing in chemostat cultures on maltose, cellobiose, and tryptone in the presence and absence of S0a
| Substrate(s) | Cell density (108 cells/ml) | Total protein (mg/liter) | QS (mmol/g/h) | QA (mmol/g/h) | QAla (mmol/g/h) | QE (mmol/g/h) | QH2 (mmol/g/h) | QH2S (mmol/g/h) | QCO2 (mmol/g/h) | QH2+H2S (mmol/g/h) | Doubling time for batch growth (min) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Maltose | 2.0 | 37 | 24.7 | 24.5 | 8.2 | ND | 63.6 | ND | 34.8 | 63.6 | 80 |
| Maltose + S0 | 4.1 | 142 | 18.1 | 24.5 | 0.1 | ND | ND | 55.1 | 31.4 | 55.1 | 40 |
| Cellobiose | 2.4 | 66 | 25.0 | 37.1 | 5.2 | 2.9 | 94.6 | ND | 47.8 | 94.6 | 90 |
| Cellobiose + S0 | 3.1 | 84 | 19.8 | 26.1 | 4.0 | 2.3 | 43.6 | 17.9 | 30.1 | 61.5 | 50 |
| Tryptone + S0 | 2.4 | 122 | NA | 6.5 | ND | ND | ND | 46.6 | 25.1 | 46.6 | 42b |
Abbreviations: QS, specific rate of consumption of substrate per gram of protein; QA, specific rate of production of acetate per gram of protein; QAla, specific rate of production of alanine per gram of protein; QE, specific rate of production of ethanol per gram of protein; QH2, specific rate of production of H2 per gram of protein; QH2S, specific rate of production of H2S per gram of protein; QCO2, specific rate of production of CO2 per gram of protein; QH+H2S, combined specific rates of production of H2 and H2S; ND, not detected; NA, not applicable.
Data from reference 50.
TABLE 2.
Stoichiometry of P. furiosus conversion of growth substratesa
| Substrate(s) | YX/S (mg protein/mmol sugar consumed) | QA/QS | QE/QS | QCO2/QS | QH2+H2S/QS | QCO2/QA | QH2+H2S/QCO2 |
|---|---|---|---|---|---|---|---|
| Theoretical | 2 | 0 | 2 | 4 | 1 | 2 | |
| Maltose | 37 | 1.0 | 0 | 1.4 | 2.6 | 1.4 | 1.8 |
| Maltose + S0 | 50 | 1.4 | 0 | 1.7 | 3.0 | 1.3 | 1.8 |
| Cellobiose | 36 | 1.5 | 0.1 | 1.9 | 3.8 | 1.3 | 2.0 |
| Cellobiose + S0 | 46 | 1.3 | 0.1 | 1.5 | 3.1 | 1.2 | 2.1 |
| Tryptone + S0 | NA | NA | NA | NA | NA | 3.8 | 1.9 |
YX/S, apparent yield coefficient. For other abbreviations, see Table 1, footnote a.
Table 2 shows the growth stoichiometry of P. furiosus, based on information in Table 1. The theoretical amounts of primary metabolic products are 4, 2, and 2 mol/mol of glucose equivalents consumed for H2 (or H2 plus H2S), acetate, and CO2, respectively. For maltose-grown cells, the amounts of acetate, CO2, and H2 (per mole of glucose) were close to the values reported previously (46). The results shown in Table 2 indicate that there were significant differences in the ways in which cellobiose and maltose were metabolized. For example, the largest amount of H2 was produced by the cellobiose-grown culture, which had a ratio of the specific rate of production of H2 (QH2) to the specific rate of consumption of glucose (QS) of 3.8. This is close to the theoretical maximum of 4, suggesting that most of the carbon flux from the sugar substrate was directed to acetate formation, which resulted in higher hydrogen levels.
In contrast, maltose-grown cultures produced only 70% as much H2, with a QH2/QS value of 2.6. With both sugars, the presence of S0 resulted in comparable ratios of the combined specific rates of production of H2 plus H2S (QH2+H2S) to QS (3.0 to 3.1) which were intermediate between the values obtained with cellobiose alone and with maltose alone. Consequently, S0 appears to have opposite effects on the metabolism of the two sugars. Its presence in cellobiose-grown cultures decreased the amount of carbon that was diverted to acetate formation (lower QH2+H2S/QS value and lower ratio of the specific rate of production of acetate [QA] to QS with S0), while more carbon derived from sugar was diverted to acetate formation in maltose-grown cells when S0 was added (higher QH2+H2S/QS and QA/QS values with S0).
Cells grown with only maltose have been reported to dispose of excess carbon as alanine (41). Here, the specific rate of accumulation of alanine (QAla) was higher in a maltose-grown culture than in a cellobiose-grown culture (8.2 versus 5.2 mmol/g/h). QAla/QA was approximately 0.33 in the maltose-grown culture and 0.14 in the cellobiose-grown culture. However, addition of S0 to the maltose-grown culture eliminated alanine production. The impact of S0 on alanine accumulation in a cellobiose-grown culture was not significant as the ratio of QAla to QA did not change (0.15). These results suggest that maltose-grown cells diverted some carbon flux into alanine and consequently reduced the amount of pyruvate processed via POR; the alanine production in a cellobiose-grown culture, on the other hand, was not affected significantly by addition of S0.
Some ethanol was produced in cellobiose-grown cells (with and without S0) (Table 1), presumably from acetaldehyde by using alcohol dehydrogenases and NADPH (31). However, the amounts represent less that 5% of the H2 and H2S produced (in terms of reductant equivalents), ruling out the possibility that ethanol is a major electron sink. Ethanol formation presumably provides a means for removing potentially toxic acetaldehyde generated as a by-product by POR in a highly reducing environment (33), although why this should be necessary in cellobiose-grown cells rather than maltose-grown cells is not clear. Extracellular pyruvate was not detected in any of the culture supernatants.
Global effect of glycoside linkage and S0 on the P. furiosus transcriptome.
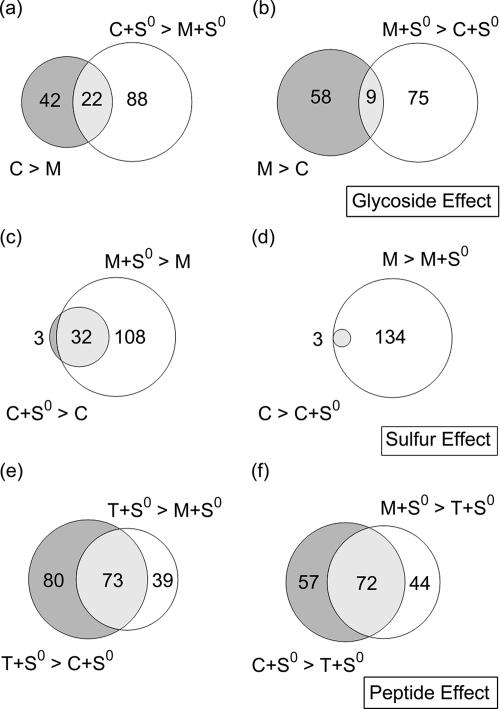
A whole-genome cDNA microarray was used to assess the transcriptional response of P. furiosus during growth on the two sugars both with and without S0. Sets of regulated genes that were common to or unique to the five growth conditions were separated into glycoside, sulfur, and peptide effects, and the results are summarized in Fig. 1.
FIG. 1.
Venn diagram analysis showing the numbers of genes regulated twofold or more (a) on cellobiose (C) (C and C+S0) compared to maltose (M) (M and M+S0), (b) on maltose (M and M+S0) compared to cellobiose (C and C+S0), (c and d) in the presence of S0, and (e and f) on tryptone (T) plus S0 (T+S0). For example, in panel a, 64 genes (in the absence of S0) and 110 genes (in the presence of S0) were up-regulated twofold or more on cellobiose compared to maltose; 22 genes were common to both comparisons. See Tables 2 to 4 and the supplemental material for the complete lists.
Glycoside effect.
