Abstract
A Saccharomyces cerevisiae strain, capable of autonomous bioluminescence, was engineered to respond to androgenic chemicals. The strain, S. cerevisiae BLYAS, contains the human androgen receptor in the chromosome and was constructed by inserting a series of androgen response elements between divergent yeast promoters GPD and ADH1 on pUTK401 that constitutively expressed luxA and luxB to create pUTK420. Cotransformation of this plasmid with a second plasmid (pUTK404), containing the genes required for aldehyde synthesis (luxCDE) and FMN reduction (frp), yielded a bioluminescent bioreporter responsive to androgenic chemicals. Using dihydrotestosterone (DHT) as a standard, the response time and the 50% effective concentration values were 3 to 4 h and (9.7 ± 4.6) × 10−9 M, respectively. The lower limit of detection in response to DHT was 2.5 × 10−9 M, and in response to testosterone it was 2.5 × 10−10 M. This strain is suitable for high-throughput screening of chemicals with potential for remote environmental monitoring systems because of the assay speed, sensitivity, and self-containment.
Many anthropogenic and naturally occurring chemicals are suspected to cause endocrine disruption in humans and wildlife, including androgens and androgen-mimicking chemicals (1, 14, 19). The agonist activity of androgens, required for normal sexual development in males, is mediated by the androgen receptor protein (AR). When the AR binds to an androgenic compound, the complex moves to the nucleus, whereupon it binds to androgen response elements (AREs) that mediate specific gene expression. Numerous biological functions are mediated by this system, including facial hair development, increased muscle mass, spermatogenesis, and vocal cord enlargement (19), but when present in excess, androgens pose potential problems for vertebrates. Evidence suggests that in wildlife, environmental exposure to low levels of androgen-mimicking chemicals may alter sex ratios and lead to endocrine dysfunction (1). Indeed, endocrine-disrupting chemicals (estrogens, androgens, progestins, etc.) have been detected in waterways and municipal wastewater effluents at concentrations up to 7.4 × 10−10 M (10, 11), and the data suggest that these chemicals alter the reproductive systems of fish. In a U.S. Geological Survey study, low concentrations of hormones were detected in more than 40% of 139 U.S. streams tested (11), with testosterone present in 2.8% and cis-androsterone present in 14.3% of 70 samples that were tested for these two compounds.
Due to growing concerns over environmental impacts of endocrine-disrupting compounds, the U.S. Congress directed the Environmental Protection Agency to issue a mandate to develop rapid, sensitive detectors for hormone-mimicking chemicals (Food Quality Protection Act, Public Law 104-170). In response, a number of androgen-responsive reporters have been developed, in both yeast and mammalian cells. For example, Purvis et al. (15) developed the Saccharomyces cerevisiae yeast androgen screen (YAS), a colorimetric (lacZ) yeast-based bioreporter containing a reporter plasmid capable of converting the chromogenic substrate chlorophenol red-β-d-galactopyranoside from yellow to red, detectable with spectrophotometry. This assay requires 3 to 5 days for color development. Another yeast-based androgen reporter utilizes firefly luciferase (luc) genes, which require the addition of an exogenous substrate (d-luciferin) for detection of the bioluminescent reaction (13). Moreover, all the mammalian cell-based androgen reporters engineered to date require at least 24 h postinduction prior to luminescence detection, as well as a series of washing steps and/or tissue culture methods for maintenance. The utility of bioluminescent (lux) bioreporters for chemical detection has been well-established in bacteria (9, 12) and more recently in yeast (6, 18) because of the ease of culture maintenance, luminescence without the addition of substrates, and the speed of the assay. Indeed, they have become as useful as, and in some ways more useful than, green fluorescent protein-based and colorimetric reporters for chemical detection (3). In this paper, the construction of a new bioluminescent yeast-based bioreporter, based on the Photorhabdus luminescens lux operon, for the detection of androgens is described which is suitable for high-throughput screening of chemicals and remote environmental monitoring.
MATERIALS AND METHODS
Strains, plasmids, and growth conditions.
Strains and plasmids used in this study are listed in Table 1. Escherichia coli TOP10 (Invitrogen, Carlsbad, CA), used as a host for plasmid construction and maintenance, once transformed, was grown in Luria-Bertani broth with 50 μg ampicillin/ml at 37°C. YPD liquid medium (1% yeast extract, 2% peptone, 2% glucose) was used for routine growth of plasmid-free S. cerevisiae. S. cerevisiae strains harboring plasmids with leucine- and uracil-selective markers were grown in modified minimal medium without leucine and uracil (YMM leu−, ura−) (16) to ensure the maintenance of plasmids within the yeast cells by complementation of leucine and uracil auxotrophies.
TABLE 1.
E. coli and S. cerevisiae strains and plasmids used in this study
| Strain or plasmid | Description | Source or reference |
|---|---|---|
| E. coli strain | ||
| TOP10 | F−mcrA Δ(mrr-hsdRMS-mcrBC) φ80lacZΔM15 ΔlacX74 recA1 araD139 Δ(ara-leu)7697 galU galK rpsL endA1 nupG Strr | Invitrogen |
| S. cerevisiae strains | ||
| YAS (hAR-lacZ) | MATα prb1-1122 pep4-3 leu2 trp1 ura3-52 gal2 | 16 |
| hAR | Strain YAS without the ARE-lacZ reporter plasmid | 16 |
| BLYRa | S. cerevisiae hAR containing pUTK401 and pUTK404 | This study |
| BLYAS1 | S. cerevisiae hAR containing pUTK417 and pUTK404 | This study |
| BLYAS2 | S. cerevisiae hAR containing pUTK418 and pUTK404 | This study |
| BLYAS3 | S. cerevisiae hAR containing pUTK419 and pUTK404 | This study |
| BLYAS4 | S. cerevisiae hAR containing pUTK420 and pUTK404 | This study |
| Plasmids | ||
| pUTK401 | pUA12B7 (pBEVY-U containing luxA and luxB) | 6 |
| pUTK404 | pLCIRESDEIRESfrp (pBEVY-L harboring luxCDEfrp) | 6 |
| pUTK416 | pUTK401 containing BglII and KasI restriction sites between GPD and ADH1 constitutive promoters | This study |
| pUTK417 | pUTK416 containing one ARE between GPD and ADH1 constitutive promoters | This study |
| pUTK418 | pUTK416 containing two AREs between GPD and ADH1 constitutive promoters | This study |
| pUTK419 | pUTK416 containing three AREs between GPD and ADH1 constitutive promoters | This study |
| pUTK420 | pUTK416 containing four AREs between GPD and ADH1 constitutive promoters | This study |
Chemicals.