The glycoside effect is shown in Fig. 1a. In cells grown on cellobiose compared to cells grown on maltose, 152 genes were up-regulated, 64 genes in the absence of S0 and 110 genes in the presence of S0; of this group, 22 genes were differentially transcribed independent of the presence of S0. On maltose compared to cellobiose, 141 genes were induced, 67 in the absence of S0 and 84 genes in the maltose-plus-S0 versus cellobiose-plus-S0 contrast. Of these 141 genes, only 9 were differentially transcribed independent of the presence of S0.
Sulfur effect.
The presence of elemental sulfur had a profound effect on the transcriptome of maltose-grown cells; the effect on these cells was much greater in terms of the number of open reading frames (ORFs) affected than the effect on cellobiose-grown cells. All but three of the differentially transcribed ORFs responding to S0 in cellobiose-grown cells (35 ORFs were up-regulated and 3 ORFs were down-regulated) also responded to S0 in maltose-grown cells. However, 277 genes were differentially transcribed (140 genes were up-regulated and 137 genes were down-regulated) when S0 was added to maltose. This difference in the magnitude of the transcriptional response for S0-based growth on the two sugars is presumably reflected in the bioenergetic parameters listed in Tables 1 and 2. The dramatic bioenergetic impact of S0 on maltose-grown cells compared to cellobiose-grown cells in terms of cell density, protein production, elimination of H2 production with concomitant H2S production, and alanine production indicates that there is a fundamental difference in how the two sugars are metabolized.
Peptide effect.
The tryptone-plus-S0 versus sugar-plus-S0 transcriptional contrast was independent of glucoside linkage (see the supplemental material). For the comparison of cells grown with tryptone plus S0 and cells grown with cellobiose plus S0, 282 genes were differentially transcribed (153 genes were up-regulated and 129 genes were down-regulated), while for the comparison of cells grown with tryptone plus S0 and cells grown with maltose and S0, 228 genes were affected (112 genes were up-regulated and 116 genes were down-regulated). When tryptone was compared to the sugars, there were 145 differentially transcribed genes in common (73 genes were up-regulated and 72 genes were down-regulated.
Cellobiose transcriptome.
Table 3 lists genes (Fig. 1) that were up-regulated in P. furiosus grown on cellobiose with and without S0 but whose expression was not significantly changed in maltose- or peptide-grown cells (with or without S0). Genes located in the corresponding genome neighborhoods are also listed. The cellobiose-affected genes included PF0496, encoding a transcriptional regulator related to the maltose/maltodextrin-regulated TrmB (30); β-glucan processing loci (PF0073 to PF0076), which encode two glycoside hydrolases (PF0073 [59] and PF0076 [56]); and genes encoding two alcohol dehydrogenases (PF0074 [31] and PF0075 [54]), which are assumed to be involved in acetaldehyde conversion to ethanol. Two putative ABC transporter binding proteins (PF0360 and PF0361) previously shown to be up-regulated during growth on chitin (19) but perhaps more likely involved in cellobiose transport responded, as did PF1696 and PF1697, encoding components of a putative ABC transporter, indicating a possible role in cellobiose uptake. Several genes related to capsular polysaccharide synthesis (PF0765 to PF0768, PF0776, and PF0777) were up-regulated, which may be related to the higher turbidity of cellobiose-grown cultures than of cultures grown on maltose (data not shown). It was interesting that PF1206-PF1207, a VapBC toxin-antitoxin locus was up-regulated, apparently in concert with up-regulation of PF1208, encoding a β-mannosidase/β-glucosidase (4). While VapBC toxin-antitoxin loci respond to heat shock in the hyperthermophile Sulfolobus solfataricus (53), this is the first indication that this system may be involved in metabolic regulation under nonstress conditions.
TABLE 3.
Transcriptional responses of selected ORFs to β-glycoside linkage (cellobiose) independent of S0 in P. furiosus
| Function | GeneID no. | Transcriptional response (fold change)a
|
Annotation | ||
|---|---|---|---|---|---|
| Cellobiose vs maltose | Cellobiose + S0 vs maltose + S0 | Cellobiose + S0 vs tryptone + S0 | |||
| Cellobiose hydrolysis | PF0073 | 35.5 | 22.7 | 8.0 | β-Glucosidase |
| PF0074 | 7.9 | 8.4 | 4.8 | Alcohol dehydrogenase, short chain | |
| PF0075 | 14.5 | 12.5 | 6.3 | Alcohol dehydrogenase, class IV | |
| PF0076 | 2.6 | 2.2 | 2.2 | β-1,3-Endoglucanase | |
| Chitin transport and utilization | PF0355 | NC | NC | NC | VapC1 |
| PF0355.1 | NA | NA | NA | VapB1 | |
| PF0356 | NC | NC | NC | GH1, β-glycosidase | |
| PF0357 | NC | NC | NC | ABC transporter, binding protein | |
| PF0358 | NC | NC | NC | ABC transporter, permease | |
| PF0359 | NC | NC | NC | ABC transporter, permease | |
| PF0360 | 7.7 | 11.3 | 6.2 | ABC transporter, ATP-binding protein | |
| PF0361 | 4.6 | 7.9 | 4.9 | ABC transporter, ATP-binding protein | |
| PF0362 | NC | NC | NC | Glucosamine-fructose-6-phosphate aminotransferase | |
| PF0363 | NC | NC | NC | Exo-β-d-glucosaminidase | |
| Transport | PF0429 | 2.9 | 3.9 | 6.1 | NA+/proline symporter |
| Carbohydrate utilization | PF0496 | 3.2 | 2.2 | 2.8 | Transcriptional regulator, TrmB homolog |
| Gluconeogenesis | PF0613 | 2.9 | 2.4 | 3.1↓ | Archaeal fructose 1,6-bisphosphatase |
| Polysaccharide synthesis | PF0765 | 5.9 | 18.4 | 6.0 | UDP-N-acetyl-d-mannosaminuronate dehydrogenase |
| PF0766 | 3.3 | 8.1 | 4.4 | Dehydrogenase | |
| PF0767 | 4.8 | 13.5 | 5.3 | Aspartate aminotransferase | |
| PF0768 | 4.6 | 11.9 | 4.5 | Acetyltransferase | |
| Regulation/polysaccharide anabolism | PF0776 | 2.2 | 3.8 | 4.6 | Nucleic acid-binding protein, PIN domain |
| PF0777 | 4.3 | 8.8 | 7.9 | Capsular polysaccharide biosynthesis | |
| β-Glucan uptake | PF1206 | 2.4 | 1.6 | 1.6 | VapC12 toxin |
| PF1207 | 5.2 | 5.0 | 3.9 | VapB12 antitoxin | |
| PF1208 | 12.5 | 8.5 | 5.5 | β-Mannosidase | |
| PF1209 | NC | NC | NC | ABC transporter, binding protein | |
| PF1210 | NC | NC | NC | ABC transporter, permease | |
| PF1211 | 2.4 | 3.1↓ | 3.0 | ABC transporter, permease | |
| PF1212 | NC | NC | NC | ABC transporter, uptake protein | |
| PF1213 | NC | NC | NC | ABC transporter, binding protein | |
| Cofactor biosynthesis | PF1674 | 7.4 | 12.0 | 5.3 | Protein-tyrosine phosphatase |
| PF1675 | NC | NC | 2.0↓ | Aspartate racemase | |
| PF1676 | 9.6 | 19.4 | 8.3 | Biotin-(acetyl-CoA carboxylase) ligase | |
| Sugar transport | PF1695 | NC | NC | NC | ABC transporter, binding protein |
| PF1696 | 15.2 | 34.1 | 10.7 | ABC transporter, ATPase | |
| PF1697 | 9.4 | 45.3 | 7.7 | ABC transporter, permease | |
| PF1698 | NC | NC | NC | ABC transporter, permease | |
NC, no change (≤1.5-fold change); NA, not available; ↓, down-regulated.
Maltose transcriptome.
Table 4 lists selected genes (Fig. 1) that were up-regulated in P. furiosus grown on maltose (with and without S0) compared to all other conditions. The transcriptional profiles of genes affected by maltose, including those belonging to the Mal II operon (D. A. Comfort and R. M. Kelly, unpublished data), are also listed for comparison. In addition, the presence of S0 affected the regulation of α-glucan-related genes differently for the two sugars. For example, PF0477 encodes an extracellular α-amylase involved in starch hydrolysis (29). Its expression was up-regulated ninefold when cells grown with maltose were compared with cells grown with cellobiose but was down-regulated fivefold when cells grown with maltose plus S0 were compared with cells grown with cellobiose plus S0 and threefold when cells grown with maltose plus S0 were compared with cells grown with tryptone plus S0. This type of response was noted previously for growth on tryptone (29, 48). The Mal I operon (PF1739 to PF1749) (62), which includes genes encoding an ABC trehalose/maltose transporter (PF1739 to PF1741) and a TrmB homolog (PF1743) known to regulate maltose uptake (30), was minimally affected by the presence of maltose. This may have been due in part to small amounts of trehalose present in yeast extract, which caused this operon to be transcribed under all growth conditions tested. It was interesting that the Mal II operon (PF1933 to PF1939) did not respond to glucoside linkage when S0 was present. A similar trend was observed for PF1109 and PF1110, which encode a single extracellular protein that binds to starch (C.J. Chou and R. M. Kelly, unpublished data) and is predicted to contain CBD9 domains (29).
TABLE 4.
Transcriptional responses of selected ORFs to α-glycoside linkage (maltose) independent of S0 in P. furiosus
| Function | GeneID no. | Transcriptional response (fold change)a
|
Annotation | ||
|---|---|---|---|---|---|
| Maltose vs cellobiose | Maltose + S0 vs cellobiose + S0 | Maltose + S0 vs tryptone + S0 | |||
| Glucan hydrolysis | PF0477 | 8.9 | 4.9↓ | 3.2↓ | Extracellular α-amylase |
| PF0478 | NC | 1.7 | 2.5 | Cycloglucantransferase | |
| PF1109 | 3.5 | NC | 5.8 | Extracellular protein, putative amylase | |
| PF1110 | 3.3 | NC | 5.1 | Extracellular protein | |
| Glucokinase pathway | PF0132 | 3.0 | NC | 1.7 | Isomaltase |
| PF0133 | 1.6 | 1.7 | 2.0 | Hypothetical protein | |
| PF0312 | 1.6↓ | NC | 2.4 | ADP-dependent glucokinase | |
| Phosphorylation pathway | PF0272 | 12.1 | 18.1 | 21.4 | 4-α-Glucanotransferase |
| PF0588 | NC | 1.6 | 1.9 | Phosphoglucose mutase | |
| PF1535 | 1.6 | 2.8 | 3.9 | Glucanophosphorylase | |
| PF1536 | NC | 2.5 | 3.7 | ADP-ribose binding module | |
| PF1537 | 1.7 | 2.5 | 3.3 | Membrane protein, DUF835 | |
| Mal I operon | PF1739 | 1.6 | 2.1 | 1.5↓ | Trehalose/maltose binding protein |
| PF1740 | NC | NC | 2.0↓ | Trehalose/maltose transport, permease | |
| PF1741 | NC | 1.8 | 1.9↓ | Trehalose/maltose transport, permease | |
| PF1742 | 1.7 | 1.9 | NC | Putative trehalose synthase | |
| PF1743 | 1.8 | 2.0 | NC | TrmB | |
| PF1744 | 1.6 | 2.0 | 1.6↓ | Trehalose/maltose transport, ATPase | |
| PF1745 | NC | 1.6 | NC | l-Fructose isomerase | |
| PF1746 | NC | 1.6 | 1.8↓ | Glycogen debranching enzyme | |
| PF1747 | NC | NC | 2.0↓ | Hypothetical protein, DUF377 | |
| PF1748 | 1.9 | 1.7 | NC | Putative sulfate transport, permease | |
| PF1749 | NC | 1.6 | NC | Putative sulfate transport, permease | |
| Mal II operon | PF1933 | 3.9 | NC | NC | Maltodextrin transport, ATPase |
| PF1934 | 8.3 | NC | 1.9 | Amylopullulanase, C-terminal | |
| PF1935 | 8.4 | NC | 2.2 | Amylopullulanase, N-terminal | |
| PF1936 | 8.1 | NC | 1.9 | Maltodextrin transport, permease | |
| PF1937 | NC | 2.1 | 2.8 | Maltodextrin transport, permease | |
| PF1938 | 9.1 | NC | 2.9 | Maltodextrin binding protein (MalE-like) | |
| PF1939 | NC | NC | NC | Intracellular cyclodextrinase, neopullulanase | |
| Stress response | PF0090 | 2.0 | 4.5 | 2.2 | Tungsten cofactor biosynthesis protein |
| PF0126 | 6.2 | 6.5 | NC | DNA repair protein Rad25 | |
| PF0347 | 3.6 | 2.1 | 1.6↓ | Next to aldehyde oxidoreductase | |
| PF0456 | 2.4 | 2.2 | 2.1↓ | Carboxypeptidase, heat shock response | |
| PF0505 | 2.1 | 3.6 | NC | Helix-turn-helix domain, next to DNA helicase | |
| Unknown | PF0560 | 3.3 | 2.2 | NC | Hypothetical protein |
| PF0561 | 2.8 | NC | NC | Hypothetical protein | |
| PF0962 | 3.0 | 2.1 | 1.7 | Contains signal peptide | |
| PF1344 | 3.0 | 2.7 | 4.9 | Putative maleate cis-trans isomerase | |
NC, no change (≤1.5-fold change); NA, not available; ↓, down-regulated.
The difference in transcription patterns between the different operons that encode maltose-processing enzymes may be related to other aspects of maltose assimilation (29). Maltose can be converted to glucose and maltotriose by α-glucanotransferase (PF0272), and the glucose is subsequently phosphorylated to glucose-1-phosphate by α-glucanophosphorylase (PF1535). Glucose-1-phosphate can then be converted into glucose-6-phosphate by phosphoglucomutase (PF0588) (3) or directed into polysaccharide biosynthesis (40). PF0272 was significantly up-regulated by maltose in all comparisons, especially in the presence of S0. On the other hand, the α-glucanophosphorylase operon (PF1535 to PF1537) was up-regulated most in the presence of S0, implying that S0 has an impact on the mechanisms by which maltose is taken into the cell. Alternatively, maltose can be hydrolyzed into glucose by a novel isomaltase (PF0132) (12) (Comfort and Kelly, unpublished data) and converted to glucose-6-phosphate by an ADP-dependent glucokinase (PF0312) (26). The transcription level of PF0132 was constitutively high under all conditions but was extraordinarily high in the culture containing only maltose. However, PF0312 and PF0588, which control the influx to the glycolytic pathway, responded indifferently to maltose and cellobiose but were down-regulated on tryptone (peptides); similar profiles were observed for other intermediate glycolytic genes (see below). Genes encoding several stress proteins were up-regulated on maltose compared with cellobiose (with and without S0), including PF0126, encoding a Rad25 DNA repair protein (61); PF0347, encoding an unknown hypothetical protein proximate to the detoxification enzyme aldehyde oxidoreductase (27); PF0456, encoding a Zn-dependent carboxypeptidase reportedly responsive to heat shock (51); and PF0505, encoding a hypothetical protein next to a DNA helicase, implicated in DNA repair. PF0090 encodes an S-adenosylmethionine-dependent tungsten cofactor biosynthesis protein, and the elevated expression level may indicate that there is an increased demand of tungsten-containing aldehyde oxidoreductases essential for cellular detoxification (5).
Peptide transcriptome.