The chemicals dihydrotestosterone (DHT; CAS no. 521-18-6; ≥99% purity), testosterone (CAS 55-22-; ≥98% purity), trenbolone (CAS 10161-33-8; ≥98% purity), 4-androstenedione (androstenedione; CAS 63-05-8; ≥98% purity), mifepristone (CAS 84371-65-3; ≥98% purity), fluoxymesterone (CAS 76-43-7), medroxyprogesterone acetate (CAS 71-58-9; ≥97% purity), norgestrel (CAS 797-63-7; ≥99% purity), spironolactone (CAS 52-01-7), 17β-estradiol (CAS 50-28-2; ≥98% purity), p-nonylphenol (CAS 104-40-5), and flutamide (CAS 13311-84-7) were purchased from Sigma-Aldrich/Fluka Chemical Company (St. Louis, MO). Stock solutions of each chemical were made in high-performance liquid chromatography-grade methanol (Fisher Scientific). Chlorophenol red-β-d-galactopyranoside was purchased from Fisher Scientific.
Molecular biology techniques.
DNA manipulations were performed according to standard protocols (17). Plasmids were transformed into chemically competent E. coli TOP10 cells (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions, while S. cerevisiae strains were made competent and transformed according to the methods of Thompson et al. (23). For S. cerevisiae strains, electroporation conditions were as follows: charging voltage of 1.5 kV, resistance of 200 Ω, capacitance of 25 μF, in an ECM600 apparatus (BTX Inc., Holliston, MA). Yeast cells (40 μl equaling approximately 4 × 108 to 8 × 108 cells) were transformed with 300 ng of each plasmid DNA, with addition of 1 ml cold 1 M sorbitol after transformation. Cells were immediately plated onto YMM (leu− ura−) Noble agar plates.
Plasmid isolation was performed using an Eppendorf Fast plasmid isolation kit (Eppendorf, Westbury, NY) or a Wizard midi-prep kit (Promega, San Luis Obispo, CA). PCR was performed in 25-μl volumes using Ready-to-Go PCR beads (Amersham Pharmacia Biotech, Inc., Piscataway, NJ), a 200 nM concentration of each primer, and 50 ng template (unless otherwise noted). Oligonucleotide primer sequences are listed in Table 2. Cycling conditions for all amplifications were as follows: 94°C for 10 min of initial denaturation, followed by 20 cycles of denaturation at 94°C for 30 s, annealing at 65°C for 30 s, and extension at 72°C for 45 s. The annealing temperature was reduced by 0.5°C in each of the first 20 cycles. This was followed by 15 cycles of denaturation at 94°C for 30 s, annealing at 55°C for 30 s, and extension at 72°C for 45 s, and then a final elongation step at 72°C for 10 min. DNA sequencing was performed with an ABI Big Dye Terminator cycle sequencing reaction kit and an ABI 3100 DNA sequencer (Perkin-Elmer, Inc., Foster City, CA). Restriction enzymes were purchased from either New England Biolabs (BglII and KasI; Beverly, MA) or Promega Corporation (BamHI and SpeI; Madison, WI).
TABLE 2.
Oligonucleotide primer sequences used for the construction of plasmids pUTK417, pUTK418, pUTK419, and pUTK420
| Designation | Primer sequence (5′ to 3′)a |
|---|---|
| GPDR | GTCGAAACTAAGTTCTTGGTGTTTTAAAAC |
| ADH1R2 | AGTTGATTGTATGCTTGGTATAGCTTGAAATATTGTGC |
| GPDBglII18ntF | GATAATGATAAACTCCCGGagatctctactactactactactag |
| 3ntBamHIGPDR | cccGGATCCGTCGAAACTAAGTTCTTGGTGTTTTAAAAC |
| 18ntKasIADH1F | tctactactactactactaggcgccATTCTTTTCTTT |
| TTTTTTCTTTTCTCTCTCC | |
| ADH1SpeI3ntR | gggACTAGTCCCGGGGAGTTGATTGTATGCTTGGTATAGCTTGAAATATTGTGC |
Underlined bases represent restriction sites; lowercase letters indicate intervening sequences added to facilitate restriction digestion.
Construction of strains BLYAS and BLYRa.
S. cerevisiae strain BLYAS contains the human AR (hAR) gene in the chromosome, the plasmid pUTK404, and either pUTK417, pUTK418, pUTK419, or pUTK420 (pUTK401 [6] containing one to four AREs; see below). The construction of pUTK404, containing the luxC, luxD, and luxE genes from Photorhabdus luminescens and the flavin oxidoreductase gene (frp) from Vibrio harveyi encoding the FMNH2 cofactor required for the bioluminescent reaction, has been described previously (6).