Table 5 lists selected genes that responded to tryptone as a carbon and energy source. Only those genes associated with glycolytic and gluconeogenesis pathways are included; amino acid anabolism genes are listed in tables in the supplemental material. The peptide transcriptome analysis was consistent with previous transcriptional response analyses comparing maltose- and peptide-grown P. furiosus in batch culture (48). In contrast to sugars, tryptone (peptides) induced transcription of genes involved in gluconeogenesis (PF0613, PF0289, and PF1874), oligopeptide transport (PF0191 to PF0195), energy conservation via acyl-CoA (PF0233, PF0532, and PF1838), fatty acid metabolism (PF0972 to PF0974), and keto acid catabolism (PF0533 to PF0534 and PF1767 to 1773). On the other hand, genes involved in glycolysis (PF0215, PF0312, PF1784, PF1956, and PF1959), amino acid anabolism, and de novo purine synthesis (see the supplemental material) were down-regulated by tryptone. The tungsten-containing aldehyde oxidoreductase, WOR5 (PF1480), was also down-regulated by tryptone. The concerted down-regulation of amino acid catabolic genes and up-regulation of amino acid anabolism genes on both maltose plus S0 and cellobiose plus S0 indicated that the surge of biomass production on maltose plus S0 may not be a direct result of utilizing peptide as a fermentation substrate, as previously suggested (2).
TABLE 5.
Transcriptional responses of selected ORFs significantly regulated by tryptone independent of glycoside type in P. furiosus
| Function | GeneID no. | Transcriptional response (fold change)a
|
Annotationb | |
|---|---|---|---|---|
| Tryptose + S0 vs cellobiose + S0 | Tryptone + S0 vs maltose + S0 | |||
| Gluconeogenesis | PF0613 | 3.1 | 7.3 | Fructose-1,6-bisphosphatase |
| PF0289 | 2.6 | 3.1 | Phosphoenolpyruvate carboxykinase | |
| PF1874 | 4.4 | 5.7 | Glyceraldehyde-3-phosphate dehydrogenase | |
| Transcriptional regulation | PF0054 | 2.6 | 2.2 | Regulator, Lrp/AsnC family |
| PF0055 | 3.5 | 2.2 | Putative HTH transcription regulator | |
| PF0056 | 2.1 | 2.2 | Putative carbohydrate binding protein | |
| Aminotransferase | PF0121 | 3.1 | 2.6 | Aromatic amino acid aminotransferase |
| PF1497 | 3.4 | 2.2 | Alanine aminotransferase | |
| Oligopeptide ABC transporter | PF0191 | 5.4 | 11.2 | Oligopeptide transporter, permease |
| PF0192 | 4.7 | 9.6 | Oligopeptide transporter, permease | |
| PF0193 | 4.5 | 10.6 | Oligopeptide transporter, ATPase | |
| PF0194 | 3.6 | 8.1 | Oligopeptide transporter, ATPase | |
| PF0195 | 5.6 | 8.6 | Hypothetical protein | |
| Acyl-CoA synthetase II | PF0233 | 2.8 | 2.3 | Acyl-CoA synthetase II homolog |
| PF1838 | 3.0 | 2.4 | Acyl-CoA synthetase II homolog | |
| PF0532 | 2.2 | 2.4 | Acyl-CoA synthetase II | |
| Acetyl-CoA synthase | PF0972 | 3.5 | 7.5 | Acyl carrier protein synthase |
| PF0973 | 3.2 | 6.3 | Acetyl-CoA synthase | |
| PF0974 | 3.3 | 7.8 | Hypothetical protein | |
| 2-Keto acid ferredoxin oxidoreductase | PF0533 | 2.0 | 1.6 | IOR, alpha subunit |
| PF0534 | 2.1 | 2.0 | IOR, beta subunit | |
| PF1767 | 15.0 | 10.8 | KGOR, delta subunit | |
| PF1768 | 14.2 | 10.5 | KGOR, gamma subunit | |
| PF1769 | 15.2 | 11.0 | KGOR, beta subunit | |
| PF1770 | 12.7 | 9.6 | KGOR, alpha subunit | |
| PF1771 | 12.9 | 8.9 | KOR, alpha subunit | |
| PF1772 | 9.1 | 7.6 | KOR, beta subunit | |
| PF1773 | 9.3 | 7.9 | KOR, gamma subunit | |
| Glycolysis | PF0215 | 2.1↓ | 3.0↓ | Enolase |
| PF0312 | 2.3↓ | 2.4↓ | ADP-dependent glucokinase | |
| PF1784 | 2.3↓ | 2.7↓ | ADP-dependent phosphofructokinase | |
| PF1956 | 2.1↓ | 2.6↓ | Fructose-1,6-bisphosphate aldolase | |
| PF1959 | 2.2↓ | 3.3↓ | Phosphoglycerate mutase, | |
| Glutamate metabolism | PF0201 | 3.4↓ | 2.4↓ | Aconitase |
| PF0202 | 3.7↓ | 2.1↓ | Isocitrate dehydrogenase | |
| PF0203 | 5.8↓ | 4.0↓ | Citrate synthase | |
| PF0204 | 2.8↓ | 3.6↓ | Glutamate synthase domain 1 | |
| PF0205 | 4.1↓ | 5.8↓ | Glutamate synthase domain 2 | |
| PF0206 | 2.9↓ | 3.2↓ | Glutamate synthase domain 3 | |
| PF0207 | 2.1↓ | 2.3↓ | Argininosuccinate synthase | |
| PF0450 | 35.9↓ | 25.2↓ | Glutamine synthetase | |
| PF1602 | 4.2 | NC | Glutamate dehydrogenase | |
| PF1852 | 1.9↓ | 6.6↓ | Glutamate synthase small subunit | |
| WOR5 | PF1480 | 5.2↓ | 4.7↓ | Aldehyde:ferredoxin oxidoreductase |
| Cofactor biosynthesis | PF1528 | 2.1↓ | 3.3↓ | Predicted glutamine amidotransferase |
| PF1529 | 3.5↓ | 5.9↓ | Pyridoxine biosynthesis enzyme | |
NC, no change (≤1.5-fold change); NA, not available; ↓, down-regulated.
HTH, helix-turn-helix; IOR, indolepyruvate ferredoxin oxidoreductase; KGOR, 2-ketoglutarate ferredoxin oxidoreductase; KOR, 2-ketoacid ferredoxin oxidoreductase.
Sulfur transcriptome.
Table 6 lists genes showing significant up-regulation in both maltose- and cellobiose-grown cells in the presence of S0. Almost all genes regulated by S0 in cellobiose-grown cultures were also present in the data set for cells grown with maltose plus S0. These genes included several purine biosynthesis genes, most members of the membrane-bound oxidoreductase operon, genes encoding MBX (PF1441 to PF1454), and three members of the MBH operon (PF1433 to PF1435). The up-regulation of purine biosynthesis genes was likely the direct result of higher protein production stimulated by S0. Two so-called sulfur-induced proteins, SipA (PF2025) and SipB (PF2026), were up-regulated by S0, consistent with previous reports (49). The fact that SipA and SipB were up-regulated to a much greater extent by S0 on maltose (101- and 22.9-fold, respectively) than on cellobiose (12.5- and 5.7-fold, respectively) suggests that there is a prominent bioenergetic role for these proteins in P. furiosus growing on the maltose.
TABLE 6.
Transcriptional responses of selected ORFs to S0 independent of glycoside type in P. furiosus
| Function | GeneID no. | Transcriptional response (fold change)a
|
Annotationb | |
|---|---|---|---|---|
| Cellobiose + S0 vs cellobiose | Maltose + S0 vs maltose | |||
| Purine metabolism | PF0153 | 4.1 | 2.1 | SAICAR synthase |
| PF0154 | 2.5 | 2.3 | Glutamine phosphoribosylpyrophosphate amidotransferase | |
| PF0426 | 4.3 | 4.0 | NCAIR synthetase | |
| PF1516 | 2.3 | 2.4 | GMP synthase, PP-ATPase domain/subunit | |
| PF1517 | 4.8 | 3.1 | Hypothetical protein | |
| Transposon | PF1366 | 4.1 | 2.0 | Transposase and inactivated derivatives |
| MBH membrane-bound hydrogenase | PF1433 | 12.2 | 7.1 | MbhK (NADH dehydrogenase subunit NuoC) |
| PF1434 | 11.0 | 6.7 | MbhL (Ni-Fe hydrogenase, NuoD) | |
| PF1435 | 11.6 | 7.2 | MbhM (NADH dehydrogenase subunit NuoH) | |
| MBX membrane-bound oxidoreductase | PF1441 | 9.7 | 7.7 | MbxN (four-cysteine proximal Fe-S binding protein) |
| PF1442 | 8.0 | 6.1 | MbxL (Ni-Fe hydrogenase, NuoD homolog) | |
| PF1443 | 8.7 | 6.1 | MbxK (NADH dehydrogenase subunit NuoC) | |
| PF1444 | 9.0 | 6.2 | MbxJ (four-cysteine proximal Fe-S binding protein) | |
| PF1445 | 9.9 | 7.9 | MbxM (NADH dehydrogenase subunit NuoH) | |
| PF1446 | 5.5 | 5.0 | MbxH′ (NADH dehydrogenase subunit NuoL) | |
| PF1448 | 7.3 | 4.7 | MbxG (membrane-bound hydrogenase, anchoring structure subunit) | |
| PF1450 | 3.3 | 2.5 | MbxD (membrane-bound hydrogenase, anchoring structure subunit) | |
| PF1451 | 7.6 | 5.4 | MbxC (membrane-bound hydrogenase, anchoring structure subunit) | |
| PF1452 | 6.0 | 4.1 | MbxB (membrane-bound hydrogenase, anchoring structure subunit) | |
| Sulfur-induced protein | PF2025 | 12.5 | 101 | SipA |
| PF2026 | 5.7 | 22.9 | SipB | |
NC, no change (≤1.5-fold change); NA, not available.
SAICAR, phosphoribosylaminoimidazole-succinocarboxamide synthase; NCAIR, phosphoribosylaminoimidazole carboxylase, ATPase subunit.
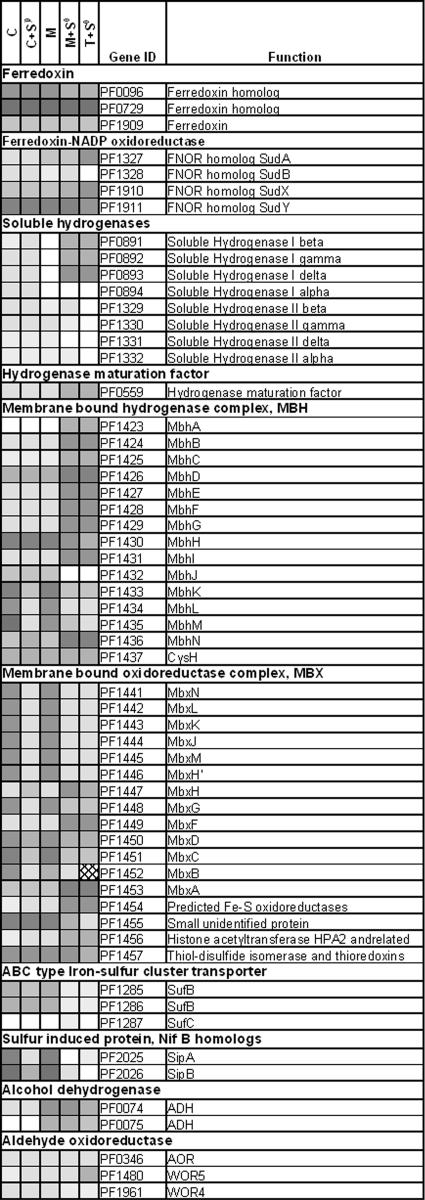
Figure 2 summarizes the expression levels (based on least-squares mean estimates from analysis of variance mixed model analysis) of genes related to energy conservation. The two cytoplasmic hydrogenases in the P. furiosus genome, SH1 and SH2, are encoded by PF0891 to PF0894 and PF1329 to PF1332, respectively. In all cases, the transcription levels of SH2 (PF1329 to PF1332) were similar, while transcription of SH1 (PF0891 to PF0894) was significantly down-regulated on maltose plus S0 and tryptone plus S0. These results support previous reports indicating that the presence of S0 represses expression of SH1 in maltose-grown cultures, although expression of SH2 was also down-regulated under the same conditions (49). However, expression of SH1 was not down-regulated by S0 in cellobiose-grown cultures. Silva et al. (52) and van Haaster et al. (55) suggested that SH1 and SH2 in P. furiosus serve as safety valves and recycling pathways for the H2 that is produced (by MBH), such that SH1 may be responsive to cellular redox status, while SH2 provides a constitutive basal capacity for these functions. The membrane-bound hydrogenase MBH (PF1423 to PF1436) and the membrane-bound oxidoreductase MBX (PF1441 to PF1453) appeared to be reciprocally regulated. While many components of MBX were positively S0 responsive, genes encoding MBH were down-regulated by S0 (Fig. 2), as previously reported for maltose-grown cells (49). This is consistent with the reciprocal regulation of MBH and MBX that occurs within minutes of addition of S0 to P. furiosus cells growing on maltose (Schut and Adams, unpublished). Similarly, contrasting transcriptional responses of MBH and MBX were also noted in response to exposure of P. furiosus to gamma irradiation (61). However, the impact of S0 on MBH expression during growth on cellobiose was much less than that during growth on maltose. This correlates with the gas profiles; H2 production, which occurs via MBH, was measured in cells grown on cellobiose plus S0 but not in cells grown on maltose plus S0 (Tables 1 and 2). These data indicate that there is a close association of MBH with H2 production and a close association of MBX with H2S production on both sugars, not just in maltose-grown cells (49). Furthermore, the pattern of regulation of SipA (PF2025) and SipB (PF2026) was similar to that of many MBX components. This is consistent with the proposal (49) that these proteins are intimately involved in the H2S production process rather than in the disposal of excess reducing equivalents through H2 generation.
FIG. 2.
Heat plot based on the least-squares means (mixed model analysis) of selected genes involved in energy conservation in P. furiosus grown on cellobiose (C), maltose (M), cellobiose plus S0 (C+S0), maltose plus S0 (M+S0), and tryptone plus S0 (T+S0). Open boxes indicate the highest expression levels, while dark gray boxes indicate the lowest expression levels. FNOR, ferredoxin NADPH oxidoreductase; ADH, alcohol dehydrogenase; AOR, aldehyde oxidoreductase.
DISCUSSION
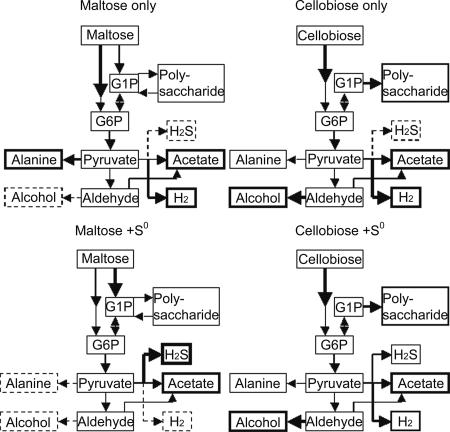
To achieve the goal of optimizing hyperthermophilic H2 production and designing metabolic engineering strategies, it is crucial to understand the physiology and regulation of hydrogenesis at high temperatures, especially as this relates to processing carbon and energy sources. Here, an anaerobic chemostat was used to obtain bioenergetics parameters and profiles of key metabolites related to the transcriptome of P. furiosus. This information is summarized in Fig. 3.
FIG. 3.
Proposed metabolic pathways for P. furiosus grown on maltose and cellobiose in the presence and absence of S0. The thickness of lines outlining boxes and of arrows reflects the significance of the product in the metabolic scheme. A dashed line indicates that the metabolite was not detected. G1P, glucose-1-phosphate; G6P, glucose-6-phosphate.