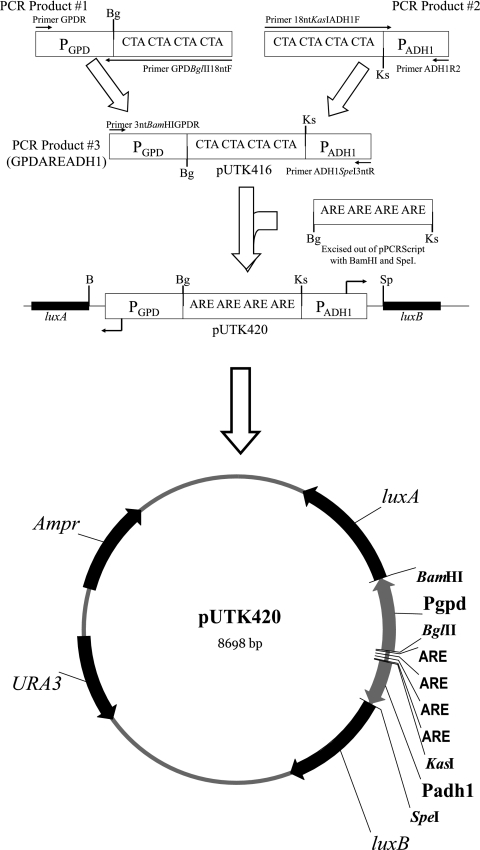
The androgen-responsive plasmids pUTK417, pUTK418, pUTK419, and pUTK420 were based on pUTK401, which contains the luxA and luxB genes constitutively expressed from the bidirectional promoter element GPD-ADH1 and has been described previously (6). The construction of the androgen-responsive luxA/luxB plasmids was mediated via pUTK416 (Fig. 1). To construct pUTK416, sequential amplifications were used to insert a fragment containing the BglII and KasI restriction sites with an intervening CTA repeat (18-mer spacer region) between the two constitutive promoters, GPD and ADH1 of pUTK401. In the first amplification, the promoter GPD was amplified using GPDBglII18ntF and GPDR primers. Primer GPDBglII18ntF contains a 5′ BglII restriction site and an 18-bp CTA repeat (Table 2). In the second amplification, promoter ADH1 was amplified using the primers 18ntKasIADH1F and ADH1R2. Primer 18ntKasIADH1F contains a 5′ KasI restriction site and an 18-bp CTA repeat (Table 2). The two amplifications were performed in quadruplicate and were each combined and gel purified (QIAquick gel extraction kit; QIAGEN Inc., Valencia, CA). Next, 75 ng of each product was used in a third round of PCR, along with 3ntBamHIGPDR and ADH1SpeI3ntR primers, to generate the fragment BamHI-GPD-BglII-18nt-KasI-ADH1-SpeI. Primer 3ntBamHIGPDR contains a 5′ BamHI restriction site with a 3-bp overhang to facilitate digestion, while primer ADH1SpeI3ntR contains a 5′ SpeI restriction site as well as a 3-bp overhang (Table 2). After gel purification, this product was double digested with BamHI and SpeI and then ligated into the BamHI and SpeI sites of pUTK401, generating pUTK416.
FIG. 1.
Cloning strategy for pUTK416 to -420. Sequential amplifications were used to generate GPDAREADH1. This construct was directionally cloned into the BamHI (B) and SpeI (Sp) site of pUTK401, generating pUTK416. A series of AREs (one to four) was directionally cloned into the BglII (Bg) and KasI (Ks) sites on pUTK416, yielding pUTK417 (one ARE), pUTK418 (two AREs), pUTK419 (three AREs), and pUTK420 (four AREs).
A series of ARE-containing fragments with one to four repeats of AGAACActaTGTTCT, bordered by 5′ BglII and 3′ KasI restriction sites, were synthesized and cloned into pPCRScript by Geneart Inc. (Toronto, Canada). Numerous ARE sequences exist in the human genome, but the ARE sequence chosen has been demonstrated to recruit more AR than the ARE of prostate-specific antigen (8). Each ARE was separated by a 12-bp CTA repeat, since this was reported to be the optimum number of bases between response elements (7). Four sequential rounds of amplification were performed on these plasmids using T3 and T7 primers, with 15 ng of target template in the initial reaction mixture and 2 μl (∼80 ng) of the previous PCR product used as the template in subsequent reactions. Cycling conditions were as described above. PCR products were double digested with BglII and KasI and cleaned with a QIAGEN nucleotide removal kit (QIAGEN Inc., Valencia, CA), which retains fragments as small as 17 bp. These products were ligated into the BglII and KasI sites on pUTK416 to generate the androgen-responsive luxA/luxB plasmids pUTK417 to pUTK420.
An S. cerevisiae androgen reporter (YAS), which contains the hAR gene on its genome and a colorimetric, androgen-inducible (leu2+) reporter plasmid, was used as the host strain (15). This strain was cured of its reporter plasmid by replica plating onto YPD medium, followed by confirmation of no growth on medium without leucine, and was designated S. cerevisiae hAR. Subsequently, S. cerevisiae hAR was transformed with pUTK401 and pUTK404 to create a constitutive bioluminescent reporter (BLYRa), which was used to monitor toxicity of chemicals. S. cerevisiae hAR was separately cotransformed with pUTK404 and either pUTK417 (one ARE), pUTK418 (two AREs), pUTK419 (three AREs), or pUTK420 (four AREs) to generate four different androgen-inducible bioluminescent bioreporters.
Bioluminescent androgen assay.
For chemical testing, 20 μl of appropriate chemical dilutions in methanol was spotted into wells of black 96-well microtiter plates (Dynex Technologies, Chantilly, VA) using a Beckman F/X automated liquid handling system. After spotting each chemical, the methanol was evaporated at room temperature. Each microtiter plate also contained dilutions of the standard DHT, a series of no-treatment control wells, and a series of methanol-only control wells. S. cerevisiae strains were grown in YMM (leu−, ura−) at 30°C with shaking overnight to an optical density at 600 nm of ∼1.0. Aliquots (200 μl) of each isolate were transferred to each well of a microtiter plate. Bioluminescence was measured every hour for 12 h in a Microbeta Plus liquid scintillation counter (Perkin-Elmer, Wellesley, MA) with an integration time of 1 s/well.
Known androgenic and estrogenic/progestagenic chemicals were tested at concentrations from 1 μM to 2.5 pM and 1 mM to 2.5 nM, respectively. In addition to S. cerevisiae BLYAS with four AREs, the toxicity of each chemical was monitored with the constitutive bioluminescence strain S. cerevisiae BLYRa.
As a preliminary test of the S. cerevisiae BLYAS and BLYRa reporters in a natural setting, the two strains were separately grown in YMM (leu−, ura−) to an optical density at 600 nm of ∼1.0 and then centrifuged and resuspended in a 2× concentrated medium. Cells (100 μl) were then added to the wells of a 96-well plate, to which 100 μl of filter-sterilized (0.22 μm) local creek water or medium plus testosterone or DHT was added (to achieve a concentration range of 1 μM to 2.5 pM), in quadruplicate.
EC50 calculations and statistical analysis.
For each chemical, bioluminescence (counts per second) versus the log of chemical concentration was plotted using luminescence values after 3 to 4 hours of incubation. A linear regression was determined using points falling on the linear portion of the curve. Each 50% effective concentration (EC50) (x axis) was calculated using the linear regression formula and the midpoint y value of the standard (DHT) concentration-response curve for the same plate. For mifepristone, whose maximum luminescence never reached the midpoint luminescence value of the DHT curve, the midpoint y value of the mifepristone curve was used to calculate the EC50. For the DHT standards, the EC50 was calculated individually for 17 independent assays, while for all other chemicals the EC50 was calculated based on three independent assays. The mean and standard deviation values were calculated from the replicate EC50 values for all chemicals to determine the variability between assays (see Table 4, below).
TABLE 4.
EC50 values of select chemicals obtained with S. cerevisiae BLYAS with four AREsa
| Chemical | nb | BLYAS EC50 (M) | BLYAS IC20c (M) |
|---|---|---|---|
| DHT | 17 | (9.7 ± 4.6) × 10−9 | NDe |
| Testosterone | 3 | (4.6 ± 0.84) × 10−9 | ND |
| Androstenedione | 3 | (1.0 ± 0.16) × 10−7 | ND |
| Trenbolone | 3 | (1.8 ± 0.11) × 10−8 | ND |
| Norgestrel | 3 | (1.6 ± 0.59) × 10−7 | ND |
| Spironolactone | 3 | (9.5 ± 3.9) × 10−6 | ND |
| 17β-Estradiol | 3 | (3.7 ± 1.8) × 10−5 | ND |
| Mifepristone (RU486) | 3 | (3.8 ± 0.97) × 10−6 | ND |
| Fluoxymesterone | 3 | (7.2 ± 2.2) × 10−8 | (4.69 ± 4.1) × 10−3 |
| MPA | 3 | (4.6 ± 2.6) × 10−6 | ND |
| Flutamide | 1 | NRd | ND |
| p-Nonylphenol | 1 | NR | 3.06 × 10−4 |
S. cerevisiae BLYAS was incubated for 3 to 4 h in a series of concentrations of each chemical, and then a linear regression was used to determine the EC50 for each. EC50 values represent the means and standard deviations of n replicates.
The number of independent replicates analyzed on different days using separate aliquots of S. cerevisiae BLYAS with four AREs.
IC20, 20% inhibitory concentration.
NR, no response.
ND, not determined because toxicity was not detected at the concentrations utilized.
For comparison between strains, induction was calculated for each of the eight isolates by dividing the maximum luminescence value by the minimum luminescence value. The average induction of each strain was compared by performing a one-way analysis of variance (SPSS v.15; Chicago, IL), with post hoc tests (Tukey analysis) and an F statistic calculation at the α = 0.05 significance level.
RESULTS
Comparison of BLYAS strains.
An androgen-inducible bioluminescent bioreporter, analogous to the recently described estrogen bioreporter S. cerevisiae BLYES (18), was constructed. Multiple androgen response elements were inserted between the two constitutive divergent promoters GPD and ADH1 on pUTK416, regulating luxA and luxB, respectively. The colorimetric androgen reporter S. cerevisiae YAS was cured of its lacZ reporter plasmid and then transformed with pUTK404 (6) and either pUTK417 (one ARE), pUTK418 (two AREs), pUTK419 (three AREs), or pUTK420 (four AREs). Each ARE is a 15-bp perfect palindrome, arranged with two hexameric, inverted repeat, half sites and a 3-bp spacer region (8). These palindromic sequences should each form a hairpin and function to repress the constitutive GPD and ADH1 promoters; therefore, with an increasing number of ARE fragments, the operon was expected to become more tightly regulated.