It was interesting that in the absence of S0, growth on maltose generated much less H2 than growth on cellobiose, despite comparable specific sugar consumption rates on the two sugars. In P. furiosus, maltose can enter the glycolytic pathway via both ADP-glucokinase (PF0312) and glucanophosphorylase (PF1535). The maltose-only transcriptome showed that relevant hydrolases and transporters were induced by this sugar substrate, although genes involved in downstream hydrogenesis pathway were not. This suggests that maltose-only cultures may have a bottleneck, leading to lower protein production and H2/CO2 ratios and greater production of alanine instead of acetate, reduced ferredoxin, and subsequently H2 (40). Thus, it appears that growth on maltose is somewhat limited because reducing power generated from substrate degradation was not involved in an H2-producing, energy-conserving process (45). In contrast, growth on cellobiose triggered transcription of two alcohol dehydrogenases implicated in ethanol production from acetaldehyde, generated from pyruvate by POR (32). It has been proposed that this reaction removes the bottleneck in energy metabolism and detoxifies the cytoplasm by removing accumulating acetaldehyde, thereby facilitating pyruvate decarboxylation (33). Transcriptional analysis also suggested that capsular polysaccharide formation is increased in cellobiose-grown cultures, indicating another possible outlet for reducing equivalents during growth on this substrate. Alteration of metabolite profiles depending on carbon and energy sources has been noted in mesophilic fermentative bacteria (14). For example, inhibition of Clostridium cellulolyticum growth was relieved when cellobiose and yeast extract were replaced by cellulose and defined nitrogen sources (15). This effect was attributed to an imbalance in NADH/NAD ratios arising from higher carbon fluxes in addition to reduced demand on biosynthesis in the presence of complex substrates (22). Whether a similar situation exists for P. furiosus remains to be seen.
The addition of S0 boosted biomass yields for both maltose- and cellobiose-grown cultures, albeit to a much greater extent for the maltose-grown cultures. This was reflected in the considerably higher number of differentially transcribed genes involved in anabolism and cellular redox management when S0 was added to maltose. The close correlation between H2S generation, biomass (protein) yield, and transcriptional levels of MBX and SipA/SipB suggests that S0 reduction is an energy-conserving process in P. furiosus and not just a mechanism for alleviating H2 inhibition. While all H2 production ceased in cultures grown on maltose plus S0, with corresponding down-regulation of MBH and SH1, only about one-third of the H2 production was replaced by H2S generation in cultures grown on cellobiose plus S0. This very surprising result shows that in cellobiose-grown cultures regulation of the expression of MBH and MBX is not the on/off mechanism that appears to be present in maltose-grown cells, where the addition of S0 causes hydrogen production and expression of the genes encoding the three hydrogenases to cease within minutes (Schut et al., unpublished data). At this point, it is not clear why P. furiosus metabolizes the two sugars so differently.
Both processes appear to use the same pathway from phosphorylated hexoses to the end products acetate, CO2, and H2 (plus H2S) since the relative amounts of these compounds (per unit of sugar utilized) are the same (Table 2). However, not only do the H2/H2S ratios differ, but there is also a dramatic difference in carbon flow. For example, on cellobiose alone, about 75% of the sugar is converted to acetate and the gaseous products (sugar/acetate/H2 ratio, ∼1:1.5:3.8), whereas only 50% of the maltose is converted (sugar/acetate/H2 ratio, 1:1:2.6). How the “extra” carbon from maltose is used is not known, but in bioenergy terms, if H2 is the required product, then cellobiose should be the carbon source rather than maltose. Conversely, in the presence of S0, the bioenergetics and end product yields and ratios are very similar (Table 2), indicating that the same pathways are utilized independent of the glycoside type. The one caveat is the production of H2 by cellobiose-grown cells. In the presence of S0, the metabolism of cellobiose in terms of end products (per unit of sugar) appears to be comparable to the metabolism of maltose (Table 2). Consequently, S0 dramatically impacts the carbon flux from sugar into acetate in cellobiose-grown cells but has less impact in maltose-grown cells. Thus, S0 affects carbon flux, in addition to its role as a reductant sink. At present, the transcriptional analyses do not provide insight into how S0 achieves this in cellobiose-grown cells.
It is possible that the profound effect of the glycoside type on the S0-dependent bioenergetics of P. furiosus extends to other hyperthermophiles, including bacteria. A homologous MBX operon can be identified in the genome of the facultative S0-reducing hyperthermophilic bacterium Thermotoga maritima, probably as a result of lateral gene transfer (7). However, the transcriptional levels of this operon were relatively low on maltose and cellobiose and were not affected by S0 (S. R. Gray and R. M. Kelly, unpublished data). A similar response was observed for two Sip homologs in T. maritima. It has been reported that S0 stimulates T. maritima growth by removing inhibitory H2 but does not impact energy conservation pathways (47).
An important outcome of this study is the realization that microbial processes aimed at high levels of H2 production must take into account the impact that substrates and other environmental influences have on cellular bioenergetics. Given the anticipated heterogeneity of biomass feedstocks that will be used for bioenergy conversion processes, a comprehensive understanding of how transcriptional regulation and metabolite production relate to the substrate pool is highly desirable. By combining traditional approaches (chemostat culture for determining bioenergetic parameters) with functional genomics tools (transcriptional response analysis), insights can be obtained and ultimately provide the basis for metabolic engineering strategies. The information provided here for P. furiosus should prove to be useful in efforts to exploit H2 production in this archaeon and other archaea for the production of biofuels.
Supplementary Material
Acknowledgments
This work was supported in part by grants from the NASA Exobiology Program (grant EXB02-0000-0050 to R.M.K.), the DOE Energy Biosciences Program (grant DE-FG02-96ER20219 to R.M.K. and grant FG05-95ER20175 to M.W.W.A.), and the NSF Biotechnology Program (grants CBET-0317886 and CBET-0617272 to M.W.W.A. and R.M.K.). D.L.L. acknowledges support from a Department of Education GAANN Fellowship.