Minimum and maximum bioluminescence changes were the only differences observed among the four variations of the ARE construct. This was demonstrated by comparing the induction (ratio of the maximum and minimum bioluminescence values) between the strains after incubation in a range of DHT concentrations (1 mM to 1 pM) (Table 3). A one-way analysis of variance was performed on the induction values of eight isolates of each strain, generating an F statistic (values of 3, 28) of 16.833 (P < 0.001). Post hoc analysis (Tukey) was used to determine whether differences in induction occurred between strains. The induction over background observed with one and two AREs was 2.1 and 2.6, respectively, but this was not significantly different (P > 0.05). However, there was a significant increase in induction with three (5.2) and four (6.6) AREs over one and two AREs (P < 0.05). In addition, there was a proportionate but nonsignificant increase (P > 0.05) in both induction and maximum luminescence with four AREs over three AREs. Moreover, the background (minimum) bioluminescence of the reporters with three and four AREs was half that of the reporters with one and two AREs. These results indicated that access to the promoters was much more restricted with three and four AREs present and provided tighter regulation of this element.
TABLE 3.
Comparison of four S. cerevisiae BLYAS strainsa
| No. of AREs in strain | Maximum luminescence | Minimum luminescence | Fold inductionb |
|---|---|---|---|
| 1 | (1.6 ± 0.51) × 105 | (7.8 ± 2.8) × 104 | 2.1 (A) |
| 2 | (2.0 ± 0.15) × 105 | (7.7 ± 0.9) × 104 | 2.6 (A) |
| 3 | (1.4 ± 0.44) × 105 | (3.0 ± 1.1) × 104 | 5.2 (B) |
| 4 | (2.4 ± 0.32) × 105 | (4.1 ± 1.7) × 104 | 6.6 (B) |
S. cerevisiae BLYAS with one to four AREs (eight isolates of each) was incubated for 4 hours in a range of DHT concentrations. Maximum and minimum luminescence values represent the means of eight isolates of each strain. Average fold induction was calculated for each strain and is indicated.
Letters in parentheses indicate statistical significance, values followed by an A were significantly different from values followed by a B.
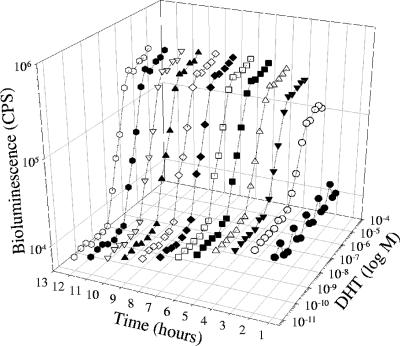
The strain with four AREs was chosen as the best androgen bioreporter, based on having isolates with both the highest induction and the highest maximum luminescence. Additional colonies of the isolate with the largest induction were screened for maximum and minimum bioluminescence and then induction values were calculated. This process was repeated twice, using colonies of the isolate with the largest induction in the next round of screening, which yielded a strain of S. cerevisiae with four AREs having a 17-fold increase in induction after 4 hours and a 30- to 40-fold increase in induction after 8 to 12 h (Fig. 2). This strain was then designated S. cerevisiae BLYAS, grown overnight (60 ml), and then frozen in glycerol to make aliquots for subsequent testing.
FIG. 2.
Three-dimensional plot of bioluminescence versus time for DHT. Initial bioluminescence was observed within 2 hours for 1.0 × 10−8 M and reached a maximum at approximately 6 hours.
Concentration-response profiles for selected chemicals.
S. cerevisiae BLYAS was incubated in a range of DHT concentrations (1 μM to 2.5 pM), and the concentration response was monitored every hour for 12 h (Fig. 2). Maximum bioluminescence was achieved by 6 h and quantifiable bioluminescence occurred within 2 h, which is similar to the estrogen reporter strain BLYES with the 17β-estradiol standard (18). At 4 h, the lower limit of detection was 2.5 × 10−10 to 5.0 × 10−10 DHT (defined as background luminescence plus 3 standard deviations), which is similar to the luminescent (luc-based) S. cerevisiae androgen reporter of Michelini et al. (13) (detection limit of 5 × 10−10). S. cerevisiae BLYAS was used to determine EC50 values for 12 chemicals (Table 4). The reproducibility of the assays was determined by calculating the EC50 values from 3 to 17 independent experiments performed on separate aliquots grown on different days.
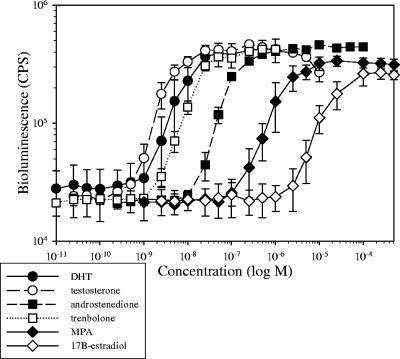
Of the 12 chemicals tested, S. cerevisiae BLYAS demonstrated a sigmoidal response to the known androgens DHT, testosterone, androstenedione, and trenbolone (Fig. 3), norgestrel, fluoxymesterone, the progesterones medroxyprogesterone acetate, mifepristone, and spironolactone (also an AR antagonist), and also to the estrogen 17β-estradiol (Table 4). In contrast, the AR antagonists flutamide and p-nonylphenol had no activity in this assay, with p-nonylphenol producing a toxic response above 10 μM, as demonstrated by decreased luminescence with the constitutive S. cerevisiae BLYRa strain (data not shown). The negative response to flutamide was expected, since it has been previously demonstrated to have no activity in another androgen agonist assay but was a potent antiandrogen in an antagonist assay (5). Spironolactone, however, displays both agonist and antagonist activity in other reporter assays (2) and showed some activity in our agonist assay, despite being considered an antiandrogen.
FIG. 3.
EC50-response profile of select chemicals using S. cerevisiae BLYAS. Data were obtained 4 hours postinduction and represent the means and standard deviations of 17 replicates for DHT and 3 replicates for all other chemicals.
A preliminary test was performed to determine how S. cerevisiae BLYAS responds in an environmental water sample relative to laboratory medium. Incubation in a filtered creek water spiked with DHT or testosterone yielded a typical concentration-response profile (data not shown), with an EC50 and detection limit of 2.0 × 10−8 M and 1.0 × 10−9 M for DHT, respectively, and 1.3 × 10−8 M and 2.5 × 10−9 M for testosterone, respectively (Table 5). These values are similar to data reported in Table 4. These limits of detection are in the concentration range of androgens found in the environment (10, 11), though further work is needed to determine if concentrating the samples would enhance detection of androgens in environmental samples.
TABLE 5.
Comparison of EC50 values in laboratory medium versus creek water
| Chemical | Laboratory medium
|
Creek water
|
||
|---|---|---|---|---|
| EC50a (M) | Limit of detection (M) | EC50 (M) | Limit of detection (M) | |
| Testosterone | (1.7 ± 0.35) × 10−8 | 2.5 × 10−9 | (1.3 ± 0.39) × 10−8 | 2.5 × 10−9 |
| DHT | (1.2 ± 0.97) × 10−8 | 5.0 × 10−10 | (2.0 ± 0.25) × 10−8 | 5.0 × 10−10 |
EC50 values represent means and standard deviations.
DISCUSSION
Androgen bioassays typically belong to one of four types: cell-free competitive binding assays (4), colorimetric (lacZ) reporter yeast-based assays (5, 20), luminescent (luc or lux) reporter yeast-based assays (13; this study), or luminescent (luc) mammalian cell-based assays (2, 21, 24). The different assays are difficult to compare because they have different mechanisms of uptake and expression times. However, even if all the assays showed equivalent responses to androgenic compounds, luminescent yeast-based assays (both luc and lux) would have an advantage of speed over both bioluminescent mammalian cell-based (luc) and colorimetric yeast-based (lacZ) assays. S. cerevisiae BLYAS quantifiable bioluminescence occurs in 1 to 2 h postinduction, with maximum luminescence occurring by 6 h (Fig. 2). In contrast, the mammalian cell-based androgen reporter assays require a 24-h incubation postinduction to process the cells for luminescence detection, and colorimetric yeast-based reporters require 1 to 5 days (5, 15) to respond to androgens. In addition, yeast-based reporters offer the distinct advantage of being easy to handle, not requiring the level of manipulation needed for the processing of mammalian cells. S. cerevisiae BLYAS cultures can be processed without additional manipulation, and luminescence detection can begin almost immediately after exposure to chemical. Moreover, luc-based (firefly luminescence) mammalian cell reporters and the luc-based yeast bioluminescent reporter (13) require the addition of the substrate luciferin to be able to detect luminescence. Therefore, yeast-based reporters that utilize bacterial luminescence (lux) genes are well suited to high-throughput testing because of both ease of use and the speed of the assays.
Recently published, perfect palindromic androgen response elements were used to construct a new bioluminescent bioreporter for the detection of androgenic compounds. Horie- Inoue et al. (8) identified and analyzed 563 exact consensus AREs present in the human genome. The ARE chosen for this study was a 15-bp perfect palindrome arranged with two hexameric inverted repeat half-sites separated by 3 bp and was selected because it recruited more AR than the proximal ARE of prostate-specific antigen (8). Further, Haendler et al. (7) reported greater induction of their reporter plasmids when two to four copies of an ARE were used and separated by a 12-bp spacer region (e.g., AGAACActaTGTTCTctactactactaAGAACActaTGTTCT). Conversely, the YAS assay of Purvis et al. (15) used three copies of a different ARE separated by a 1-bp spacer. In this study, multiple copies of the perfect palindromic ARE were tested by constructing bioluminescent S. cerevisiae androgen reporters containing one to four AREs. It was expected that the number of AREs would affect either the response sensitivity (EC50) or the level of induction in response to DHT. Theoretically, each ARE should form a hairpin, thereby decreasing access to the constitutive GPD and ADH1 promoters. Among the four versions tested, induction was variable within each strain, as has been reported in other studies (21, 24). More importantly, induction, but not sensitivity, was affected by the tighter regulation of the increasing number of AREs. Similarly, Sonneveld et al. (21) noted that changes in induction with the AR CALUX reporter did not influence their ability to calculate the androgenic potency of a chemical; also, the changes in induction were mainly due to small differences in the low-range background having a large effect on the induction calculation. Since S. cerevisiae BLYAS with four AREs had isolates with the highest induction and maximum luminescence observed, it was chosen for further testing.
The response of S. cerevisiae BLYAS to the known androgen agonist DHT was first tested by comparison with the colorimetric S. cerevisiae YAS using a range of DHT concentrations. Despite a change in both the number and sequence of AREs on the reporter plasmid and a change from a 1-bp spacer between AREs to 12 bases, the EC50 values calculated for S. cerevisiae BLYAS and S. cerevisiae YAS were similar, (9.7 ± 4.6) × 10−9 and (7.4 ± 2.1) × 10−8, respectively. S. cerevisiae BLYAS performed similarly to many other androgen-responsive reporters described in the literature, having mid-range EC50 values for tested androgenic, estrogenic, and progestagenic chemicals (Tables 4 and 6). For most chemicals tested, the variability of this assay was similar to other assays, which typically range from 22 to 23% among the groups reporting this variability (Table 4) (13, 21). In preliminary tests, most androgen reporters responded positively to known androgens like testosterone, DHT, and trenbolone, as did our reporter, and had low EC50 values (Table 6). In addition, most groups have demonstrated that estrogens such as 17β-estradiol have a low response in these assays and progestins have an intermediate response. In terms of relative potency, i.e., the ratio of the EC50 for the tested chemical to the EC50 for testosterone, S. cerevisiae BLYAS was better able to distinguish between androgenic and estrogenic chemicals than many other reporters (Table 6). In addition, based on comparison of the relative potencies of 17β-estradiol to DHT and testosterone, which were 30 times more active (than 17β-estradiol) in the S. cerevisiae YAS assay (20), they were ∼3,800-fold and ∼8,000-fold more active, respectively, in the S. cerevisiae BLYAS assay as reported here, indicating that the assay is highly specific for androgenic chemicals (Table 6). This property makes BLYAS suitable for use with environmental samples, where it might be used to distinguish if a sample is actually positive for androgens or if the reporter is responding to other types of steroid hormones.