Footnotes
Published ahead of print on 7 September 2007.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Adams, M. W. 1993. Enzymes and proteins from organisms that grow near and above 100°C. Annu. Rev. Microbiol. 47:627-658. [DOI] [PubMed] [Google Scholar]
- 2.Adams, M. W., J. F. Holden, A. L. Menon, G. J. Schut, A. M. Grunden, C. Hou, A. M. Hutchins, F. E. Jenney, Jr., C. Kim, K. Ma, G. Pan, R. Roy, R. Sapra, S. V. Story, and M. F. Verhagen. 2001. Key role for sulfur in peptide metabolism and in regulation of three hydrogenases in the hyperthermophilic archaeon Pyrococcus furiosus. J. Bacteriol. 183:716-724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Akutsu, J., Z. Zhang, M. Tsujimura, M. Sasaki, M. Yohda, and Y. Kawarabayasi. 2005. Characterization of a thermostable enzyme with phosphomannomutase/phosphoglucomutase activities from the hyperthermophilic archaeon Pyrococcus horikoshii OT3. J. Biochem. 138:159-166. [DOI] [PubMed] [Google Scholar]
- 4.Bauer, M. W., E. J. Bylina, R. V. Swanson, and R. M. Kelly. 1996. Comparison of a beta-glucosidase and a beta-mannosidase from the hyperthermophilic archaeon Pyrococcus furiosus. Purification, characterization, gene cloning, and sequence analysis. J. Biol. Chem. 271:23749-23755. [DOI] [PubMed] [Google Scholar]
- 5.Bevers, L. E., E. Bol, P. L. Hagedoorn, and W. R. Hagen. 2005. WOR5, a novel tungsten-containing aldehyde oxidoreductase from Pyrococcus furiosus with a broad substrate specificity. J. Bacteriol. 187:7056-7061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blumentals, I. I., M. Itoh, G. J. Olson, and R. M. Kelly. 1990. Role of polysulfides in reduction of elemental sulfur by the hyperthermophilic archaebacterium Pyrococcus furiosus. Appl. Environ. Microbiol. 56:1255-1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Calteau, A., M. Gouy, and G. Perriere. 2005. Horizontal transfer of two operons coding for hydrogenases between bacteria and archaea. J. Mol. Evol. 60:557-565. [DOI] [PubMed] [Google Scholar]
- 8.Chin, H. L., Z. S. Chen, and C. P. Chou. 2003. Fedbatch operation using Clostridium acetobutylicum suspension culture as biocatalyst for enhancing hydrogen production. Biotechnol. Prog. 19:383-388. [DOI] [PubMed] [Google Scholar]
- 9.Claassen, P. A. M., J. B. van Lier, A. M. L. Contreras, E. W. J. van Niel, L. Sijtsma, A. J. M. Stams, S. S. de Vries, and R. A. Weusthuis. 1999. Utilisation of biomass for the supply of energy carriers. Appl. Microbiol. Biotechnol. 52:741-755. [Google Scholar]
- 10.Conners, S. B., C. I. Montero, D. A. Comfort, K. R. Shockley, M. R. Johnson, S. R. Chhabra, and R. M. Kelly. 2005. An expression-driven approach to the prediction of carbohydrate transport and utilization regulons in the hyperthermophilic bacterium Thermotoga maritima. J. Bacteriol. 187:7267-7282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Converti, A., and P. Perego. 2002. Use of carbon and energy balances in the study of the anaerobic metabolism of Enterobacter aerogenes at variable starting glucose concentrations. Appl. Microbiol. Biotechnol. 59:303-309. [DOI] [PubMed] [Google Scholar]
- 12.Costantino, H. R., S. H. Brown, and R. M. Kelly. 1990. Purification and characterization of an alpha-glucosidase from a hyperthermophilic archaebacterium, Pyrococcus furiosus, exhibiting a temperature optimum of 105 to 115°C. J. Bacteriol. 172:3654-3660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cui, X., M. K. Kerr, and G. A. Churchill. 2003. Transformations for cDNA microarray data. Stat. Appl. Genet. Mol. Biol 2:Article 4. [DOI] [PubMed]
- 14.Desvaux, M. 2006. Unravelling carbon metabolism in anaerobic cellulolytic bacteria. Biotechnol. Prog. 22:1229-1238. [DOI] [PubMed] [Google Scholar]
- 15.Desvaux, M., E. Guedon, and H. Petitdemange. 2001. Carbon flux distribution and kinetics of cellulose fermentation in steady-state continuous cultures of Clostridium cellulolyticum on a chemically defined medium. J. Bacteriol. 183:119-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Desvaux, M., E. Guedon, and H. Petitdemange. 2001. Metabolic flux in cellulose batch and cellulose-fed continuous cultures of Clostridium cellulolyticum in response to acidic environment. Microbiology 147:1461-1471. [DOI] [PubMed] [Google Scholar]
- 17.de Vos, W. M., S. W. Kengen, W. G. Voorhorst, and J. van der Oost. 1998. Sugar utilization and its control in hyperthermophiles. Extremophiles 2:201-205. [DOI] [PubMed] [Google Scholar]
- 18.Fiala, G., and K. O. Stetter. 1986. Pyrococcus furiosus sp. nov. represents a novel genus of marine heterotrophic archaebacteria growing optimally at 100°C. Arch. Microbiol. 145:56-61. [Google Scholar]
- 19.Gao, J., M. W. Bauer, K. R. Shockley, M. A. Pysz, and R. M. Kelly. 2003. Growth of hyperthermophilic archaeon Pyrococcus furiosus on chitin involves two family 18 chitinases. Appl. Environ. Microbiol. 69:3119-3128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gray, K. A., L. Zhao, and M. Emptage. 2006. Bioethanol. Curr. Opin. Chem. Biol. 10:141-146. [DOI] [PubMed] [Google Scholar]
- 21.Gruyer, S., E. Legin, C. Bliard, S. Ball, and F. Duchiron. 2002. The endopolysaccharide metabolism of the hyperthermophilic archeon Thermococcus hydrothermalis: polymer structure and biosynthesis. Curr. Microbiol. 44:206-211. [DOI] [PubMed] [Google Scholar]
- 22.Guedon, E., M. Desvaux, S. Payot, and H. Petitdemange. 1999. Growth inhibition of Clostridium cellulolyticum by an inefficiently regulated carbon flow. Microbiology 145:1831-1838. [DOI] [PubMed] [Google Scholar]
- 23.Hallenbeck, P. C. 2005. Fundamentals of the fermentative production of hydrogen. Water Sci. Technol 52:21-29. [PubMed] [Google Scholar]
- 24.Kadar, Z., T. de Vrije, G. E. van Noorden, M. A. Budde, Z. Szengyel, K. Reczey, and P. A. Claassen. 2004. Yields from glucose, xylose, and paper sludge hydrolysate during hydrogen production by the extreme thermophile Caldicellulosiruptor saccharolyticus. Appl. Biochem. Biotechnol. 113-116:497-508. [DOI] [PubMed] [Google Scholar]
- 25.Kanai, T., H. Imanaka, A. Nakajima, K. Uwamori, Y. Omori, T. Fukui, H. Atomi, and T. Imanaka. 2005. Continuous hydrogen production by the hyperthermophilic archaeon, Thermococcus kodakaraensis KOD1. J. Biotechnol. 116:271-282. [DOI] [PubMed] [Google Scholar]
- 26.Kengen, S. W., J. E. Tuininga, F. A. de Bok, A. J. Stams, and W. M. de Vos. 1995. Purification and characterization of a novel ADP-dependent glucokinase from the hyperthermophilic archaeon Pyrococcus furiosus. J. Biol. Chem. 270:30453-30457. [DOI] [PubMed] [Google Scholar]
- 27.Kletzin, A., S. Mukund, T. L. Kelley-Crouse, M. K. Chan, D. C. Rees, and M. W. Adams. 1995. Molecular characterization of the genes encoding the tungsten-containing aldehyde ferredoxin oxidoreductase from Pyrococcus furiosus and formaldehyde ferredoxin oxidoreductase from Thermococcus litoralis. J. Bacteriol. 177:4817-4819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kumar, N., and D. Das. 2001. Continuous hydrogen production by immobilized Enterobacter cloacae IIT-BT 08 using lignocellulosic materials as solid matrices. Enzyme Microb. Technol. 29:280-287. [Google Scholar]
- 29.Lee, H. S., K. R. Shockley, G. J. Schut, S. B. Conners, C. I. Montero, M. R. Johnson, C. J. Chou, S. L. Bridger, N. Wigner, S. D. Brehm, F. E. Jenney, Jr., D. A. Comfort, R. M. Kelly, and M. W. Adams. 2006. Transcriptional and biochemical analysis of starch metabolism in the hyperthermophilic archaeon Pyrococcus furiosus. J. Bacteriol. 188:2115-2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee, S. J., C. Moulakakis, S. M. Koning, W. Hausner, M. Thomm, and W. Boos. 2005. TrmB, a sugar sensing regulator of ABC transporter genes in Pyrococcus furiosus exhibits dual promoter specificity and is controlled by different inducers. Mol. Microbiol. 57:1797-1807. [DOI] [PubMed] [Google Scholar]