TABLE 6.
Relative potencies of androgenic and estrogenic chemicals for different types of assaysa
| Test chemical | Relative potency (EC50) for assay type (reference)
|
||||
|---|---|---|---|---|---|
| Yeast based, lux (this study) | Cell-free binding assay (4) | Mammalian cell based, luc (22) | Yeast based, lacZ (5) | Yeast based, luc (13) | |
| Testosterone | 1.0 (4.6 × 10−9) | 1.0 (2.2 × 10−9) | 1.0 (6.3 × 10−10) | 1.0 (4.7 × 10−9) | 1.0 (1 × 10−8) |
| DHT | 2.1 (9.7 × 10−9) | 7.3 (1.6 × 10−8) | 0.2 (1.3 × 10−10) | 0.7 (3.5 × 10−9) | NDb |
| Androstenedione | 21.7 (1.0 × 10−7) | 591 (1.3 × 10−6) | 6.3 (4.0 × 10−9) | ND | 50 (5 × 10−7) |
| 17β-Estradiol | 8,044 (3.7 × 10−5) | 186 (4.1 × 10−7) | 5,008 (3.2 × 10−6) | 24.6 (8.6 × 10−8) | 5 (5 × 10−8) |
Relative potencies in reference to testosterone (EC50 of the chemical/EC50 of testosterone) were compared for the four different assay types and demonstrated that 17β-estradiol is ∼8,044 times less potent than testosterone with the S. cerevisiae BLYAS assay. EC50 values (concentration of chemical at which the assay is half-maximally active) are given in parentheses.
ND, not determined.
Ongoing studies will continue to address the detection of androgens in environmental samples at their relevant concentrations. While the results presented here are very preliminary, sigmoidal concentration-response curves were observed in creek water for both DHT and testosterone, with limits of detection of 5.0 × 10−10 M for DHT and 2.5 × 10−9 M for testosterone, which are approaching environmentally relevant concentrations (10, 11). Future work will also include testing of mixtures of compounds to determine how the reporter would react in a complex environmental sample, which will include mixtures of androgens, antiandrogens, and toxic chemicals.
Four bioluminescent bioreporter assays for screening the hormonal activity of chemicals have been developed to date: S. cerevisiae strains BLYAS and BLYRa for the detection of androgens and their associated toxicity, respectively, and S. cerevisiae BLYES and BLYR for the detection of estrogens and their associated toxicity, respectively. The primary advantages of these assays include ease of use, efficiency of gathering data, and inclusion of multiple assays per microtiter plate. In addition, these strains, when combined with appropriate photodetection technology, may be used for rapid, remote monitoring of industrial and municipal waste effluents carrying hormonally active compounds.
Acknowledgments
We express appreciation to J. P. Sumpter for permission to use the YAS strain, to Otakuye Conroy and David Quanrud for supplying the strain, and to Cynthia Hackney at the University of Tennessee's Statistical Consulting Center for statistical consultation.
This research was supported by the U.S. Environmental Protection Agency's STAR program through grant RD-831302.
Although the research described in this article has been funded wholly or in part by the U.S. Environmental Protection Agency through a grant/cooperative agreement (RD-831302), it has not been subjected to the Agency's required peer and policy review and therefore does not necessarily reflect the views of the Agency, and no official endorsement should be inferred.
Footnotes
Published ahead of print on 3 August 2007.
REFERENCES
- 1.Andersen, L., R. Goto-Kazeto, J. M. Trant, J. P. Nash, B. Korsgaard, and P. Bjerregaard. 2006. Short-term exposure to low concentrations of the synthetic androgen methyltestosterone affects vitellogenin and steroid levels in adult male zebrafish (Danio rerio). Aquat. Toxicol. 76:343-352. [DOI] [PubMed] [Google Scholar]
- 2.Blankvoort, B. M. G., E. M. de Groene, A. P. van Meeteren-Kreikamp, R. F. Witkamp, R. J. Rodenburg, and J. M. Aarts. 2001. Development of an androgen reporter gene assay (AR-LUX) utilizing a human cell line with an endogenously regulated androgen receptor. Anal. Biochem. 298:93-102. [DOI] [PubMed] [Google Scholar]
- 3.Daunert, S., G. Barrett, J. S. Feliciano, R. S. Shetty, S. Shrestha, and W. Smith-Spencer. 2000. Genetically engineered whole-cell sensing systems: coupling biological recognition with reporter genes. Chem. Rev. 100:2705-2738. [DOI] [PubMed] [Google Scholar]
- 4.Fang, H., W. Tong, W. S. Branham, C. L. Moland, S. L. Dial, H. Hong, Q. Xie, R. Perkins, W. Owens, and D. M. Sheehan. 2003. Study of 202 natural, synthetic, and environmental chemicals for binding to the androgen receptor. Chem. Res. Toxicol. 16:1338-1358. [DOI] [PubMed] [Google Scholar]
- 5.Gaido, K. W., L. S. Leonard, S. Lovell, J. C. Gould, D. Babaï, C. J. Portier, and D. P. McDonald. 1997. Evaluation of chemicals with endocrine modulating activity in a yeast-based steroid hormone receptor gene transcription assay. Toxicol. Appl. Pharmacol. 143:205-212. [DOI] [PubMed] [Google Scholar]