- 31.Ma, K., and M. W. Adams. 1999. An unusual oxygen-sensitive, iron- and zinc-containing alcohol dehydrogenase from the hyperthermophilic archaeon Pyrococcus furiosus. J. Bacteriol. 181:1163-1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ma, K., A. Hutchins, S. J. Sung, and M. W. Adams. 1997. Pyruvate ferredoxin oxidoreductase from the hyperthermophilic archaeon, Pyrococcus furiosus, functions as a CoA-dependent pyruvate decarboxylase. Proc. Natl. Aca. Sci. USA 94:9608-9613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ma, K., H. Loessner, J. Heider, M. K. Johnson, and M. W. Adams. 1995. Effects of elemental sulfur on the metabolism of the deep-sea hyperthermophilic archaeon Thermococcus strain ES-1: characterization of a sulfur-regulated, non-heme iron alcohol dehydrogenase. J. Bacteriol. 177:4748-4756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ma, K., R. N. Schicho, R. M. Kelly, and M. W. Adams. 1993. Hydrogenase of the hyperthermophile Pyrococcus furiosus is an elemental sulfur reductase or sulfhydrogenase: evidence for a sulfur-reducing hydrogenase ancestor. Proc. Natl. Acad. Sci. USA 90:5341-5344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ma, K., R. Weiss, and M. W. Adams. 2000. Characterization of hydrogenase II from the hyperthermophilic archaeon Pyrococcus furiosus and assessment of its role in sulfur reduction. J. Bacteriol. 182:1864-1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Musfeldt, M., M. Selig, and P. Schonheit. 1999. Acetyl coenzyme A synthetase (ADP forming) from the hyperthermophilic archaeon Pyrococcus furiosus: identification, cloning, separate expression of the encoding genes, acdAI and acdBI, in Escherichia coli, and in vitro reconstitution of the active heterotetrameric enzyme from its recombinant subunits. J. Bacteriol. 181:5885-5888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nath, K., and D. Das. 2004. Improvement of fermentative hydrogen production: various approaches. Appl. Microbiol. Biotechnol. 65:520-529. [DOI] [PubMed] [Google Scholar]
- 38.Ogino, H., T. Miura, K. Ishimi, M. Seki, and H. Yoshida. 2005. Hydrogen production from glucose by anaerobes. Biotechnol. Prog. 21:1786-1788. [DOI] [PubMed] [Google Scholar]
- 39.Pysz, M. A., S. B. Conners, C. I. Montero, K. R. Shockley, M. R. Johnson, D. E. Ward, and R. M. Kelly. 2004. Transcriptional analysis of biofilm formation processes in the anaerobic, hyperthermophilic bacterium Thermotoga maritima. Appl. Environ. Microbiol. 70:6098-6112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rinker, K. D., and R. M. Kelly. 2000. Effect of carbon and nitrogen sources on growth dynamics and exopolysaccharide production for the hyperthermophilic archaeon Thermococcus litoralis and bacterium Thermotoga maritima. Biotechnol. Bioeng 69:537-547. [DOI] [PubMed] [Google Scholar]
- 41.Rinker, K. D., and R. M. Kelly. 1996. Growth physiology of the hyperthermophilic archaeon Thermococcus litoralis: development of a sulfur-free defined medium, characterization of an exopolysaccharide, and evidence of biofilm formation. Appl. Environ. Microbiol. 62:4478-4485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Roy, R., and M. W. Adams. 2002. Characterization of a fourth tungsten-containing enzyme from the hyperthermophilic archaeon Pyrococcus furiosus. J. Bacteriol. 184:6952-6956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rupprecht, J., B. Hankamer, J. H. Mussgnug, G. Ananyev, C. Dismukes, and O. Kruse. 2006. Perspectives and advances of biological H2 production in microorganisms. Appl. Microbiol. Biotechnol. 72:442-449. [DOI] [PubMed] [Google Scholar]
- 44.Sakuraba, H., S. Goda, and T. Ohshima. 2004. Unique sugar metabolism and novel enzymes of hyperthermophilic archaea. Chem. Rec. 3:281-287. [DOI] [PubMed] [Google Scholar]
- 45.Sapra, R., K. Bagramyan, and M. W. Adams. 2003. A simple energy-conserving system: proton reduction coupled to proton translocation. Proc. Natl. Acad. Sci. USA 100:7545-7550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schicho, R. N., K. Ma, M. W. Adams, and R. M. Kelly. 1993. Bioenergetics of sulfur reduction in the hyperthermophilic archaeon Pyrococcus furiosus. J. Bacteriol. 175:1823-1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schroder, C., M. Selig, and P. Schonheit. 1994. Glucose fermentation to acetate, CO2 and H2 in the anaerobic hyperthermophilic eubacterium Thermotoga maritima—involvement of the Embden-Meyerhof pathway. Arch. Microbiol. 161:460-470. [Google Scholar]
- 48.Schut, G. J., S. D. Brehm, S. Datta, and M. W. Adams. 2003. Whole-genome DNA microarray analysis of a hyperthermophile and an archaeon: Pyrococcus furiosus grown on carbohydrates or peptides. J. Bacteriol. 185:3935-3947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schut, G. J., J. Zhou, and M. W. Adams. 2001. DNA microarray analysis of the hyperthermophilic archaeon Pyrococcus furiosus: evidence for an new type of sulfur-reducing enzyme complex. J. Bacteriol. 183:7027-7036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shockley, K. R. 2004. Functional genomics investigation of microbial physiology in the hyperthermophilic microorganism Pyrococcus furiosus and Thermotoga maritima. Ph. D. thesis. North Carolina State University, Raleigh.
- 51.Shockley, K. R., D. E. Ward, S. R. Chhabra, S. B. Conners, C. I. Montero, and R. M. Kelly. 2003. Heat shock response by the hyperthermophilic archaeon Pyrococcus furiosus. Appl. Environ. Microbiol. 69:2365-2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Silva, P. J., E. C. van den Ban, H. Wassink, H. Haaker, B. de Castro, F. T. Robb, and W. R. Hagen. 2000. Enzymes of hydrogen metabolism in Pyrococcus furiosus. Eur. J. Biochem. 267:6541-6551. [DOI] [PubMed] [Google Scholar]
- 53.Tachdjian, S., and R. M. Kelly. 2006. Dynamic metabolic adjustments and genome plasticity are implicated in the heat shock response of the extremely thermoacidophilic archaeon Sulfolobus solfataricus. J. Bacteriol. 188:4553-4559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.van der Oost, J., W. G. Voorhorst, S. W. Kengen, A. C. Geerling, V. Wittenhorst, Y. Gueguen, and W. M. de Vos. 2001. Genetic and biochemical characterization of a short-chain alcohol dehydrogenase from the hyperthermophilic archaeon Pyrococcus furiosus. Eur. J. Biochem. 268:3062-3068. [DOI] [PubMed] [Google Scholar]
- 55.van Haaster, D. J., P. L. Hagedoorn, J. A. Jongejan, and W. R. Hagen. 2005. On the relationship between affinity for molecular hydrogen and the physiological directionality of hydrogenases. Biochem. Soc. Trans. 33:12-14. [DOI] [PubMed] [Google Scholar]
- 56.van Lieshout, J., M. Faijes, J. Nieto, J. van der Oost, and A. Planas. 2004. Hydrolase and glycosynthase activity of endo-1,3-beta-glucanase from the thermophile Pyrococcus furiosus. Archaea 1:285-292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.van Niel, E. W., P. A. Claassen, and A. J. Stams. 2003. Substrate and product inhibition of hydrogen production by the extreme thermophile, Caldicellulosiruptor saccharolyticus. Biotechnol. Bioeng 81:255-262. [DOI] [PubMed] [Google Scholar]
- 58.Van Ooteghem, S. A., S. K. Beer, and P. C. Yue. 2002. Hydrogen production by the thermophilic bacterium Thermotoga neapolitana. Appl. Biochem. Biotechnol. 98-100:177-189. [DOI] [PubMed] [Google Scholar]
- 59.Voorhorst, W. G., R. I. Eggen, E. J. Luesink, and W. M. de Vos. 1995. Characterization of the celB gene coding for beta-glucosidase from the hyperthermophilic archaeon Pyrococcus furiosus and its expression and site-directed mutation in Escherichia coli. J. Bacteriol. 177:7105-7111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ward, D. E., S. W. Kengen, J. van Der Oost, and W. M. de Vos. 2000. Purification and characterization of the alanine aminotransferase from the hyperthermophilic archaeon Pyrococcus furiosus and its role in alanine production. J. Bacteriol. 182:2559-2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Williams, E., T. M. Lowe, J. Savas, and J. Diruggiero. 2007. Microarray analysis of the hyperthermophilic archaeon Pyrococcus furiosus exposed to gamma irradiation. Extremophiles 11:19-29. [DOI] [PubMed] [Google Scholar]
- 62.Xavier, K. B., R. Peist, M. Kossmann, W. Boos, and H. Santos. 1999. Maltose metabolism in the hyperthermophilic archaeon Thermococcus litoralis: purification and characterization of key enzymes. J. Bacteriol. 181:3358-3367. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.