- 6.Gupta, R. K., S. S. Patterson, S. Ripp, M. L. Simpson, and G. S. Sayler. 2003. Expression of the Photorhabdus luminescens lux genes (luxA, B, C, D, E) in Saccharomyces cerevisiae. FEMS Yeast Res. 4:305-313. [DOI] [PubMed] [Google Scholar]
- 7.Haendler, B., I. Schuttke, and W.-D. Schleuning. 2001. Androgen receptor signaling: comparative analysis of androgen response elements and implication of heat shock 90 and 14-3-3n. Mol. Cell. Endocrinol. 173:63-73. [DOI] [PubMed] [Google Scholar]
- 8.Horie-Inoue, K., H. Bono, Y. Okazaki, and S. Inoue. 2004. Identification and functional analysis of consensus androgen response elements in human prostate cancer cells. Biochem. Biophys. Res. Commun. 325:1312-1317. [DOI] [PubMed] [Google Scholar]
- 9.King, J. M. H., P. M. DiGrazia, B. Applegate, R. Burlage, J. Sanseverino, P. Dubar, F. Larimer, and G. S. Sayler. 1990. Rapid, sensitive bioluminescent reporter technology for naphthalene exposure and biodegradation. Science 249:778-781. [DOI] [PubMed] [Google Scholar]
- 10.Kolodziej, E. P., J. L. Gray, and D. L. Sedlak. 2003. Quantification of steroid hormones with pheromonal properties in municipal wastewater effluent. Environ. Toxicol. Chem. 22:2622-2629. [DOI] [PubMed] [Google Scholar]
- 11.Kolpin, D. W., E. T. Furlong, M. T. Meyer, E. M. Thurman, S. D. Zaugg, L. B. Barber, and H. T. Buxton. 2002. Pharmaceuticals, hormones, and other organic wastewater contaminants in US streams, 1999-2000: a national reconnaissance. Environ. Sci. Technol. 36:1201-1211. [DOI] [PubMed] [Google Scholar]
- 12.Layton, A. C., M. Muccini, M. M. Ghosh, and G. S. Sayler. 1998. Construction of a bioluminescent reporter strain to detect polychlorinated biphenyls. Appl. Environ. Microbiol. 64:5023-5026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Michelini, E., P. Leskinen, M. Virta, M. Karp, and A. Roda. 2005. A new recombinant cell-based bioluminescent assay for sensitive androgen-like compound detection. Biosensors Bioelectron. 20:2261-2267. [DOI] [PubMed] [Google Scholar]
- 14.Orlando, E. F., A. S. Kolok, G. A. Binzcik, J. L. Gates, M. K. Horton, C. S. Lambright, L. E. Gray, A. M. Soto, and L. J. Guillette. 2004. Endocrine-disrupting effects of cattle feedlot effluent on an aquatic sentinel species, the fathead minnow. Environ. Health Perspect. 112:353-358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Purvis, I. J., D. Chotai, C. W. Dykes, D. B. Lubahn, F. S. French, E. M. Wilson, and A. N. Hobden. 1991. An androgen-inducible expression system for Saccharomyces cerevisiae. Gene 106:35-42. [DOI] [PubMed] [Google Scholar]
- 16.Routledge, E. J., and J. P. Sumpter. 1996. Estrogenic activity of surfactants and some of their degradation products assessed using a recombinant yeast screen. Environ. Toxicol. Chem. 15:241-248. [Google Scholar]
- 17.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Press, Cold Spring Harbor, NY.
- 18.Sanseverino, J., R. K. Gupta, A. C. Layton, S. S. Patterson, S. A. Ripp, L. Saidak, M. L. Simpson, T. W. Schultz, and G. S. Sayler. 2005. Use of Saccharomyces cerevisiae BLYES expressing bacterial bioluminescence for rapid, sensitive detection of estrogenic compounds. Appl. Environ. Microbiol. 71:4455-4460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Singh, S. M., S. Gauthier, and F. Labrie. 2000. Androgen receptor antagonists (antiandrogens): structure-activity relationships. Curr. Med. Chem. 7:211-247. [DOI] [PubMed] [Google Scholar]
- 20.Sohoni, P., and J. P. Sumpter. 1998. Several environmental oestrogens are also anti-androgens. J. Endocrinol. 158:327-339. [DOI] [PubMed] [Google Scholar]
- 21.Sonneveld, E., H. J. Jansen, J. A. C. Riteco, A. Brouwer, and B. van der Burg. 2005. Development of androgen- and estrogen-responsive bioassays, members of a panel of human cell line-based highly selective steroid-responsive bioassays. Toxicol. Sci. 83:136-148. [DOI] [PubMed] [Google Scholar]
- 22.Sonneveld, E., J. A. C. Riteco, H. J. Jansen, B. Pieterse, W. G. Schoonen, and B. van der Burg. 2006. Comparison of in vitro and in vivo screening models for androgenic and estrogenic activities. Toxicol. Sci. 89:173-187. [DOI] [PubMed] [Google Scholar]
- 23.Thompson, J. R., E. Register, J. Curotto, M. Kurtz, and R. Kelly. 1998. An improved protocol for the preparation of yeast cells for transformation by electroporation. Yeast 14:565-571. [DOI] [PubMed] [Google Scholar]
- 24.Willemsen, P., M.-L. Scippo, G. Kausel, J. Figueroa, G. Maghuin-Rogister, J. A. Martial, and M. Muller. 2004. Use of reporter cell lines for detection of endocrine-disrupting activity. Anal. Bioanal. Chem. 378:655-663. [DOI] [PubMed] [Google Scholar]